Abstract
Pyruvate dehydrogenase complex (PDHc) defect is a well-known cause of mitochondrial disorders (MD) with at least six responsible genes (PDHA1, PDHB, DLAT, DLD, PDHX, PDP1). The aim of this work was to assess the diagnostic value of biochemical methods in recognition of PDHc defect in Polish patients with suspicion of MD.
In the first step, Western blot of the E1α subunit was performed on 86 archive muscle bioptates with suspicion of MD. In the second step, Sanger PDHA1 sequencing was performed in 21 cases with low E1α expression. In the third step, 7 patients with negative results of PDHA1 sequencing were subjected to whole-exome sequencing (WES). This protocol revealed 4 patients with PDHA1 and one with DLD mutations. Four additional probands were diagnosed outside the protocol (WES or Sanger sequencing).
The molecular characterization of PDHc defect was conducted in a total of 9 probands: 5 according to and 4 off the protocol. Additionally, two affected relatives were recognized by a family study. Altogether we identified seven different PDHA1 changes, including two novel variants [c.464T > C (p.Met155Thr) and c.856_859dupACTT (p.Arg288Leufs*10)] and one DLD variant.
The lactate response to glucose load in the PDHA1 subset was compared to a subset of non PDHc-related MD. Opposite responses were observed, with an increase of 23% and decrease of 27%, respectively.
The results show that determining lactate response to glucose load and muscle E1α expression may contribute to distinguishing PDHc-related and other MD, however, WES is becoming the method of choice for MD diagnostics.
Keywords: Pyruvate dehydrogenase complex deficiency, PDHc, PDHA1, DLD, Novel pathogenic variants, Whole-exome sequencing
1. Introduction
Pyruvate dehydrogenase complex (PDHc) deficiency is a frequent cause of mitochondrial disorders. Progressive neurological symptoms usually start in infancy, but may be evident at birth or in later childhood, and adult onset is very rare. They may include developmental delay, brain malformations, microcephaly, poor muscle tone, seizures, intermittent ataxia, West syndrome, and Leigh-like syndrome.
There are six forms of PDHc deficiency, depending on the genetic background and damaged subunit of the enzyme complex. A few cases are known to result from mutations in genes encoding subunits: E1β (PDHB), E2 (DLAT), E3 (DLD), and E3BP (PDHX) or PDH phosphatase (PDP1). The most common causes are mutations in X-linked PDHA1, encoding the E1α subunit [1]. PDHA1 maps to the Xp22.1 region and consists of eleven exons. The majority of mutations in this gene occur de novo. Hemizygous males are generally symptomatic, whereas heterozygous females present variable expression of the mutant and normal genes in different tissues as a result of the X-inactivation pattern [2].
This is the first genetic study of PDHc deficiency in Poland and we report novel pathogenic variants and recurrent causal mutations in the genes PDHA1 and DLD. Our aim was also to check the utility of Western blotting of the PDHc E1α subunit in muscle and of fasting/postprandial lactate/pyruvate ratio specificity in the recognition of PDHc defect when pyruvate oxidation in fresh muscle cannot be measured [3].
2. Methods
2.1. Patients
Muscle specimens of 86 patients with suspicion of mitochondrial disorder (MD) were selected from 570 muscle biopsies performed at our nation-wide referral center (CMHI) in the period from 1996 to 2015. Open muscle biopsy was done and histochemical and spectrophotometric techniques were applied for assessment of oxidative phosphorylation system function (OXPHOS) as described previously [4].
The inclusion criteria were: (1) OXPHOS activity normal or inconclusive, (2) chronic/progressive encephalopathy with lactic acidosis, brain malformations, and Leigh syndrome, (3) lactate elevation in response to glucose/carbohydrate load, (4) negative screening for common mtDNA mutations (m.8993T > C and m.8993T > G in MTATP6, m.3243A > G in MTTL1, m.8344A > G in MTTK) and nDNA mutations (c.311_312insAT312_321del10 and c.845_846delCT in SURF1) responsible for LS (5), muscle sample and/or DNA availability, (6) informed consent of the patient's parents or guardians.
2.2. Western blot analysis
Tissue lysates (30 μg protein) from muscle biopsies (patients and healthy controls) were separated electrophoretically in 10% SDS polyacrylamide gels and transferred onto PVDF membranes (BioRad). The membranes were blocked and proteins were detected using anti-subunit E1α pyruvate dehydrogenase (PDH) WB Antibody (Abcam) followed by appropriate secondary AP-conjugated antibodies (1:1000, Bio-Rad). The level of the E1α subunit of the pyruvate dehydrogenase complex was analyzed densitometrically and the levels of individual subunits were calculated as ratios to the corresponding subunit in control tissue. Reference values were established in 27 muscle samples from patients with other (non-mitochondrial) diseases. The deficiency threshold was arbitrarily assumed as below of 50% of the reference mean. When important inclusion criteria coexisted (LS, X-linked inheritance, normal lactate/pyruvate ratio), a higher threshold was applied (about 60%).
