Abstract
Published data on the association between the coiled-coil domain-containing 26 (CCDC26) rs4295627 polymorphism and the risk of glioma have been inconclusive. To further investigate this association, a meta-analysis was performed. By a comprehensive literature search using PubMed and EMBASE databases, a total of 16 case-control studies were identified for inclusion in the meta-analysis. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess this association. Our results confirmed that the risk with allele G was higher compared with that with allele T for glioma. The results indicated that the allele G of rs4295627 polymorphism in the CCDC26 gene was associated with increased risk of glioma in the homozygote model (GG vs. TT, OR=1.936, 95 %CI: 1.500–2.658, P<0.001), the heterozygote model (GT vs. TT, OR=1.323, 95% CI: 1.241–1.412, P=0.206), the dominant model (GG+GT vs. TT, OR=1.375, 95% CI: 1.256–1.505, P=0.026), the recessive model (GG vs. GT+TT, OR=1.769, 95% CI: 1.302–2.403, P<0.001) and the allele model (G vs. T, OR=1.310, 95% CI: 1.185–1.448, P<0.001). Current evidence suggests that the rs4295627 polymorphism in the CCDC26 gene may contribute to glioma susceptibility. However, further case-control studies are required to confirm our results.
Keywords: glioma, polymorphism, coiled-coil domain-containing 26, meta-analysis
Introduction
Glioma is the most common type of central nervous system (CNS) tumor in adults, accounting for ~80% of CNS tumors, with a dismal prognosis (1). However, gliomagenesis is yet to be understood. Exposure to ionizing radiation is the only identified environmental risk factor for glioma supported by strong evidence (2). However, not all individuals exposed to radiation ultimately develop glioma, suggesting that other causes, including host genetic factors, may play a critical role in the process of gliomagenesis.
In recent years, several genes have been identified as potential glioma susceptibility genes, including RAD51, TP53, telomerase reverse transcriptase and regulator of telomere elongation helicase 1 (3–8). Another important gene is coiled-coil domain-containing 26 (CCDC26), which increases the apoptosis of glioblastoma cells by modulating differentiation and apoptosis following induction by retinoic acid treatment (9). The CCDC26 gene is located on chromosome 8q24.21. The rs4295627 polymorphism is located in intron 3 of the CCDC26 gene. The G→A change in this single-nucleotide polymorphism (SNP) has been reported by several studies to affect CCDC26 expression and glioma risk (8,10–14). However, the results were inconclusive, partially due to the possible small effect of the polymorphism on glioma risk and the relatively small sample size in each of published studies. Therefore, a meta-analysis was performed to evaluate the association between the rs4295627 polymorphism and the risk of glioma.
Materials and methods
Publication search
A computerized literature search was conducted through PubMed and EMBASE databases to identify the relevant studies evaluating the association between the CCDC26 rs4295627 polymorphism and the risk of glioma, using the following search terms: ‘CCDC26’ or ‘rs4295627’, ‘polymorphism’ or ‘variation’ and ‘glioma’. The search was limited to human studies. The last search was performed on March 28th, 2015. All selected studies were retrieved and their bibliographies were checked for other relevant publications. References from related articles were also reviewed for additional eligible studies. Only published studies with full-text articles were included.
Inclusion criteria
The inclusion criteria were as follows: i) Evaluation of the CCDC26 and glioma risk; ii) case-control studies; and iii) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI).
Data extraction
Information was carefully extracted from all eligible publications by two authors independently, and any disagreements were resolved by discussion. Conflicting evaluations were resolved by discussion and by consulting a senior reviewer to reach a final decision. The following data were collected from each study: First author, publication year, country of origin, ethnicity, total number of cases and controls, and genotype frequency of cases and controls. We did not define any minimum number of patients for inclusion in our meta-analysis.
