Abstract
The primary root plays essential roles in root development, nutrient absorption, and root architectural establishment. Primary root growth is generally suppressed by phosphate (P) deficiency in A. thaliana; however, the underlying molecular mechanisms are largely elusive to date. We found that AtOPR3 specifically inhibited primary root growth under P deficiency via suppressing root tip growth at the transcriptional level, revealing an important novel function of AtOPR3 in regulating primary root response to the nutrient stress. Importantly, AtOPR3 functioned to down-regulate primary root growth under P limitation mostly by its own, rather than depending on the Jasmonic acid signaling pathway. Further, AtOPR3 interacted with ethylene and gibberellin signaling pathways to regulate primary root growth upon P deficiency. In addition, the AtOPR3’s function in inhibiting primary root growth upon P limitation was also partially dependent on auxin polar transport. Together, our studies provide new insights into how AtOPR3, together with hormone signaling interactions, modulates primary root growth in coping with the environmental stress in Arabidopsis.
Initiated during embryo development, the primary root is the fundamental part of a root system that absorbs mineral nutrients and provides mechanical support for shoot growth. The primary root plays important roles in nutrient uptake during the early period of plant development and displays a surprising capacity of nutrient uptake in the later developmental stage, too. The maize rtcs (rootless for crown and seminal roots) mutant only with a functional primary root is able to finish its life cycle and generates progeny as a normal plant does1, suggesting that the primary root, with great growth plasticity in response to internal and external stimuli, is sufficient to support whole plant growth in terms of nutrient and water uptake. Root growth adapts to environmental changes in soil composition, and water and mineral nutrient availability via developmental and configurational alterations2. In the agricultural ecosystem, nutrient insufficiency becomes a major limiting factor for plant growth, development, and productivity, which, together with intrinsic developmental programs, reshapes root architectural patterning for nutrient favorable root morphogenesis3.
Phosphorus (P) deficiency is a very common abiotic stress that inhibits plant growth and reduces crop productivity due to poor mobility and low availability of phosphate in soils4. In contrast to inconsistent effects of low P on primary root growth in different maize inbred lines5,6,7, low P inhibits cell division in the meristematic region and promotes premature cell differentiation within the root tip, resulting in severe suppression of primary root growth in Arabidopsis8,9. Several genes have been reported to be involved in mediating primary root responses to low P in Arabidopsis. The PHOSPHATE DEFICIENCY RESPONSE 2 gene (PDR2) encodes a P5-type ATPase regulating expression of SCARECROW (SCR), a key regulator of root patterning and stem-cell niche maintenance in roots under P deficiency, and the absence of PDR2 protein further reduced primary root growth under the low P condition10,11. The other two genes, PLDζ(1,2) (Phospholipase Ds) and PRD (Phosphate root development), also positively regulate primary root growth under P deficiency12,13, while LPR (Low phosphate root) has a negative regulatory role14. Interestingly, both PDR2 and LPR1 are expressed in the stem-cell niche and distal root meristem and collaboratively modulate root meristem activities in response to external P in an ER-resident pathway11. Beyond these regulators, hormones play critical roles in root patterning under low P conditions. Ethylene modulates cell division in the quiescent center during root development15 and plays a role in restricting primary root growth in response to low P in Arabidopsis9. P deficiency can also lead to lower concentrations of bioactive gibberellins (GA) that may promote DELLA protein accumulation which, in turn, restricts primary root growth in Arabidopsis16. Exogenous application of GA can restore primary root growth in Arabidopsis under low P conditions, while DELLA-deficient mutants are less responsive to P deficiency in terms of primary root growth16. Different from ethylene and GA signaling, auxin regulates root growth under low P conditions via its redistribution17. Higher auxin concentrations in the root meristem caused by HPS4 (hypersensitive to phosphate starvation 4) mutation or blockage of auxin polar transport by 2,3,5-triiodobenzoic acid (TIBA) inhibits primary root elongation in Arabidopsis under P deficiency18,19.
Jasmonic acid (JA), a vital hormone mediating plant defense and development20,21,22,23,24, has no molecular link with primary root growth suppression in Arabidopsis under P deficiency, although exogenous application of JA is able to suppress primary root growth of Arabidopsis seedlings by reducing root meristematic activity and promoting abnormal quiescent center division under sufficient P conditions9,25,26,27. AtOPR3 is the only gene responsible for JA biosynthesis among six OPR genes in Arabidopsis28,29,30. Loss-of-function of AtOPR3 or its maize ortholog causes male sterility which is reversible by JA spray23,28, revealing a vital role of AtOPR3 in mediating flower development. Interestingly, enhanced primary root growth under low P stress in the lpi4 (low P insensitive 4) mutant is correlated with down-regulation of AtOPR3 expression9. This correlation, together with a recent report that AtOPR3 is involved in lateral root development31, implies that AtOPR3 may be a potential player regulating primary root growth in P-deficient Arabidopsis. In spite of above advances, molecular and genetic mechanisms of growth suppression of the primary root by P limitation are still largely elusive. It is particularly interesting to investigate the potential functions of AtOPR3, if any, in regulating primary root growth in Arabidopsis upon P deficiency and to reveal the underlying molecular mechanisms. Considering the limitation of P resources and environment pressure of P fertilization32,33, it is also economically imperative to explore adaptive mechanisms of plants with insufficient P supplies.