2.3. Molecular analysis
Genomic DNA was isolated from different tissues (blood, muscles, fibroblasts) using standard phenol-chloroform or automatic DNA extraction (MagNA Pure LC 2.0, Roche).
Scanning of PDHA1 molecular variants was performed by Sanger sequencing and long-range polymerase chain amplification as a gene deletion assay. The sizes of wild-type or mutant (shorter) PCR fragments were determined by agarose (1%)-gel electrophoresis. In PDHA1-unsolved patients, whole-exome sequencing (WES) was further conducted.
WES was performed on a HiSeq 1500 using an Exome Enrichment Kit (Illumina) according to a published protocol [5]. The average read depth was 120 with > 90% of the targeted regions covered at least 20-fold. Generated reads were aligned to the hg19 reference human genome. Alignments were viewed with Integrative Genomics Viewer v.2.3.40. The detected variants were annotated using Annovar and converted to MS Access format for final manual analysis. Variants were filtered to exclude changes with an average frequency higher than 0.01 (for AR inheritance model) and 0.001 (for AD inheritance model) in different exome sequencing project databases (e.g. project ESP 6500, ExAC 60,706), and POL 400 (in-house-project of 400 exomes of Polish individuals with unrelated diseases). Molecular variants were assessed by pathogenicity prediction tools (CADD, MetaSMV, PolyPhen2 HDIV and HVAR, Mutation Assessor, LRT, MetaLR, SIFT, FATHMM and MutationTaster softwares). The websites were simultaneously consulted using dedicated Alamut Interactive Biosoftware. The nomenclature of molecular variants follows the Human Genome Variation Society guidelines (HGVS, www.hgvs.org/mutnomen) and referral to the cDNA sequences follows the Human Gene Mutation Database (HGMD, www.hgmd.cf.ac.uk).
Molecular analysis was performed after obtaining informed consent from the patients' parents or legal guardians.
2.4. Biochemical analysis
Lactate response to glucose loading was assessed during a routine intravenous glucose tolerance test (IVGTT) in cases with suspicion of PDHc defect. The plasma lactate level was measured at 0′, 15′, 30′, 60′, 90′, and 120′ and the pyruvate level, at 0′, 60′, and 120′. Control values were obtained from 25 non-mitochondrial patients in whom IVGTT was performed during differential diagnosis. The reference group included 8 patients with various molecularly confirmed mitochondrial disorders not related to PDHc defect (ACAD9, SCO2, SURF1 and DGUOK pathogenic changes).
3. Results
3.1. Western blot findings
In the first step of the study, the amount of E1α was assessed in 86 muscle specimens. Its content ranged from 0 to 272% of the mean reference. In 33 of 83 muscle bioptates, low amounts of E1α (below 50% of the mean reference value) were found, and in another 6 patients, amounts below 60% were determined.
3.2. Molecular findings
In the second step of the study, Sanger sequencing of PDHA1 was performed in a total of 21 unrelated patients with low muscle E1α. Pathogenic variants in PDHA1 were revealed in four of these probands (19%). In one of them (P4 in Table 1), with a number of data pointing to a PDHc defect, the E1α amount was 52.5% (above 50% of the reference value).
Table 1.
Clinical, biochemical and molecular data of 11 patients with pathogenic variants in PDHc related genes.
| Data and symptom(s) |
Patient KI (p) |
Patient KW (b) |
Patient SzO (p) |
Patient BF (p) |
Patient KBS (m) |
Patient PM (p) |
Patient GP (p) |
Patient KG (p) |
Patient PM (p) |
Patient ZJ (p) |
Patient PZ (p) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient no. |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
| Family no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| Sex | M | M | F | M | F | M | F | F | F | F | F |
| Age of onset | 2 y | ND | Neonatal | Neonatal | 2 y | 15 m | 3 m | 7 m | Birth | 4 m | 1.5 y |