Statistical analysis
Crude ORs with 95% CIs were used to assess the strength of the association between the CCDC26 polymorphism and glioma risk. The pooled ORs were performed with the additive model (GT vs. TT; GG vs. TT), dominant model (GT+GG vs. TT), recessive model (GG vs. GT+TT) and allele model (G vs. T). Heterogeneity was assessed by the Chi-square-based Q test. If the P-value was >0.10, indicating a lack of heterogeneity among studies, the pooled OR estimate of the each study was calculated by the fixed-effects model. Otherwise, the random-effects model (DerSimonian-Laird method) was applied (15). A sensitivity analysis was performed to assess the stability of the results. A single study involved in the meta-analysis was deleted each time to reflect the effect of the individual data set on the pooled OR. Funnel plots, Begg's tests and Egger's tests were performed to diagnose potential publication bias (P<0.05 was considered representative of statistically significant publication bias) (16). All the statistical tests were performed with Stata software, version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Study characteristics
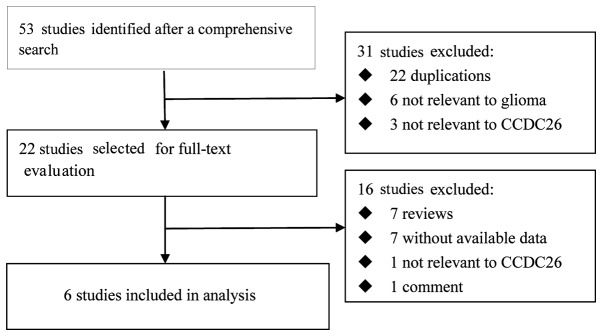
A total of 6 publications including 16 case-control studies met the inclusion criteria (8,10–14). The study selection process is outlined in Fig. 1. These studies were published between 2009 and 2015. Two studies were conducted in America, one in China, one in Sweden and two were multi-country studies. In total, 7,290 cases and 11,630 controls were used in the pooled analyses. In the 16 studies, the sample size ranged from 95 to 1,579 subjects. Of the 16 studies, 15 included a Caucasian population and 1 study included an Asian population. The main characteristics of the studies are summarized in Table I.
Figure 1.
Flow diagram of the study selection process. CCDC26, coiled-coil domain-containing 26.
Table I.
Characteristics of studies included in the present meta-analysis.
| Simple size | Case genotype | Control genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First Author | Year | Country | Ethnicity | Cases | Controls | GG | GT | TT | GG | GT | TT | Source of controls | Genotyping method | (Refs.) |
| Shete (France) | 2009 | USA | Caucasian | 1,374 | 1,579 | 0 | 418 | 885 | 25 | 421 | 1,133 | PB | Illumina | (14) |
| Shete (German) | 2009 | USA | Caucasian | 498 | 571 | 30 | 185 | 283 | 13 | 144 | 414 | PB | Illumina | (14) |
| Shete (Sweden) | 2009 | USA | Caucasian | 643 | 775 | 27 | 223 | 393 | 36 | 247 | 492 | PB | Illumina | (14) |
| Shete (UK) | 2009 | USA | Caucasian | 631 | 1,434 | 29 | 216 | 386 | 48 | 410 | 976 | PB | Illumina | (14) |
| Shete (US) | 2009 | USA | Caucasian | 1,246 | 2,235 | 60 | 451 | 735 | 72 | 667 | 1,496 | PB | Illumina | (14) |
| Schoemaker (Denmark) | 2010 | UK | Caucasian | 123 | 147 | 7 | 40 | 76 | 3 | 46 | 98 | PB | PCR | (13) |
| Schoemaker (Finland) | 2010 | UK | Caucasian | 97 | 95 | 16 | 34 | 47 | 6 | 31 | 58 | PB | PCR | (13) |
| Schoemaker (Sweden) | 2010 | UK | Caucasian | 199 | 372 | 6 | 63 | 130 | 14 | 117 | 241 | PB | PCR | (13) |
| Schoemaker (UK-North) | 2010 | UK | Caucasian | 375 | 617 | 16 | 122 | 237 | 27 | 156 | 434 | PB | Illumina, PCR | (13) |
| Schoemaker (UK-South) | 2010 | UK | Caucasian | 232 | 396 | 12 | 83 | 137 | 11 | 119 | 266 | PB | Illumina, PCR | (13) |
| Wang | 2011 | USA | Caucasian | 332 | 817 | 24 | 121 | 187 | 19 | 242 | 556 | HB | Illumina | (12) |
| Li | 2012 | China | Asian | 225 | 254 | 12 | 92 | 121 | 25 | 102 | 127 | HB | MassArray | (11) |
| Safaeian (NCI) | 2013 | USA | Caucasian | 322 | 385 | 17 | 123 | 182 | 11 | 107 | 267 | HB | Illumina | (10) |
| Safaeian (NIOSH) | 2013 | USA | Caucasian | 300 | 538 | 14 | 107 | 179 | 23 | 158 | 357 | PB | Illumina | (10) |
| Safaeian (PLCO) | 2013 | USA | Caucasian | 133 | 855 | 10 | 40 | 83 | 23 | 248 | 584 | PB | Illumina | (10) |
| Wibom | 2015 | Sweden | Caucasian | 560 | 560 | 36 | 174 | 350 | 6 | 163 | 391 | HB | PCR | (8) |
PB, population-based; HB, hospital-based; PCR, polymerase chain reaction; NCI, National Cancer Institute; NIOSH; National Institute for Occupational Safety and Health; PLCO, prostate, lung, colorectal and ovarian cancer screening trial.