Results
AtOPR3 knockout plants had a longer primary root than WT seedlings only under the low P condition among three macronutrient deficiencies
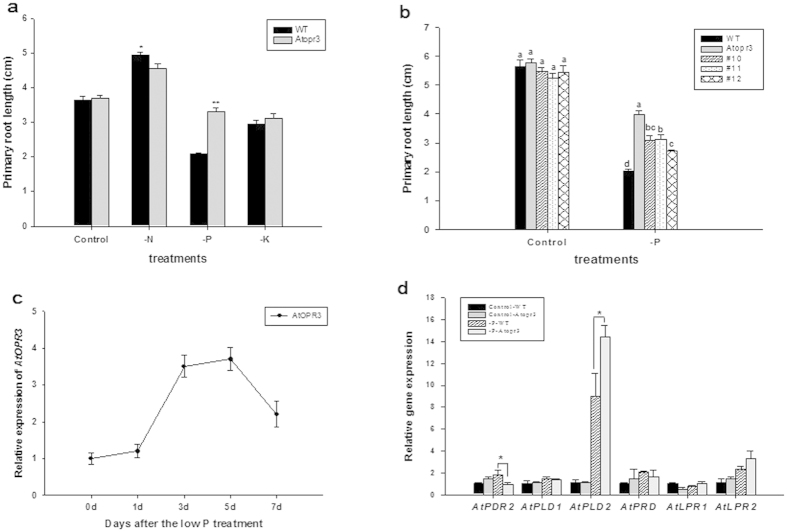
Roots respond to three macronutrient deficiencies via distinct morphological modifications. In Arabidopsis, low nitrogen (N) or P stimulates overall root growth to enhance nutrient uptake, while low potassium (K) suppresses entire root growth34,35,36. Within a root system, low N promotes lateral root growth with little effect on primary root growth36; whereas low P hinders primary root growth and induces compensatory growth of lateral roots34,37. In our results, primary root growth was inhibited by low P or K supply in sharp contrast to a significant stimulatory effect of low N. Primary root length under low N, P, and K was respectively 1.4, 0.6, 0.8 times as that of control plants (Table 1). Surprisingly, AtOPR3 knockout mutants had a 40% longer primary root than wild type (WT) plants under the low P condition (Fig. 1a, Table 1). Primary roots of Atopr3 plants remained suppressed under the low K condition and showed no significant difference compared with WT plants (Fig. 1a). These data suggested that AtOPR3 is specifically required to inhibit primary root growth under P deficiency.
Table 1. The relative length of the primary root in wild type (WT) and the Atopr3 mutant plants (Atopr3) under the low nitrogen, phosphorus, or potassium condition or under phosphorus deficiency with various chemical additives.
| Treatment | WT | Atopr3 |
|---|---|---|
| -Na | 136.0% | 123.2% |
| -Pa | 57.7% | 88.9% |
| -Ka | 81.3% | 84.4% |
| -P + 1.5 μM JAb | 89.7% | 81.0% |
| -P + 5 μM IBUb | 101.9% | 124.8% |
| -P + 5 μM DIECAb | 106.6% | 139.3% |
| -P + 5 μM AgNO3b | 135.7% | 109.0% |
| -P + 1.25 μM AVGb | 132.9% | 100.7% |
| -P + 5 μM TIBAb | 77.0% | 67.0% |
| -P + 7.5 μM GAb | 135.7% | 104.2% |
| -P + 7.5 μM PACb | 76.5% | 62.0% |
| -P + 5 μM Ancyb | 74.2% | 59.5% |
aindicated comparison of the macronutrient deficient treatment with the full nutrient treatment. “-N”, “-P”, and “-K” represented treatments of low nitrogen, phosphorus, and potassium, respectively.
bindicated comparison of the treatment with that without chemical additives under P deficiency.
Figure 1. AtOPR3 specifically inhibited primary root growth in Arabidopsis under P deficiency.
(a) Five-day-old seedlings were transferred to full nutrient (Control), low N (-N), low P (-P), or low K (-K) conditions respectively for 7 days. WT, wild type; Atopr3, the mutant line. Results were presented as means (n = 30) with error bars (standard deviation), and asterisks indicated significant differences as determined by a t-test analysis (*P < 0.05; **P < 0.01). (b) Comparision of primary root length among WT, Atopr3 and three representative AtOPR3 complementary lines (#10, #11 and #12). Results were presented as means (n = 30) with error bars (standard deviation), and different letters indicated significant differences between different lines within the same treatment (P < 0.05). (c) The relative expression level of AtOPR3 in WT during a 7-day low P treatment. Primary roots were sampled at 0d, 1d, 3d, 5d and 7d after transfer, and expression levels of AtOPR3 were determined by RT-qPCR. Data represented as means and SD (standard deviation) of three independent biological replicates. (d) Relative expression levels of six known genes regulating root response to low P in WT and Atopr3 mutant plants under the whole nutrient (Control) or low P (-P) treatment. Root samples were harvested 7d after transfer, and mRNA abundance was determined by RT-qPCR. Error bars represented SD of three independent biological replicates.
To confirm that enhanced primary root growth in Atopr3 under low P was indeed due to AtOPR3 knockout, the AtOPR3 coding sequence was expressed in the Atopr3 mutant driven by the AtUbiquitin promoter. Three independent transgenic lines were chosen for phenotypic analysis. As expected, transgenic plants showed reduced primary root growth compared to the Atopr3 mutant (Fig. 1b). Notably, gene transformation was unable to fully restore the inhibitory effect probably due to imperfect drive of a non-native promoter.
To better understand whether AtOPR3 mediates primary root growth at the transcriptional level, we analyzed relative abundance of AtOPR3 transcripts over a 7-day low P treatment. Low P stimulated AtOPR3 expression with a clear peak on day 5 after the treatment, followed by a gradual decline to the control level in WT seedlings (Fig. 1c). The low P responsive expression curve of AtOPR3 revealed that AtOPR3, as a negative regulator, may hamper primary root growth via transcriptional regulation. Notably, P1BS and P1BS-like elements are PHR1 (Phosphate Starvation Response 1)/PHLs (PHR1-Like)-bound sequences involved in regulation of P deficiency responses38,39,40. We found one putative P1BS element (GAATATAC-1897) and one putative P1BS-like (AAATATCC-910) element in the 5′-upstream region of AtOPR3 (Fig. S1). We further analyzed expression levels of previously reported genes (PDR2, PLDζ1, PLDζ2, PRD, LPR1 and LPR2) regulating primary root growth in response to P deficiency, and found down-regulation of PDR2 expression and up-regulation of PLDζ2 expression in the Atopr3 mutant root under P deficiency (Fig. 1d), although there was no significant difference in transcript accumulation of LPR1, LPR2, PRD, and PLDζ1 between WT and Atopr3 seedlings under the same condition (Fig. 1d).