| Age at diagnosis | 8.5 | ND | 2 | 4 y | ND | 8 y | 2.5 y | 4.5 y | 25 y | 2 y | 9 y |
| Neurological findings | |||||||||||
| Psychomotor retardation/developmental delay | No | No | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Developmental delay (> 3 y) | No | No | Yes | No | ND | Yes | Yes | Yes | Yes | Yes | |
| Dysarthria | No | No | Nd | Yes | Yes | Yes | ND | ND | ND | ND | Yes |
| Microcephaly | No | ND | No | ND | No | No | Yes | Yes | Yes | Yes | No |
| Seizures | No | ND | No | No | No | No | Yes | Yes | No | Yes | No |
| Ataxia | No | ND | Yes | Yes | Yes | Yes | ND | No | ND | No | No |
| Hypotonia/hypertonia | No | ND | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Peripheral neuropathy | Yes | ND | ND | ND | Yes | Yes | ND | ND | ND | ND | No |
| Brain malformations | |||||||||||
| Brain atrophy | ND | ND | No | Yes | No | ND | Yes | ND | Yes | Yes | Yes |
| Corpus callosum hypoplasia | ND | ND | No | ND | No | ND | Yes | Yes | ND | Yes | Yes |
| Cerebellum atrophy | ND | ND | No | No | No | No | ND | No | ND | VH | VH |
| Demyelinisation | ND | ND | No | Yes | No | ND | ND | ND | ND | ND | Yes |
| Basal ganglia abnormalities | ND | ND | Yes | ND | No | ND | ND | ND | ND | ND | Yes |
| Brain stem involvement | ND | ND | Yes | ND | No | ND | ND | ND | ND | ND | ND |
| Ocular findings | |||||||||||
| Nystagmus | No | No | Yes | No | No | No | ND | No | Yes | No | No |
| Ptosis | No | No | No | No | No | No | ND | No | Yes | No | Yes |
| Oculomotor apraxia | No | No | Yes | No | No | Yes | ND | No | Yes | No | No |
| Biochemical findings | |||||||||||
| Blood lactate (fasting) [mg/dL] | 42.66 | NA | 26 | 20.8 | 47.4 | 62 | 36.2–65.6 | 39 | 42 | 54 | 104 |
| Blood pyruvate (fasting) [mg/dL] | 2.2 | NA | 2 | 1.7 | 2.3 | 3.7 | NA | 2.8 | NA | NA | NA |
| Blood lactate (after carbohydrate) [mg/dL] | 39 | NA | 25 | 29.6 | NA | 92 | NA | NA | 51 | 53 | NA |
| Blood pyruvate (after carbohydrate) [mg/dL] | 2.6 | NA | 1.5 | 2.1 | NA | 4.1 | NA | NA | NA | 4.5 | NA |
| Blood lactate/pyruvate ratio | 18 | NA | 13 | 12 | 20.7 | 17 | NA | 14 | 7.5 | 12 | NA |
| CSF lactate [mg/dL] | 40 | NA | NA | NA | NA | 67 | 75 | 47 | 87 | NA | 58.8 |
| Alanine [μmol/dL] | NA | NA | 559 | 180 | NA | 238 | 952 | NA | 222–1338 | 806 | 407 |
| GC/MS urine | LA, PA, 2-KGA | NA | NA | 2-KGA, LA | NA | 2-KGA, LA | LA, 2-KGA | NA | 2-KGA, LA | 2-KGA, LA, MMA | Normal |
| pH | NA | NA | 7.43 | 7.49 | NA | 7.46 | NA | 7.374 | 7,368 | 7.3 | 7.39 |
| pO2 [mmHg] | NA | NA | 117 | 86.5 | NA | 83.5 | NA | 81.6 | NA | 121 | 92.3 |
| pCO2 [mmHg] | NA | NA | 26,3 | 21.5 | NA | 26.7 | NA | 25.6 | 35.7 | 18.7 | 32.1 |
| Muscle biochemistry | |||||||||||
| Complex I | NA | NA | NA | NA | NA | 10 | NA | 12.2 | 11.6 | < 3.0 | 5.9 |
| Complex II | NA | NA | NA | NA | NA | 7.6 | NA | 6.5 | 3.5 | 7.3 | 3.4 |
| Complex II + III | NA | NA | NA | NA | NA | 4.7 | NA | 4 | 5.5 | < 3.0 | 3.6 |
| Complex III | NA | NA | NA | NA | NA | 44.7 | NA | 69.2 | 47.5 | 71.9 | 72 |
| Complex IV | NA | NA | NA | NA | NA | 10 | NA | 6.1 | 19.5 | 5.2 | 3.9 |
| SC | NA | NA | NA | NA | NA | 275 | NA | 211.4 | 122.6 | 513.8 | 268.2 |
| E1α [% of mean reference] | NA | NA | NA | 52.5 | NA | 0 | NA | 38.5 | NA | 20.9 | 59.7 |
| Muscle histology and histochemistry | |||||||||||
| Variability of fiber size | No | ND | ND | ND | ND | Yes | ND | Yes | ND | No | Yes |
| Lipid accumulation | No | ND | ND | ND | ND | Yes | ND | No | Yes | Yes | No |
| Predominance of fibers type I | No | ND | ND | ND | ND | No | ND | Yes | ND | Yes | Yes |
| Clinical diagnosis | |||||||||||
| Clinical diagnosis prior to genetic testing | Guillain-Barre syndrome |
Family study |
Family study |
PDHc deficiency |
Ataxia | PDHc deficiency |
Leigh syndrome |
Mitochondrial encephalopathy | Mitochondrial encephalomyopathy | PDHc deficiency |