Meta-analysis results
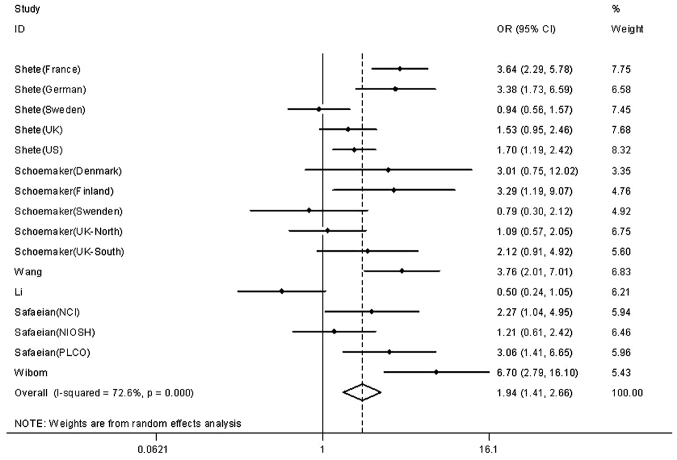
The main results of the meta-analysis are listed in Table II. Our results confirmed that the risk for glioma with allele G was higher compared with that with allele T. The results indicated that the allele G of rs4295627 polymorphism in the CCDC26 gene was associated with increased risk of glioma in the homozygote model (GG vs. TT, OR=1.936, 95% CI: 1.500–2.658, P<0.001), the heterozygote model (GT vs. TT, OR=1.323, 95% CI: 1.241–1.412, P=0.206), the dominant model (GG+GT vs. TT, OR=1.375, 95% CI: 1.256–1.505, P=0.026), the recessive model (GG vs. GT+TT, OR=1.769, 95% CI: 1.302–2.403, P<0.001) and the allele model (G vs. T, OR=1.310, 95% CI: 1.185–1.448, P<0.001). Subgroup analyses by ethnicity and source of controls were not performed, as there was only one Asian case-control study with a small sample size and a small hospital-based population. The forest plot for the CCDC26 rs4295627 polymorphism in the homozygote model is presented in Fig. 2.
Table II.
Summary of total pooled results in different comparative genetic models.
| Genetic models | OR (95% CI) | Z | P-value | I2 (%) | Effects model |
|---|---|---|---|---|---|
| GG vs. TT | 1.936 (1.500–2.658) | 4.08 | <0.001 | 72.60 | Random |
| GT vs. TT | 1.323 (1.241–1.412) | 8.49 | 0.206 | 21.80 | Fixed |
| GG+GT vs. TT | 1.375 (1.256–1.505) | 6.91 | 0.026 | 45.10 | Random |
| GG vs. GT+TT | 1.769 (1.302–2.403) | 3.64 | <0.001 | 71.30 | Random |
| G vs. T | 1.310 (1.185–1.448) | 5.27 | <0.001 | 68.20 | Random |
OR, odds ratio; CI, confidence interval.
Figure 2.
Forest plot for the coiled-coil domain-containing 26 rs4295627 polymorphism (homozygote model). OR, odds ratio; CI, confidence interval.
Sensitivity analysis
A sensitivity analysis was performed to evaluate the stability of the overall results by sequential omission of individual studies. The results of sensitivity analysis demonstrated that any single study did not affect the overall results qualitatively, indicating robustness and reliability of our results (data not shown).