Morphological analysis of primary roots showed that AtOPR3 inhibited elongation growth of the root tip
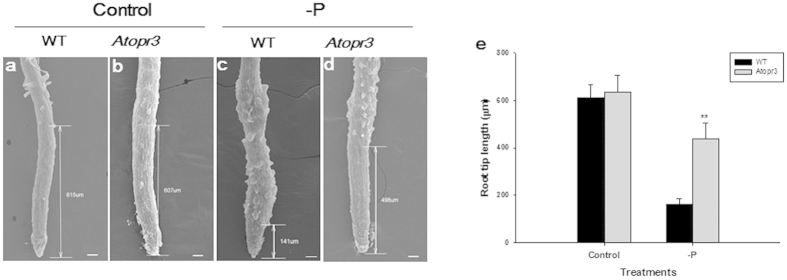
Longitudinal growth of the root tip is a prerequisite for fast root growth41. To investigate whether AtOPR3 modulates root tip growth, we analyzed in-depth morphological variation in root tips using scanning electron microscopy (Fig. 2a–d). In WT plants, root tip growth was suppressed by P limitation (Fig. 2a,c,e), and average root tip length (from the very root tip to the position where the first root hair emerges) was reduced from 613 (±52, n = 10) μm to 164 (±22, n = 10) μm. Root tip length (635 ± 70, n = 10) of Atopr3 mutant plants was similar to that of WT plants with sufficient P supply (Fig. 2a,b,e). However, Atopr3 mutant plants had approximately 2.7-fold long root tips (437 ± 66, n = 10) compared to those of WT plants (164 ± 22, n = 10) under P deficiency (Fig. 2c–e), indicating that AtOPR3 negatively mediates primary root growth, at least partially, via inhibiting longitudinal growth of the root tip.
Figure 2. AtOPR3 negatively mediates primary root growth via inhibiting longitudinal growth of the root tip.
(a–d) Scan electron microscopy analysis of root tip length in WT and Atopr3 mutant plants under the whole nutrient (Control) or low P (-P) treatment. (e) Statistical analysis of root tip length between WT and Atopr3 mutant plants. Results were presented as means (n = 10) with error bars (standard deviation), and asterisks indicated significant differences as determined by a t-test analysis (**P < 0.01).
Exogenous application of JA or JA inhibitors did not eliminate the significant difference in primary root growth between WT and Atopr3 mutant plants under P deficiency
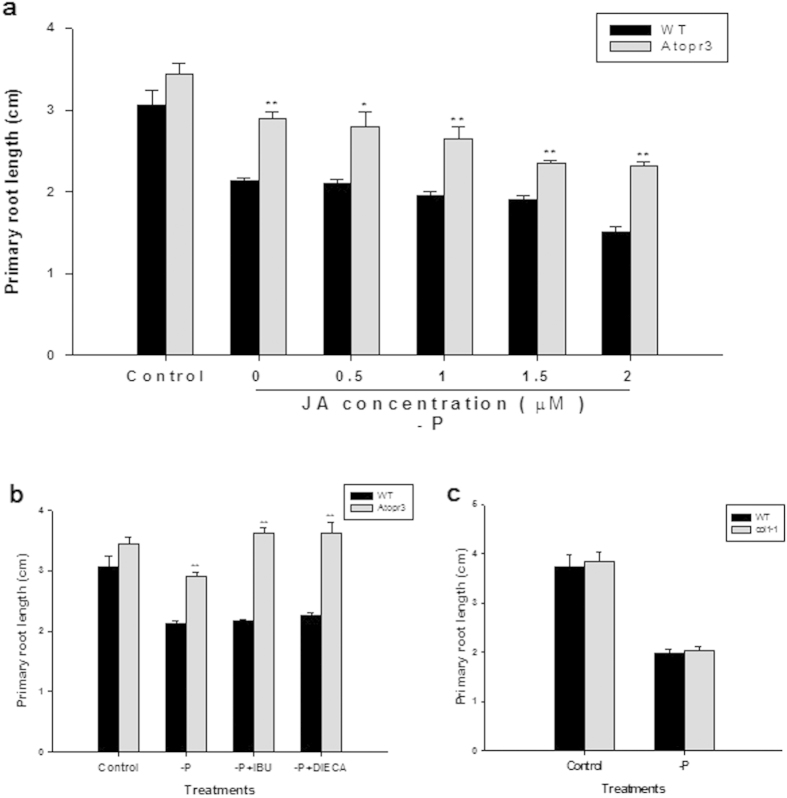
Given that AtOPR3 is a critical enzyme for JA biosynthesis, AtOPR3 may mediate root growth under low P conditions via the JA signaling pathway. We treated plants under P deficiency with exogenous JA to reduce primary root growth in Atopr3 plants. Primary roots of Atopr3 plants were still 1.3 times longer than that of WT plants although JA application reduces primary root growth in WT and Atopr3 mutants at different ratios (Table 1). The primary root is 19.0% shorter in Atopr3 mutants and 10.3% shorter in WT plants after the JA treatment (Table 1), indicating a larger effect of the JA treatment on primary root growth of the Atopr3 mutant plants. Next, we adjusted JA concentrations in a reasonable range to further minimize the length difference in the primary root of the Atopr3 mutant and WT plants under P deficiency. However, the JA treatment at other concentrations failed to eliminate the significant difference in primary root length between Atopr3 mutant and WT plants, either (Fig. 3a). We then applied JA biosynthesis inhibitors ((S)-(+)-Ibuprofen, IBU and Diethyldithiocarbamic acid, DIECA) to remove restriction of primary root growth presumably exerted by AtOPR3 mediated JA synthesis and signaling in WT plants under P deficiency. Unexpectedly, JA biosynthesis inhibitor IBU stimulated 24.8% more primary root growth in Atopr3 mutant plants compared to non-IBU treated mutant plants, by contrast to only 1.9% stimulation in WT plants (Fig. 3b, Table 1). Another JA biosynthesis inhibitor DIECA also had a larger stimulatory effect on primary root growth of Atopr3 mutant plants as compared to that in WT plants (Fig. 3b, Table 1). Together, these results suggested that the AtOPR3’s function in negatively mediating primary root growth under P deficiency is likely independent of JA biosynthesis.
Figure 3. AtOPR3’s function in mediating primary root growth under low P is independent of JA biosynthesis and signaling.
Primary root length of wild type and mutant lines (Atopr3 and Ws; coi1-1 and Col-0) were analyzed under various treatments. Five-day-old seedlings were transferred to whole nutrient (Control) or low P (-P) solutions in the presence or absence of various chemicals for 7 days. IBU and DIECA were used to block JA biosynthesis. (a) Effects of various concentrations of JA on primary root growth. (b) Effects of 5 μM IBU (-P + IBU) or 5 μM DIECA (-P + DIECA) on primary root growth. (c) Comparison of primary root growth between Col-0 and coi1-1 under control and low P conditions. Results were presented as means (n = 30) with error bars (standard deviation). Asterisks indicated significant differences as determined by a t-test analysis (*P < 0.05; **P < 0.01).
To further differentiate the short primary root phenotype of Atopr3 under P deficiency from JA signaling, we took advantage of the coi1-1 (coronatine insensitive 1) mutant line to analyze whether this mutant line has a longer primary root under P deficiency. In contrast to an obviously longer primary root in the Atopr3 mutant line than that in WT plants upon P deficiency, coi1-1 mutant plants had as short primary roots as WT plants (Fig. 3c), suggesting that blockage of JA signaling itself is not able to promote primary root growth either with sufficient P supply or under P deficiency.