Leigh syndrome |
| Nijmegen score | 4 | 3 | 5 | 3 | 2 | 3 | 4 | 3 | 6 | 3 | 6 |
| Type of PDHc | PDH E1α subunit |
PDH E1α subunit | PDH E1α subunit | PDH E1α subunit | PDH E1α subunit | PDH E1α subunit | PDH E1α subunit | PDH E1α subunit |
PDH E1α subunit |
PDH E1α subunit |
PDH E3 subunit |
| Neurological phenotype (final assigning) | |||||||||||
| Neonatal encephalopathy with lactic acidosis | x | x | x | x | |||||||
| Basal ganglia abnormalities (Leigh-like) | x | x | |||||||||
| Chronic/progressive neurologic deterioration | x | x | x | ||||||||
| Intermittent ataxia | x | x | |||||||||
| Molecular genetics | |||||||||||
| DNA source (tissue) | Blood | Blood | Blood | Blood | Blood | Blood | Blood | Muscle | Muscle | Muscle | Muscle |
| Molecular test | WES | Sanger | Sanger | Sanger | Sanger | Sanger | WES | Sanger | WES | Sanger | WES |
| Mutated gene | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | PDHA1 | DLD |
| Nucleotide exchange | c.262C > T | c.262C > T | c.464T > C | c.787C > G | c.787C > G | c.787C > G | c.856_859dupACTT | c.904C > T | c.933_935delAAG | c. 934_940delAGTAAGA | c.1123G > A |
| Amino acid exchange | p.R88C | p.R88C | p.M155 T | p.R263G | p.R263G | p.R263G | p.Arg288Leufs*10 | p.R302C | p.Arg311del | p.S312Vfs*12 | p.E375K |
| Localization of mutation | Exon 3 | Exon 3 | Exon 5 | Exon 8 | Exon 8 | Exon 8 | Exon 9 | Exon 10 | Exon 10 | Exon 10 | Exon 11 |
| Mutation type | Missense | Missense | Missense | Missense | Missense | Missense | Frameshift | Missense | Inframe deletion | Frameshift | Missense |
| Mutation status | Known | Known | Novel | Known | Known | Known | Novel | Known | Known | Known | Known |
| Mutation inheritance | Maternal | Maternal | Maternal | Maternal | Na | Na | De novo | De novo | De novo | Na | Parental |
| Familial history | |||||||||||
| Affected relative | Brother | Brother | No | Mother, uncle | Son, brother | NA | No | No | No | NA | No |
p - proband; b - brother; m - mother; y - year; m - month; 2-KGA - ketoglutaric acid; LA - lactic acid; MMA - methylmalonic acid, NA - not analysed; ND - no data; PA - pyruvic acid, Sanger - Sanger sequencing; VH - vermis hypoplasia; WES – whole-exome sequencing.
In the third step of the study, WES was performed in 7 patients with negative results of PDHA1 sequencing. Pathogenic variants were found only in not PDHc-related genes (FBXL4, MTND1, MTND5, PGAP2) in four of seven patients with muscle E1α amounts below 50% of the mean reference value. One PDHc-related defect detected by WES was a homozygous DLD variant found in the patient (P11 in Table 1) with LS brain changes seen on MRI (another inclusion criterion).
Molecular characteristics conducted together for all PDHc defects diagnosed in Poland encompassed five patients identified under the study protocol and four others diagnosed outside the protocol. In the latter patients, muscle biopsy was not performed nor was the amount of E1α assessed. They were identified by WES carried out due to general MD suspicion (3 cases). One patient was properly diagnosed through a positive family history and pathogenic variant was identified in PDHA1 by Sanger sequencing.
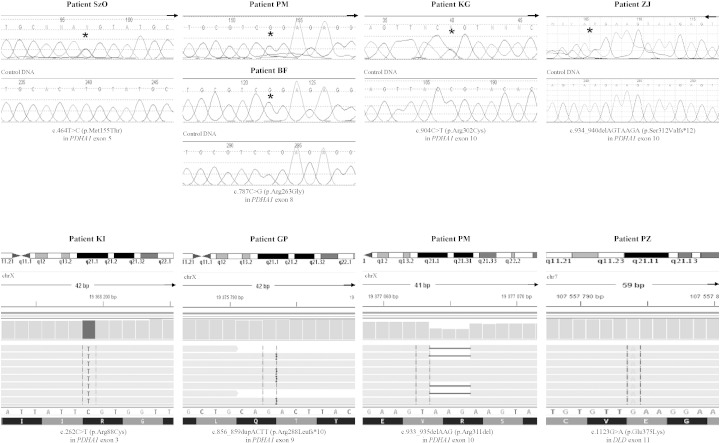
Molecular analysis revealed pathogenic variants in two PDHc-related genes in nine probands and two relatives. Two novel changes [c.464T > C (p.Met155Thr), c.856_859dupACTT (p.Arg288Leufs*10)] and five known variants [c.787C > G (p.Arg263Gly), c.904C > T (p.Arg302Cys), c.934_940delAGTAAGA (p.Ser312Valfs*12), c.262C > T (p.Arg88Cys), c.933_935delAAG (p.Arg311del)] in PDHA1, as well as one homozygous known mutation, c.1123G > A (p.Glu375Lys), in DLD were identified (Fig. 1, Table 1). Long-range PCR of the entire PDHA1 gene excluded major intragenic rearrangements, e.g., large deletions or insertions, in patients without a pathogenic variant.