Publication bias
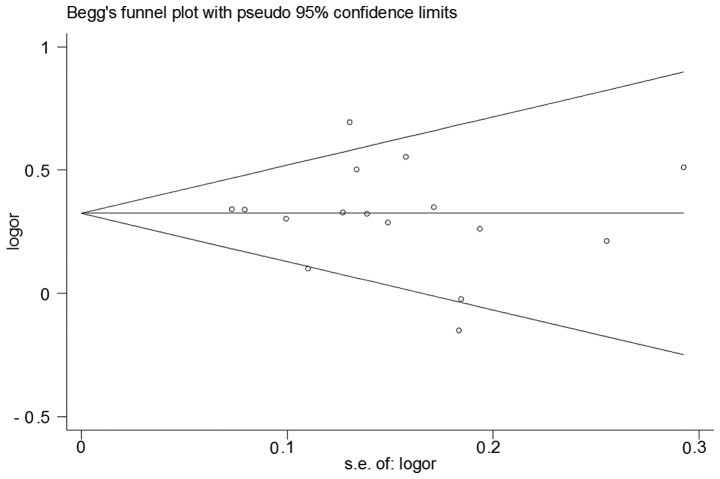
The shape of the funnel plot did not reveal any obvious asymmetry (Fig. 3). In addition, the results of Egger's test did not suggest any evidence of publication bias (P=0.667 for GG vs. TT; P=0.578 for GT vs. TT; P=0.669 for GG+GT vs. TT; P=0.605 for GG vs. GT+TT; and P=0.670 for G vs. T).
Figure 3.
Begg's funnel plot with pseudo 95% confidence limits for publication bias among selected studies on the coiled-coil domain-containing 26 rs4295627 polymorphism (dominant model). s.e., standard error; or, odds ratio.
Discussion
Although glioma is a rare type of cancer, it accounts for ~80% of malignant brain tumors in adults (17). However, the risk factors for glioma remain largely unknown. Gene susceptibility for glioma has been suggested in numerous case-control studies (18). The majority of studies conducted to date have evaluated a number of polymorphisms in a certain pathway, such as those affecting methylation, carcinogen metabolism, DNA repair, cell cycle, or inflammation. Polymorphisms may affect protein function, promoter activity, messenger RNA stability and splice variants and, therefore, may result in a change in the cellular ability to cope with DNA damage, which contributes to altered disease susceptibility (19).
Recently, the association between CCDC26 rs4295627 polymorphism and risk of glioma has been reported in a number of studies, and CCDC26 has been identified as a potential cancer susceptibility gene. The CCDC26 gene is located on chromosome 8q24.21. The rs4295627 polymorphism is located in intron 3 of the CCDC26 gene and encodes a retinoic acid modulator of differentiation and apoptosis (20). Retinoic acid induces caspase-8 transcription through phosphorylation of cyclic adenosine monophosphate response element binding, and increases apoptosis in glioblastoma cells via downregulation of telomerse activity (9). Variations of the 8q24.21 have been implicated in the risk of a number of common tumors, including colorectal, breast, bladder and prostate cancer (21–23).
To the best of our knowledge, this is the first comprehensive meta-analysis investigating the association between the CCDC26 rs4295627 polymorphism and risk of glioma. The present meta-analysis, including 7,290 cases and 11,630 controls, investigated the association between the CCDC26 rs4295627 polymorphism and risk of glioma and the results indicated that the CCDC26 rs4295627 polymorphism is associated with increased risk of glioma (GG vs. TT, OR=1.936, 95% CI: 1.500–2.658, P<0.001; GT vs. TT, OR=1.323, 95% CI: 1.241–1.412, P=0.206; GG+GT vs. TT, OR=1.375, 95% CI: 1.256–1.505, P=0.026; GG vs. GT+TT, OR=1.769, 95% CI: 1.302–2.403, P<0.001; and G vs. T, OR=1.310, 95% CI: 1.185–1.448, P<0.001). These findings may be biologically plausible. A subgroup analysis by ethnicity was not performed, as only one Asian case-control study with a small sample size was included. In previous studies, different results in the CCDC26 gene have been found in different populations (French results: OR=1.36; German results: OR=1.73; UK results: OR=1.26; and Asian results: OR=1.32) (4,14). However, the ethnic specifity of this SNP requires further investigation and analysis.
Several limitations in this meta-analysis should be mentioned. First, only published studies in the selected databases were included in the meta-analysis, whereas some relevant published studies or unpublished studies with null results were not identified, which may bias our results. Second, heterogeneity is a potential problem in all meta-analyses. In the present meta-analysis, we found moderate to large heterogeneity in most genetic models, indicating that caution should be taken when interpreting our results. Third, more accurate ORs should be adjusted for age, gender, smoking, alcohol consumption and other factors associated with cancer risk (24). In addition, the small sample size (<100 cases and controls) may overestimate the true association, due to deficiencies in statistical power.
In conclusion, the currently available evidence suggests that the rs4295627 polymorphism in the CCDC26 gene may contribute to glioma susceptibility; however, further case-control studies are required to confirm our results.