AtOPR3 interacted with ethylene signaling to mediate primary root growth under the low P condition
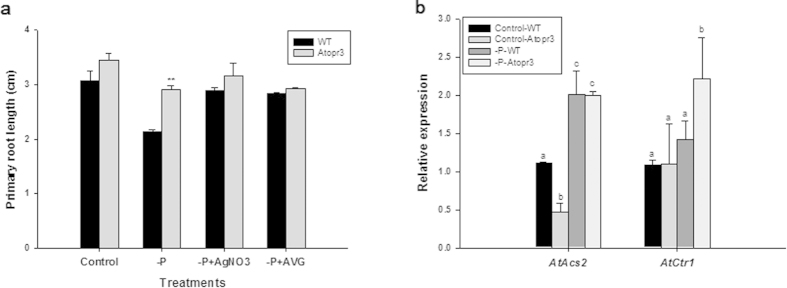
Transcriptomic analysis shows that expression of genes controlling ethylene biosynthesis are up-regulated in Arabidopsis under low P, and inhibition of ethylene biosynthesis is able to maintain normal meristem organization and activity in low P growth medium9,27,42. To investigate whether AtOPR3 interacts with ethylene signaling in regulating primary root growth in response to P deficiency, inhibitors of ethylene signaling (AgNO3) and biosynthesis (Aminoethoxyvinyl glycine hydrochloride, AVG) were separately applied into growth media. Both AgNO3 and AVG treatments had a larger stimulatory effect on primary root growth in WT plants as compared to that in the Atopr3 mutant (Table 1). Primary root length of WT plants treated with AgNO3 increased to 1.4 times when compared to non-AgNO3-treated plants under P deficiency, successfully closing the length gap between primary roots of WT and Atopr3 mutant plants (Fig. 4a, Table 1). AVG addition also nearly restored primary root growth of WT plants under P deficiency, making it statistically indistinguishable from that of Atopr3 plants (Fig. 4a, Table 1). These results indicated that AtOPR3 likely interacts with ethylene signaling to control primary root growth in response to P limitation.
Figure 4. The role of ethylene signaling in mediating primary root growth in Atopr3 and WT plants.
Seedlings were grown as described in Fig. 3. (a) Effects of 5 μM AgNO3 (-P + AgNO3) or 1.25 μM AVG (-P + AVG) on primary root growth. Results were presented as means (n = 30) with error bars (standard deviation). Asterisks indicated significant differences as determined by a t-test analysis (**P < 0.01). (b) Relative expression levels of critical genes mediating ethylene biosynthesis or signaling (determined by RT-qPCR) in WT and Atopr3 mutant plants. Data represented means and SD of three independent biological replicates. Different letters indicated means with significant differences (P < 0.05).
Ethylene biosynthesis is regulated by ACC synthase (ACS), a rate-limiting enzyme that catalyzes synthesis of the ethylene precursor ACC43. Downstream of ethylene signaling transduction is a Raf-like Ser/Thr kinase CTR1 that negatively regulates ethylene signaling44. We analyzed the expression level of AtACS2 and AtCTR1 to further characterize molecular interaction of AtOPR3 with ethylene signaling. Reverse transcription-quantitative real time PCR (RT-qPCR) analysis showed no significant difference in the expression level of AtACS2 between WT and mutant plants (Fig. 4b), whereas the expression level of AtCTR1 in the Atopr3 mutant was 1.5-fold as that in WT plants under P deficiency (Fig. 4b), suggesting that AtOPR3 may interact with ethylene signaling by restraining up-regulation of AtCTR1 expression in WT plants under P deficiency.
The GA signaling pathway was also involved in AtOPR3-mediated primary root growth under P deficiency
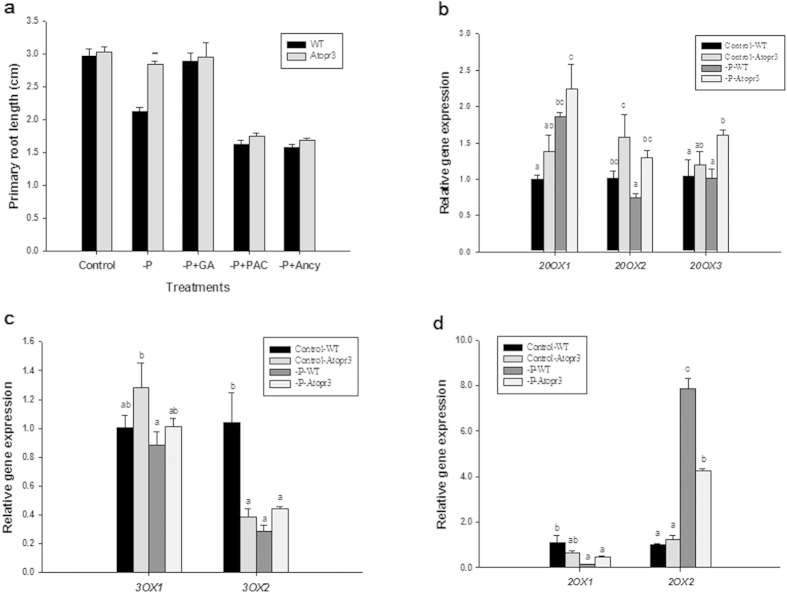
GA is a key player regulating root development and growth under P limitation16. To detect the potential interaction of AtOPR3 with the GA signaling pathway, various concentrations of GA and GA biosynthesis inhibitors (Paclobutrazol, PAC and Ancymidol, Ancy) were separately applied to growth media (Table S1). Under low P conditions, GA application promoted more primary root growth in WT plants than in Atopr3 plants so that primary root length had no significant difference between the mutant and WT plants (Fig. 5a, Table 1); GA inhibitor (PAC and Ancy) also eliminated the length gap between WT and Atopr3 plants (Fig. 5a). These results suggested that regulation of primary root growth by AtOPR3 under P deficiency is dependent on GA signaling.
Figure 5. The role of GA signaling in regulating primary root growth in Atopr3 and WT plants.