Fig. 1.
The results of genetic analysis showing novel variants in PDHA1 and DLD identified by Sanger (A) and whole-exome (B) sequencing.
Available parents and family members were also investigated. Carrier status of the PDHA1 variant was confirmed in two mothers (one of them symptomatic) and in one affected brother, while two remaining changes occurred de novo (Table 1). Both parents and one sister of the patient with the homozygous DLD variant were found to be asymptomatic carriers.
3.3. Clinical findings of PDHA1 patients
Detailed clinical, biochemical and molecular characteristics of ten patients (P1–P10) with PDHA1 defects are summarized in Table 1. In all patients the course of disease corresponds to a major extent to the literature data [3]. The neurological phenotype was: neonatal encephalopathy with lactic acidosis in four patients, basal ganglia abnormalities in two, chronic progressive neurologic deterioration in three, and intermittent ataxia in two patients (Table 1). The onset of symptoms ranged from the neonatal period to 2 years. All patients survived; the oldest symptomatic carrier is the affected mother (P5 in Table 1), now aged 34 years.
Vitamin B1 (thiamine) responsiveness is considered in five patients (from four families) only on the basis of subjective parents' observation. It is not, however, evidence based. We tried a loading test to objectively verify the responsiveness by use of lactate response for i.v. glucose loading and successfully demonstrated it in one affected individual.
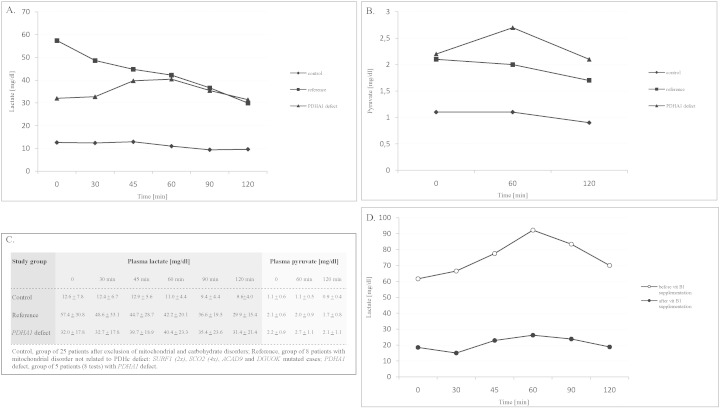
The test was assessed 8 times in four PDHA1-mutated patients and in the reference group of eight non PDHc-related mitochondrial disease cases (Fig. 2). The plasma lactate response to the glucose bolus was antithetical in the compared groups. The mean lactate concentration increased by 23% in the PDHA1 group and decreased by 27% in the non PDHc-related mitochondrial disease group (Fig. 2, panel A). The difference was statistically significant (P = 0.0354).
Fig. 2.
The results of response to intravenous glucose loading in PDHc-affected patients and reference groups. Mean value of lactate concentration (A), pyruvate concentration (B), mean value of lactate and pyruvate concentrations with standard deviation (C), effect of vitamin B1 supplementation (300 mg/day) in patient PM (D).
In the above-mentioned PDHA1-affected patient (bearing the p.Arg263Gly variant), the test performed before and during vitamin B1 supplementation at the maximum dose of 300 mg/day showed complete normalization of the lactate response to glucose loading (Fig. 2, panel D).
3.4. Clinical course of the DLD patient
In our lipoamide dehydrogenase-deficient patient with the DLD mutation, the expression of PDHc E2/E3BP and E1α subunits was reduced (45.2% and 59.7%, respectively) (Table 1). The patient with the DLD mutation responsible for dihydrolipoamide dehydrogenase deficiency (subunit E3 of PDHc) was the second child with Leigh syndrome in this study. This girl was born by cesarean section with a birth weight of 3250 g and Apgar score of 9 points. Her psychomotor development in infancy was moderately delayed. At the age of 12 months ptosis and involuntary movements developed. Markedly elevated lactate concentrations in plasma and cerebrospinal fluid were found (104 and 58.8 mg/dl) and basal ganglia changes were revealed in brain MRI scans. Muscle biopsy showed a generalized OXPHOS defect. At the age of 6 years her condition was stable. Liver function was never impaired. The girl is not able to walk, but her physical growth is otherwise normal, mental development is mildly impaired, and plasma lactate remains high borderline.
4. Discussion
Molecular analysis revealed PDHc-causing molecular variants in eleven patients from nine unrelated families. Seven different variants were identified in PDHA1 and one homozygous mutation affected DLD. Six of these changes were reported previously and two novel heterozygous variants in PDHA1 were found in female patients.