Seedlings were grown as described in Fig. 3. PAC and Ancy were applied to inhibit GA biosynthesis. (a) Effects of 7.5 μM GA (-P + GA), 7.5 μM PAC (-P + PAC), 5 μM Ancy (-P + Ancy) on primary root growth. Results were presented as means (n = 30) with error bars (standard deviation). Asterisks indicated significant differences as determined by a t-test analysis (**P < 0.01). (B-D) Relative expression levels of genes mediating GA biosynthesis (determined by RT-qPCR) in WT and Atopr3 mutants. (b) GA 20-oxidases, (c) GA 3-oxidases, and (d) GA 2-oxidases. Data represented means and SD of three independent biological replicates. Different letters indicated means with significant differences (P < 0.05).
The bioactive GA level is modulated by transcriptional up-regulation of GA 20-oxidases (GA20OX) and GA 3-oxidases (GA3OX) or transcriptional down-regulation of GA 2-oxidases (GA2OX)45,46,47. We analyzed expression levels of GA20OX, GA3OX, and GA2OX via RT-qPCR to further characterize molecular interaction of AtOPR3 with GA metabolism. We found significantly higher expression levels of GA20OX2 and GA20OX3 in Atopr3 than in WT plants under low P supply (Fig. 5b). There was no significant difference in GA3OX transcription between WT and Atopr3 mutant plants under P limitation, in spite of the higher expression level of GA3OX2 in WT plants with sufficient P supply (Fig. 5c). On the other hand, Atopr3 mutants had a significantly lower expression level of GA2OX2 than WT plants under low P conditions (Fig. 5d). Taken together, Atopr3 mutants had more GA biosynthesis and less degradation, resulting in higher bioactive GA levels in the mutant line than in WT plants under low P conditions, which stimulated root growth under P limitation.
In addition, auxin plays an essential role in controlling root growth via asymmetric distribution48. Inhibition of auxin polar transport by TIBA reduced primary root growth by 33.0% and 23.0% respectively in Atopr3 mutant and WT plants under P deficiency compared to non-TIBA-treated plants, implying that Atopr3 mutant plants are likely more sensitive to TIBA than WT seedlings (Table 1, Fig. S2). However, the Atopr3 plants still had significantly longer primary roots than WT seedlings regardless of various concentrations of TIBA treatments (Table S1). Although auxin preconditions root growth, our data showed that auxin signaling was not a major player in AtOPR3 mediated primary root growth under P deficiency.
Discussion
The primary root plays essential roles in nutrient uptake, and is sufficient for plants to finish their lifecycle1. Low P bioavailability in the soil makes P deficiency one of the most limiting factors for fast plant growth, reproduction, and food production32. A conspicuous change in P deficient Arabidopsis is arrested primary root growth34. Although several genes and hormone signaling have been reported to be involved in regulating primary root growth upon P deficiency49, molecular and genetic mechanisms of growth inhibition of the primary root by P deficiency in Arabidopsis remain fundamental questions to be elucidated in plant stress physiology.
AtOPR3 specifically inhibits primary root growth under the low P condition by restraining longitudinal growth of the root tip in Arabidopsis
Macronutrient deficiencies alter root architecture in certain common ways in Arabidopsis. P and K deficiencies inhibit primary root growth, and N and P deficiencies enhance lateral root growth34,35,36. It is particularly important to dissect these commonalities and identify nutrient-specific features at the molecular level. AtOPR3, as a critical enzyme in JA biosynthesis, has crucial biological functions in flower development and defense response28,50. Here, Atopr3 mutant plants showed compensatory primary root growth under N deprivation and no effect on primary root growth under K privation compared to WT plants under the same condition (Fig. 1a). Importantly, the primary root of Atopr3 plants showed continuous growth, rather than arrested by P deficiency as shown in WT plants (Fig. 1a). Our results demonstrated a novel function of AtOPR3 in regulating primary root response to abiotic stresses: AtOPR3 inhibited primary root growth only under P deficiency among three macronutrient deficiencies (Fig. 1a).
Up-regulation of AtOPR3 expression in wild-type plants under P deficiency confirmed that it is a negative regulator functioning at the transcriptional level (Fig. 1c). The presence of the P1BS and P1BS-like elements in the AtOPR3 5′-upstream region indicates that AtOPR3 could potentially interact with PHR1/PHLs in regulation of root responses to P deficiency38,39,40 (Fig. S1). Many genes are common regulators in root response to N, P, and K deficiencies. Identification of AtOPR3 as a P specific root growth regulator not only helps interpret contrasting performance of the primary root under macronutrient deficiencies, but also provides a powerful molecular marker in P nutritional diagnosis. Further, AtOPR3 negatively regulates primary root growth partially by inhibiting longitudinal growth of the root tip (Fig. 2a–e). We speculated that the inhibitory effect of AtOPR3 on primary root growth was due to a dramatic decrease either in cell number or cell length in elongation and apical meristematic zones. This is consistent with previous report that low P inhibits primary root growth in Arabidopsis through arresting cell division and promoting cell differentiation in these two zones8. Six genes (PDR2, PLDζ1, PLDζ2, PRD, LPR1 and LPR2) are previously reported to modulate primary root growth under P deficiency10,11,12,13,14,51; however, four (PLDζ1, PRD, LPR1, LPR2) of them had no significantly altered expression in Atopr3 mutant plants upon P deficiency (Fig. 1d). Down-regulation of PDR2 is expected to have a negative effect on primary root growth10,11, in contrast to stimulatory root growth in the Atopr3 mutant plant under low P; 1.5-fold up-regulation of PLDζ2 expression probably causes no effect on root growth given that only simultaneous knockout of PLDζ1 and PLDζ2 leads to a shorter primary root12. Thus, we concluded that AtOPR3 inhibits primary root growth under the low P condition likely independent of these reported molecular pathways, revealing a novel molecular mechanism of root growth regulation in Arabidopsis in response to the P stress.