The first novel variant is a missense substitution c.464T > C (p.Met155Thr) in exon 5. Although there is only a moderate physicochemical difference between hydrophobic methionine and polar, uncharged threonine, this change is predicted to be disease causing by Mutation Taster and SIFT software. Methionine 155 is a very highly conserved residue across species (Table 2), and the change to threonine at this residue is predicted to be deleterious also by the Grantham score, which categorizes codon replacements into classes of increasing chemical dissimilarity (Grantham score = 81, moderately radical change). This variant was not found in the ClinVar, dbSNP, LOVD, and PubMed databases, or in the collection of POL 400, indicating its rarity.
Table 2.
Protein alignment of ten species shows the methionine 155 residue is evolutionary conserved.
| Species | Match | Gene | AA | Alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient SzO | Not conserved | 155 | G | C | A | K | G | K | G | G | S | M | H | T | Y | A | K | N | F | Y | G | G | N | G | I | |
| Human | ENST00000422285 | 155 | G | C | A | K | G | K | G | G | S | M | H | M | Y | A | K | N | F | Y | G | G | N | G | I | |
| P.troglodytes | All identical | ENSPTRG00000021717 | 155 | G | C | A | K | G | K | G | G | S | M | H | M | Y | A | K | N | F | Y | G | G | N | G | I |
| F.catus | All identical | ENSFCAG00000012903 | 155 | G | C | A | K | G | K | G | G | S | M | H | M | Y | A | |||||||||
| M.musculus | All identical | ENSMUSG00000031299 | 155 | G | C | A | K | G | K | G | G | S | M | H | M | Y | A | K | N | F | Y | G | G | N | G | I |
| G.gallus | All identical | ENSGALG00000016430 | 162 | G | C | A | K | G | K | G | G | S | M | H | M | Y | T | K | N | F | Y | G | G | N | G | I |
| T.rubripes | All identical | ENSTRUG00000016917 | 163 | G | I | A | K | G | K | G | G | S | M | H | M | Y | C | K | H | F | Y | G | G | N | G | I |
| D.rerio | All identical | ENSDARG00000012387 | 158 | G | I | A | K | G | K | G | G | S | M | H | M | Y | T | K | H | F | Y | G | G | N | G | I |
| D. melanogaster | All identical | FBgn0028325 | 203 | G | C | A | R | G | K | G | G | S | M | H | M | Y | A | P | N | F | Y | G | G | N | G | I |
| C.elegans | All identical | T05H10.6 | 166 | G | S | M | H | M | Y | T | K | N | F | Y | G | G | N | G | I | |||||||
| X.tropicalis | All identical | ENSXETG00000006212 | 184 | S | M | H | M | Y | A | K | N | F | Y | G | G | N | G | I | ||||||||
AA: Amino acid number corresponds to the mutated position p.Met155Thr of PDHA1 gene. Data obtained from http://www.mutationtaster.org
The second novel pathogenic variant, c.856_859dupACTT (p.Arg288Leufs*10), is a de novo duplication of 4 bps in exon 9 that creates a frameshift starting at codon Arg288 and results in premature termination nine positions downstream. The mRNA product is probably targeted for nonsense-mediated decay. A different duplication, but resulting in the same translation effect, has been reported previously [6].
Worldwide, at least 337 patients with PDHA1 mutations [3], [7] and > 160 molecular variants have been reported [8]. Most of them are missense changes, similar to the findings in this study. Among them, the c.787C > G (p.Arg263Gly) variant is considered the most frequent mutation in PDHA1 since it is repeated in 19 published cases [2], [8]. The recurrent p.Arg263Gly variant (as well as the second, p.Arg88Cys) was reported to occur in vitamin B1 responders [9], [10].
The p.Arg263Gly variant was also found in our molecularly confirmed PDHA1 group: in two unrelated boys and in the mother of one of them. The woman has presented symptoms of mild neuropathy since the age of 3 years. Leigh syndrome described in association with this substitution [2] was not observed in any of our p.Arg263Gly mutated cases. The families observed vitamin B1 efficiency in doses of 50–150 mg per day, but this should be treated with caution. The appropriate way of determining vitamin B1 responsiveness has not yet been described in the literature. In one of the patients with the p.Arg263Gly variant we managed to demonstrate that supplementation with vitamin B1 at a dose of 300 mg per day not only improved his clinical condition, but resulted in a decreased fasting lactate concentration, as well as in a decrease in lactate/pyruvate response to glucose load (Fig. 2, panel D). We speculate that measuring lactate during an IVGTT test may be a useful way to assess vitamin B1 responsiveness in PDHc defect. We have used the glucose loading test in the differential diagnostics of PDHc defect in practice. The intravenous, not oral, glucose load was arbitrarily chosen to simplify the interpretation. We began to use the test in the 1990s when access to molecular methodology was limited in Poland. We took advantage of the pioneering studies of Fernandes on glucose homeostasis in GSD type I and GSD type III/VI/IX and applied them to MD diagnostics. We assumed that the PDHc defect would behave like GSD type III/VI/IX, whereas the other mitochondrial defects would be more like GSD type I [11]. The results proved in our hands to be extremely useful and we continue to apply the test even today at the bedside. We believe that the use of this test to assess responsiveness of the PDHA1 defect to vitamin B1 may find wider application. This assumption requires future confirmation, however.