The AtOPR3’s function in regulating primary root growth under low P conditions is mostly independent of JA signaling
AtOPR3 regulates flower development and pathogen defense, and AtOPR3 knockout causes male sterility and seriously weakens plant resistance to pathogen attack23,28. These functional defects in the Atopr3 mutant line are fully restored by exogenous JA application, suggesting that AtOPR3 functions via the JA signaling pathway. Similarly, AtOPR3 down-regulates primary root growth under P deficiency probably via JA signaling, too. However, under P deficiency, JA addition was unable to suppress compensatory growth of the primary root in the Atopr3 mutant, and JA inhibitors failed to restore primary root growth of WT plants (Fig. 3a,b), clearly suggesting that stimulated primary root growth in the Atopr3 mutant under P deficiency is primarily not a major consequence of down-regulation of AtOPR3 mediated-JA synthesis, but a more direct result derived from functional knockout of AtOPR3 itself. Further, the JA signaling mutant coi1-1 exhibited growth arrest of the primary root in response to low P just as WT seedlings did (Fig. 3c), providing strong genetic evidence that AtOPR3 down-regulates primary root growth under low P conditions mostly independent of the JA signaling pathway, similar to many other genes with multiple functions depending on biological contexts. TRH1 primarily functions as a potassium transporter, mediates auxin transport, and is required for morphogenesis of root hairs52,53,54. The potassium transporter KUP2 is also essential for cell expansion in the shoot55. Although exogenous JA application inhibits primary root growth to different extents in Arabidopsis26, our data implied that JA is not a dominant player in AtOPR3 mediated primary root growth in response to P deficiency. However, we are not ruling out the possibility that JA may function in this process via unidentified interactions with other hormone signaling pathways.
AtOPR3 interacts with ethylene and GA signaling pathways to regulate root growth under low P conditions
Although it is well established that ethylene plays an essential role in modulating root development15, it remains largely unclear how ethylene is involved in regulation of primary root growth under P deficiency. Upregulated expression of ethylene biosynthesis and signaling related genes under low P supports a potential role of ethylene in mediating plant response to P limitation42, which is further strengthened by the recent finding that AVG and AgNO3 treatments are able to maintain meristem organization and activity under P deficiency, while ACC addition disrupts meristem functions in the primary root under low or high P supply9. In our study, ethylene signaling and biosynthesis inhibitors fully restored primary root growth in WT and Atopr3 plants under low P conditions and closed the large length gap in primary roots between WT and mutant plants (Fig. 4a). Thus, we speculated that AtOPR3 inhibited primary root growth via interacting with the ethylene signaling pathway in Arabidopsis. We further found that CTR1, negatively regulating ethylene signaling as a downstream receptor44,56, had significantly higher expression levels under low P in the Atopr3 mutant seedlings than in the WT plants (Fig. 4b). Up-regulation of AtCTR1 expression in Atopr3 mutant plants alleviated inhibitory effects of P-deficiency triggered ethylene signaling on primary root growth.
Not just ethylene signaling, the decrease in bioactive GA levels and subsequent DELLA accumulation under low P conditions result in growth arrest of the primary root and trigger other P starvation responses16. In our studies, GA inhibitors (PAC and Ancy) had a larger inhibitory effect on primary root growth of Atopr3 plants than on WT, and exogenous GA promoted more primary root growth in WT plants than in Atopr3 mutants under low P conditions (Fig. 5a). Either treatment eliminated the significant difference in primary root length between the mutant and WT plants exerted by the P stress, suggesting that AtOPR3 also interacts with GA signaling to mediate primary root growth in response to P limitation. In P-deficient WT plants, AtOPR3 may down-regulate accumulation of bioactive GAs to reduce primary root growth, putatively via down-regulating GA20OX2 and GA20OX3 transcription and up-regulating GA2OX2 transcription (Fig. 5b,d), although specific mechanisms need further investigation. Functional knockout of AtOPR3 may result in higher bioactive GA levels that in turn promote root growth in adaptation to external stimulus of P stress (Fig. 5a,b,d).
Although auxin is also an essential growth regulator, and exogenous auxin application restricts primary root growth17,57, blockage of auxin polar transport by TIBA failed to eliminate the significant length difference between WT and Atopr3 mutant plants in our studies (Table 1, Fig. S2). Therefore, we conclude that AtOPR3 mostly interacts with ethylene and GA signaling pathways to modulate root growth, with auxin signaling as a secondary interaction when coping with the P stress.
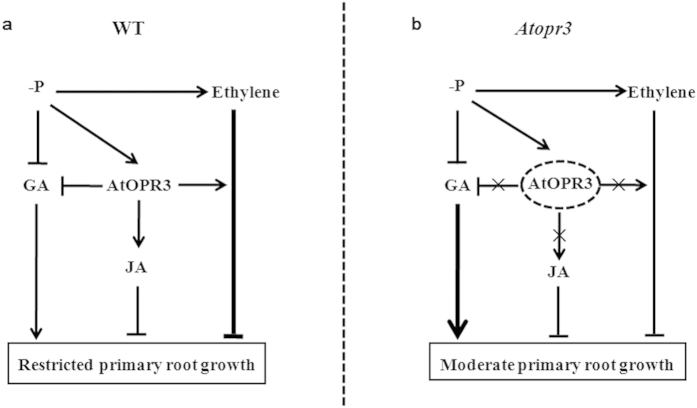
Therefore, we proposed a mechanistic model of how AtOPR3 mediates primary root growth via interaction with ethylene and GA signaling pathways (Fig. 6). With sufficient P supply, the primary root maintains its normal growth rhythm in WT plants; blockage of JA biosynthesis itself has no any inhibitory or significant stimulatory effect on primary root growth in the Atopr3 mutant (Fig. 3b,c). Under P limitation, up-regulation of AtOPR3 expression in the WT plants promotes JA biosynthesis, enhances P stress-triggered ethylene signaling potentially via down-regulating CTR1 expression, and attenuates GA signaling by reducing bioactive GA contents, which collectively suppresses primary root growth (Fig. 6a). Although our observation did not support that JA is a major player in AtOPR3 mediated-primary root growth (Fig. 3), an increase in JA content, caused by up-regulation of AtOPR3 expression, may reduce root growth to a certain extent or has other complicated effects via unknown hormone signaling interactions. By contrast, knockout of AtOPR3 in the mutant line indirectly up-regulates the level of bioactive GA, reduces strength of P-stress directed ethylene signaling, and terminates JA biosynthesis under P deficiency (Fig. 6b). All these modulations alleviate suppression effects of the P stress on primary root growth, resulting in a significantly longer primary root in the Atopr3 mutant plants than in WT plants, although underlying molecular mechanisms remain to be further explored.
Figure 6. A schematic model of how AtOPR3 regulates primary root growth under low P conditions, putatively via interacting with ethylene and GA signaling pathways in Arabidopsis.
When supplied with sufficient P, both WT and Atopr3 mutant plants show normal root growth due to absence of low P stress signals. (a) In the WT plants, AtOPR3 expression is stimulated by low P supply. Up-regulation of AtOPR3 causes three biological consequences: stimulation of JA biosynthesis, enhancement of ethylene signaling, and down-regulation of the bioactive GA content, which collectively suppresses primary root growth under P deficiency. (b) In AtOPR3 knockout mutant plants, absence of functional AtOPR3 transcripts blocks JA biosynthesis, reduces strength of P-stress directed ethylene signaling, and indirectly up-regulates the level of bioactive GA. All these alterations result in continuous primary root growth as a whole in spite of P limitation.
Materials and Methods
Materials
Arabidopsis ecotype Wassilewskija (Ws), Columbia (Col), T-DNA insertional mutant Atopr328 in which AtOPR3-mediated JA biosynthesis is blocked, and coi1-1 mutant58 in which JA signaling is blocked were used in our experiments.
Plasmid construction and plant transformation
AtOPR3 coding sequence was cloned into the T-vector pMD19 (Takara), and then inserted into a pUT-hyg vector59 (with the AtUbiquitin promoter and hygromycin resistance) using restriction sites SalI and SpeI for functional complementation. The resulting plasmid was transformed into Agrobacterium tumefaciens GV3101 by electroporation and further delivered into the Atopr3 plants using the standard floral dip method60. Homozygous lines of the T3 generation were used for phenotypic analysis.
Arabidopsis growth
Seeds were first imbibed in water in the dark at 4 °C for 2 days to break dormancy. Then seeds were surface sterilized (75% ethanol (V/V) for 1 minute and 2% NaClO (V/V) for 2 minutes, followed by six rinses in sterile water) and sown on 1/2MS (Murashige and Skoog Stock) plate with 1% sugar and 0.8% agar. Plates were placed vertically in a standard plant growth chamber (Kooland, China) under the following condition: 22 °C, illumination 100 μmol photons m−2 s−1, 16/8 h light/dark, 60% relative humidity. For coi1-1 mutant screen, seeds were sown on the plate with 25 μM Methyl jasmonate for three days to screen for homozygous plants which were then transferred to 1/2 MS for continuous growth. After five days, the uniform seedlings were transferred to plates with whole nutrient, N deficient (5 μM N), P deficient (10 μM P) or K deficient MS medium (5 μM K) respectively. The whole nutrient medium contained 21 mM NH4NO3, 19 mM KNO3, 1.25 mM KH2PO4, 3 mM CaCl2, 1.5 mM MgSO4·7H2O, 0.005 mM KI, 0.1 mM MnSO4·H2O, 0.03 mM ZnSO4·H2O, 0.001 mM Na2MoO4·2H2O, 0.0001 mM CuSO4·5H2O, 0.0001 mM CoCl2·6H2O, 0.1003 mM H3BO3, 0.1 mM EDTA-Fe. The pH maintains at 5.8. To make the low N and low P media, NH4NO3, KNO3, KH2PO4 were replaced by KCl in the nutrient solution. To make low K media, KNO3 and KH2PO4 were replaced by NH4H2PO4 in the nutrient solution.
The plates were supplemented with or without various hormones or inhibitors. All experiments had six technical replicates, five biological replicates. Wild-type and mutant plants were transferred and grown on different sides of the same plate. Seedlings were harvested seven days after transfer. For each treatment, primary roots of 30 12-day-old seedlings were measured from the root tip to the hypocotyl base with a ruler.
Hormone treatments
Before making plates, appropriate amount of individual hormone solutions were added into the culture medium around 50 °C. Jasmonic acid (Sigma), (S)-(+)-Ibuprofen (Sigma), Paclobutrazol (Sigma), Gibberellic acid (Sigma), Ancymidol (Sigma) were dissolved in ethanol. Aminoethoxyvinyl glycine hydrochloride (Sigma), AgNO3 (Sinopharm Chemical Reagent Co., LTD), 2,3,5-triiodobenzoic acid (Sigma) and Diethyldithiocarbamic acid (Sigma) were dissolved in ddH2O.
Transcriptional analyses
Root samples were quickly harvested and immediately frozen in liquid nitrogen. Arabidopsis total RNA was extracted with the RNAprep pure Plant kit (TIANGEN, Beijing). cDNA synthesis and RT-qPCR was carried out following manufacturer’s instructions61. The Arabidopsis TUB4 gene was used as a positive internal control. The primers used for RT-qPCR analysis and gene cloning are listed in Table S2.
Preparations of Arabidopsis root samples for scanning electron microscopy (SEM)
We followed a standard protocol slightly modified after Burgess and Linstead62. Briefly, 0.5 mm root tip samples were fixed with 2.5% Glutaraldehyde and 1% osmic acid sequentially. After wash in PBS (PH 7.2, 0.1M) buffer, fixed root samples were dehydrated with six gradient ethanol (30%–50%–70%–80%–90%–100%) and isoamyl acetate three times, followed by critical-point drying (HITACHI HCP-2) and ion sputtering (EIKO IB-3). The samples were than analyzed on the scanning electron microscope (HITACHI S-3400N) according to the standard instructions. The root tip referred to the section from the very tip point to the point where the first root hair primordium initiated.
Additional Information
How to cite this article: Zheng, H. et al. AtOPR3 specifically inhibits primary root growth in Arabidopsis under phosphate deficiency. Sci. Rep. 6, 24778; doi: 10.1038/srep24778 (2016).
Supplementary Material
Acknowledgments
This work was supported by the NSFC grant (31471928), Program for New Century Excellent Talents in University (NCET-12-0521), and the Innovative Group Grant of the National Natural Science Foundation of China (31421092). The authors thank Dr. John Browse for providing the Atopr3 mutant line and Dr. Daoxin Xie for providing the coi1-1 mutant line.
Footnotes
Author Contributions X.L., H.Z. and P.L. designed research; H.Z., X.P., Y.D. and H.W. performed research; X.L. and H.Z. analyzed data; H.Z. and X.L. wrote the paper.