Two small deletions and one small duplication were identified in our group of PDHA1 pathogenic variants. These defects of the PDHA1 gene are rare (20% of cases) and mainly associated with a frameshift effect that leads to premature termination of translation [1], [8]. The literature also contains descriptions of gross rearrangements (22 cases) that may be masked by the presence of the wild-type allele. Long-range polymerase chain amplification of the entire PDHA1 gene [12] revealed no major intragenic rearrangements in our female patients.
Lipoamide dehydrogenase deficiency (E3 subunit defect) found in one of our patients is a very rare cause of PDHc deficiency that has been reported to date in only 29 cases [3]. The phenotype of the disorder is poorly characterized, and ranges from neonatal distress to paroxysmal myoglobinuria and recurrent liver failure [13]. The patient presented in this study (homozygous for c.1123G > A (p.Glu375Lys) in DLD) shows a clear Leigh syndrome phenotype, as once reported earlier [14].
It should be noted that all of the patients with PDHA1 deficiency reported in this study survived and more than half of them seem to be vitamin B1 responders, whereas in previously described cohorts nearly half of the patients died in infancy or early childhood [15] and the percentage of vitamin B1 responsiveness did not exceed 10% [3]. This may, in part, be due to the children with a severe, fatal course dying without a diagnosis of PDHc deficiency because specific diagnostic methods were unavailable in Poland for a long time. Another explanation can be better selection of patients suspected of PDHc deficit in our material due to routine inclusion of lactate response to i.v. glucose loading in differential diagnostics.
Our results indicate limited usefulness of E1α expression assessed in muscle homogenate for the recognition of PDHc deficiency. Unexpectedly, among 21 cases with reduced E1α expression we confirmed only four patients with a PDHA1 pathogenic variant. On the other hand, exome sequencing revealed a significant number of genetic defects (including mitochondrial) in this group. Admittedly, secondary PDHc deficiency was described in single cases of such disorders as MELAS or MERRF [16], [17], but definitive confirmation of such a relationship or its absence requires more data and observations.
The paper describes a study on archival muscle bioptates stored when PDHA1 sequencing was not available in our lab. At present we recommend starting from PDHA1 sequencing in each patient with clinical and biochemical features of PDHc defect (including normal lactate/pyruvate ratio, abnormal response of lactate to i.v. glucose load, vit B1 responsiveness, typical MRI changes, normal OXPHOS, low E1α in muscle, X-linked inheritance). In our opinion, the next step is WES which enables detection of variants in a vast range of known genes as well as in candidates.
5. Conclusion
The results of the study suggest that a specific lactate/pyruvate response to glucose load may provide a marker of responsiveness of PDHA1 deficiency to vitamin B1 administration that is dependent on the type of PDHA1 pathogenic variant.
Assessment of E1α expression using Western blotting may contribute to the differential diagnosis of PDHc only to a small extent. It should be stressed that this method reveals not only primary PDHc defects caused by mutations in genes coding for individual subunits of the complex, but also may reflect its secondary dysfunction that is not yet sufficiently recognized.
Both methods can be useful in diagnostics, but whole-exome sequencing definitely prevails in mitochondrial diagnostics and is becoming the method of choice also for the diagnostics of PDHc defects.
Acknowledgments
We thank all of the physicians who referred affected children to our medical center. The study was supported by CMHI projects no. S134/13, no. S136/13, no. 216/12, and by grants from the National Science Centre Harmonia 4 No. UMO-2013/08/M/NZ5/00978 and EU Structural Funds Project POIG.02.01.00-14-059/09.
References
- 1.Brown R.M., Head R.A., Boubriak I.I., Leonard J.V., Thomas N.H., Brown G.K. Mutations in the gene for the E1beta subunit: a novel cause of pyruvate dehydrogenase deficiency. Hum. Genet. 2004;115(2):123–127. doi: 10.1007/s00439-004-1124-8. [DOI] [PubMed] [Google Scholar]
- 2.Lissens W., De Meirleir L., Seneca S., Liebaers I., Brown G.K., Brown R.M., Ito M., Naito E., Kuroda Y., Kerr D.S., Wexler I.D., Patel M.S., Robinson B.H., Seyda A. Mutations in the X-linked pyruvate dehydrogenase (E1) alpha subunit gene (PDHA1) in patients with a pyruvate dehydrogenase complex deficiency. Hum. Mutat. 2000;15(3):209–219. doi: 10.1002/(SICI)1098-1004(200003)15:3<209::AID-HUMU1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 3.Sperl W., Fleuren L., Freisinger P., Haack T.B., Ribes A., Feichtinger R.G., Rodenburg R.J., Zimmermann F.A., Koch J., Rivera I., Prokisch H., Smeitink J.A., Mayr J.A. The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis. 2015;38(3):391–403. doi: 10.1007/s10545-014-9787-3. [DOI] [PubMed] [Google Scholar]
- 4.Piekutowska-Abramczuk D., Popowska E., Pronicki M., Karczmarewicz E., Tylek-Lemanska D., Sykut-Cegielska J., Szymanska-Dembinska T., Bielecka L., Krajewska-Walasek M., Pronicka E. High prevalence of SURF1 c.845_846delCT mutation in Polish Leigh patients. Eur. J. Paediatr. Neurol. 2009;13(2):146–153. doi: 10.1016/j.ejpn.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Ploski R., Pollak A., Muller S., Franaszczyk M., Michalak E., Kosinska J., Stawinski P., Spiewak M., Seggewiss H., Bilinska Z.T. Does p.Q247X in TRIM63 cause human hypertrophic cardiomyopathy? Circ. Res. 2014;114(2):e2–e5. doi: 10.1161/CIRCRESAHA.114.302662. [DOI] [PubMed] [Google Scholar]
- 6.Cameron J.M., Levandovskiy V., Mackay N., Tein I., Robinson B.H. Deficiency of pyruvate dehydrogenase caused by novel and known mutations in the E1alpha subunit. Am. J. Med. Genet. A. 2004;131(1):59–66. doi: 10.1002/ajmg.a.30287. [DOI] [PubMed] [Google Scholar]
- 7.Castiglioni C., Verrigni D., Okuma C., Diaz A., Alvarez K., Rizza T., Carrozzo R., Bertini E., Miranda M. Pyruvate dehydrogenase deficiency presenting as isolated paroxysmal exercise induced dystonia successfully reversed with thiamine supplementation. Case report and mini-review. Eur. J. Paediatr. Neurol. 2015;19(5):497–503. doi: 10.1016/j.ejpn.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 8.http://www.hgmd.cf.ac.uk.
- 9.van Dongen S., Brown R.M., Brown G.K., Thorburn D.R., Boneh A. Thiamine-responsive and non-responsive patients with PDHC-E1 deficiency: a retrospective assessment. JIMD Rep. 2015;15:13–27. doi: 10.1007/8904_2014_293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachmann-Gagescu R., Merritt J.L., 2nd, Hahn S.H. A cognitively normal PDH-deficient 18-year-old man carrying the R263G mutation in the PDHA1 gene. J. Inherit. Metab. Dis. 2009;32(Suppl. 1) doi: 10.1007/s10545-009-1101-4. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya K. Investigation and management of the hepatic glycogen storage diseases. Transl. Pediatr. 2015;4(3):240–248. doi: 10.3978/j.issn.2224-4336.2015.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brivet M., Moutard M.L., Zater M., Venet L., Chenel C., Mine M., Legrand A. First characterization of a large deletion of the PDHA 1 gene. Mol. Genet. Metab. 2005;86(4):456–461. doi: 10.1016/j.ymgme.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Odievre M.H., Chretien D., Munnich A., Robinson B.H., Dumoulin R., Masmoudi S., Kadhom N., Rotig A., Rustin P., Bonnefont J.P. A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of alpha-ketoglutarate dehydrogenase deficiency. Hum. Mutat. 2005;25(3):323–324. doi: 10.1002/humu.9319. [DOI] [PubMed] [Google Scholar]
- 14.Quinonez S.C., Leber S.M., Martin D.M., Thoene J.G., Bedoyan J.K. Leigh syndrome in a girl with a novel DLD mutation causing E3 deficiency. Pediatr. Neurol. 2013;48(1):67–72. doi: 10.1016/j.pediatrneurol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBrosse S.D., Okajima K., Zhang S., Nakouzi G., Schmotzer C.L., Lusk-Kopp M., Frohnapfel M.B., Grahame G., Kerr D.S. Spectrum of neurological and survival outcomes in pyruvate dehydrogenase complex (PDC) deficiency: lack of correlation with genotype. Mol. Genet. Metab. 2012;107(3):394–402. doi: 10.1016/j.ymgme.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Wilichowski E., Korenke G.C., Ruitenbeek W., De Meirleir L., Hagendorff A., Janssen A.J., Lissens W., Hanefeld F. Pyruvate dehydrogenase complex deficiency and altered respiratory chain function in a patient with Kearns-Sayre/MELAS overlap syndrome and A3243G mtDNA mutation. J. Neurol. Sci. 1998;157(2):206–213. doi: 10.1016/s0022-510x(98)00068-9. [DOI] [PubMed] [Google Scholar]
- 17.Buda P., Piekutowska-Abramczuk D., Karkucinska-Wieckowska A., Jurkiewicz E., Chelstowska S., Pajdowska M., Migdal M., Ksiazyk J., Kotulska K., Pronicka E. "Drop attacks" as first clinical symptoms in a child carrying MTTK m.8344A > G mutation. Folia Neuropathol. 2013;51(4):347–354. doi: 10.5114/fn.2013.39726. [DOI] [PubMed] [Google Scholar]