References
- Hetz W., Hochholdinger F., Schwall M. & Feix G. Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant Journal 10, 845–857 (1996). [Google Scholar]
- Malamy J. E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77 (2005). [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Cruz-Ramírez A. & Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6, 280–287 (2003). [DOI] [PubMed] [Google Scholar]
- Vonuexkull H. R. & Mutert E. Global extent, development and economic-impact of acid soils. Plant Soil 171, 1–15 (1995). [Google Scholar]
- Anghinoni I. & Barber S. A. Phosphorus influx and growth-characteristics of corn roots as influenced by phosphorus supply. Agronomy Journal 72, 685–688 (1980). [Google Scholar]
- Khamis S., Chaillou S. & Lamaze T. CO2 assimilation and partitioning of carbon in maize plants deprived of orthophosphate. J. Exp. Bot. 41, 1619–1625 (1990). [Google Scholar]
- Mollier A. & Pellerin S. Maize root system growth and development as influenced by phosphorus deficiency. J. Exp. Bot. 50, 487–497 (1999). [Google Scholar]
- Sanchez-Calderon L. et al. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant & cell physiology 46, 174–184 (2005). [DOI] [PubMed] [Google Scholar]
- Chacón-López A., Ibarra-Laclette E., Sánchez-Calderón L., Gutiérrez-Alanis D. & Herrera-Estrella L. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signaling & Behavior 6, 382–392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi C. A., Delatorre C. A., Lahner B., Salt D. E. & Abel S. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant Journal 37, 801–814 (2004). [DOI] [PubMed] [Google Scholar]
- Ticconi C. A. et al. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106, 14174–14179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Qin C., Welti R. & Wang X. Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140, 761–770 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Cristobal J. J. et al. PRD, an Arabidopsis AINTEGUMENTA-like gene, is involved in root architectural changes in response to phosphate starvation. Planta 228, 511–522 (2008). [DOI] [PubMed] [Google Scholar]
- Svistoonoff S. et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nature genetics 39, 792–796 (2007). [DOI] [PubMed] [Google Scholar]
- Ortega-Martinez O., Pernas M., Carol R. J. & Dolan L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317, 507–510 (2007). [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N. P. & Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol 145, 1460–1470 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P. et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138, 2061–2074 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J. et al. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129, 244–256 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. L., Luo N., Sun L. C. & Liu D. HPS4/SABRE regulates plant responses to phosphate starvation through antagonistic interaction with ethylene signalling. J. Exp. Bot. 63, 4527–4538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P., Shockey J., Levesque C. A., Cook R. J. & Browse J. A role for jasmonate in pathogen defense of Arabidopsis. Proc Natl Acad Sci USA 95, 7209–7214 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P. M. et al. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D. F. & Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128, 876–884 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 24, 1420–1436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P. et al. JAV1 Controls Jasmonate-Regulated Plant Defense. Molecular Cell 50, 504–515 (2013). [DOI] [PubMed] [Google Scholar]
- Staswick P. E., Su W. P. & Howell S. H. Methyl jasmonate inhibition of root-growth and induction of a leaf protein are decreased in an arabidopsis-thaliana mutant. Proc Natl Acad Sci USA 89, 6837–6840 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W. et al. Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol 141, 1400–1413 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23, 3335–3352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A. & Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97, 10625–10630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breithaupt C. et al. Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization. Proc Natl Acad Sci USA 103, 14337–14342 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon E. R. et al. The role of oxophytodienoate reductases in the detoxification of the explosive 2,4,6-trinitrotoluene by Arabidopsis. Plant Physiol 151, 253–261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ma J. & Liu P. OPR3 is expressed in phloem cells and is vital for lateral root development inArabidopsis. Canadian Journal of Plant Science 93, 165–170 (2013). [Google Scholar]
- Vance C. P., Uhde-Stone C. & Allan D. L. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447 (2003). [DOI] [PubMed] [Google Scholar]
- Conley D. J. et al. ECOLOGY Controlling Eutrophication: Nitrogen and Phosphorus. Science 323, 1014–1015 (2009). [DOI] [PubMed] [Google Scholar]
- Williamson L. C., Ribrioux S., Fitter A. H. & Leyser H. M. O. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 126, 875–882 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R. & Schachtman D. P. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101, 8827–8832 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T. et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140, 909–921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. A Comprehensive Differential Proteomic Study of Nitrate Deprivation in Arabidopsis Reveals Complex Regulatory Networks of Plant Nitrogen Responses. J. Proteome Res. 11, 2301–2315 (2012). [DOI] [PubMed] [Google Scholar]
- Bustos R. et al. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. Plos Genetics 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobkowiak L. et al. The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Frontiers in Plant Science 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Song L., Zhang Y., Zheng Z. & Liu D. Arabidopsis PHL2 and PHR1 Act Redundantly as the Key Components of the Central Regulatory System Controlling Transcriptional Responses to Phosphate Starvation. Plant Physiol. 170, 499–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G. T. S. & Baskin T. I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud M. C. et al. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. The Plant journal : for cell and molecular biology 64, 775–789 (2010). [DOI] [PubMed] [Google Scholar]
- Yang S. F. & Hoffman N. E. Ethylene biosynthesis and its regulation in higher-plants. Annual Review of Plant Physiology and Plant Molecular Biology 35, 155–189 (1984). [Google Scholar]
- Kieber J. J. The ethylene response pathway in arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology 48, 277–296 (1997). [DOI] [PubMed] [Google Scholar]
- Chiang H. H., Hwang I. & Goodman H. M. Isolation of the arabidopsis GA4 locus. Plant Cell 7, 195–201 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. L. et al. The GA5 locus of arabidopsis-thaliana encodes a multifunctional gibberellin 20-oxidase-molecular-cloning and functional expression. Proc Natl Acad Sci USA 92, 6640–6644 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. G., Phillips A. L. & Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96, 4698–4703 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K. & Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology 24, 55–80 (2008). [DOI] [PubMed] [Google Scholar]
- Peret B., Clement M., Nussaume L. & Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16, 442–450 (2011). [DOI] [PubMed] [Google Scholar]
- Brioudes F. et al. Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. The Plant Journal : for Cell and Molecular Biology 60, 1070–1080 (2009). [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon L. et al. Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140, 879–889 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S. et al. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13, 139–151 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Agullo F. et al. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40, 523–535 (2004). [DOI] [PubMed] [Google Scholar]
- Desbrosses G. et al. AKT1 and TRH1 are required during root hair elongation in Arabidopsis. J Exp Bot 54, 781–788 (2003). [DOI] [PubMed] [Google Scholar]
- Elumalai R. P., Nagpal P. & Reed J. W. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14, 119–131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. L., Larsen P. B., Wang X. X. & Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors (vol 95, pg 5401, 1998). Proc Natl Acad Sci USA 95, 9060–9060 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J. et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol 137, 681–691 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D. X., Feys B. F., James S., Nieto-Rostro M. & Turner J. G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998). [DOI] [PubMed] [Google Scholar]
- Gu R. et al. Characterization of AMT-Mediated High-Affinity Ammonium Uptake in Roots of Maize (Zea mays L.). Plant Cell Physiol. 54, 1515–1524 (2013). [DOI] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Burgess J. & Linstead P. J. Scanning electron-microscopy of cell-wall formation around isolated plant protoplasts. Planta 131, 173–178 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.