References
- 1.Ahmed R, Oborski MJ, Hwang M, Lieberman FS, Mountz JM. Malignant gliomas: Current perspectives in diagnosis, treatment and early response assessment using advanced quantitative imaging methods. Cancer Manag Res. 2014;6:149–170. doi: 10.2147/CMAR.S54726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il'yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, et al. Brain Tumor Epidemiology Consortium: Brain tumor epidemiology: Consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113(7 Suppl):S1953–S1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Bian Y, Zhu W, Zou P, Tang G. Regulator of telomere elongation helicase 1 (RTEL1) rs6010620 polymorphism contribute to increased risk of glioma. Tumour Biol. 2014;35:5259–5266. doi: 10.1007/s13277-014-1684-8. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y, Tong X, Tang LL, Zhou K, Zhong CH, Jiang S. Associations between the rs6010620 polymorphism in RTEL1 and risk of glioma: A meta-analysis of 20,711 participants. Asian Pac J Cancer Prev. 2014;15:7163–7167. doi: 10.7314/APJCP.2014.15.17.7163. [DOI] [PubMed] [Google Scholar]
- 5.Kayani MA, Khan S, Baig RM, Mahjabeen I. Association of RAD 51 135 G/C, 172 G/T and XRCC3 Thr241Met gene polymorphisms with increased risk of head and neck cancer. Asian Pac J Cancer Prev. 2014;15:10457–10462. doi: 10.7314/APJCP.2014.15.23.10457. [DOI] [PubMed] [Google Scholar]
- 6.Du SL, Geng TT, Feng T, Chen CP, Jin TB, Chen C. The RTEL1 rs6010620 polymorphism and glioma risk: A meta-analysis based on 12 case-control studies. Asian Pac J Cancer Prev. 2014;15:10175–10179. doi: 10.7314/APJCP.2014.15.23.10175. [DOI] [PubMed] [Google Scholar]
- 7.Liang HJ, Yan YL, Liu ZM, Chen X, Peng QL, Wang J, Mo CJ, Sui JZ, Wu JR, Zhai LM, et al. Association of XRCC3 Thr241Met polymorphisms and gliomas risk: Evidence from a meta-analysis. Asian Pac J Cancer Prev. 2013;14:4243–4247. doi: 10.7314/APJCP.2013.14.7.4243. [DOI] [PubMed] [Google Scholar]
- 8.Wibom C, Späth F, Dahlin AM, Langseth H, Hovig E, Rajaraman P, Johannesen TB, Andersson U, Melin B. Investigation of established genetic risk variants for glioma in pre-diagnostic samples from a population-based nested case-control study. Cancer Epidemiol Biomarkers Prev. 2015;24:810–816. doi: 10.1158/1055-9965.EPI-14-1106. [DOI] [PubMed] [Google Scholar]
- 9.Das A, Banik NL, Ray SK. Differentiation decreased telomerase activity in rat glioblastoma C6 cells and increased sensitivity to IFN-gamma and taxol for apoptosis. Neurochem Res. 2007;32:2167–2183. doi: 10.1007/s11064-007-9413-y. [DOI] [PubMed] [Google Scholar]
- 10.Safaeian M, Rajaraman P, Hartge P, Yeager M, Linet M, Butler MA, Ruder AM, Purdue MP, Hsing A, Beane-Freeman L, et al. Joint effects between five identified risk variants, allergy and autoimmune conditions on glioma risk. Cancer Causes Control. 2013;24:1885–1891. doi: 10.1007/s10552-013-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Jin T, Zhang J, Lou H, Yang B, Li Y, Chen C, Zhang Y. Polymorphisms of TREH, IL4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol. 2012;36:283–287. doi: 10.1016/j.canep.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Wang SS, Hartge P, Yeager M, Carreón T, Ruder AM, Linet M, Inskip PD, Black A, Hsing AW, Alavanja M, et al. Joint associations between genetic variants and reproductive factors in glioma risk among women. Am J Epidemiol. 2011;174:901–908. doi: 10.1093/aje/kwr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoemaker MJ, Robertson L, Wigertz A, Jones ME, Hosking FJ, Feychting M, Lönn S, McKinney PA, Hepworth SJ, Muir KR, et al. Interaction between 5 genetic variants and allergy in glioma risk. Am J Epidemiol. 2010;171:1165–1173. doi: 10.1093/aje/kwq075. [DOI] [PubMed] [Google Scholar]
- 14.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Egger M, Smith Davey G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J, Liu Y, Kyritsis AP, Bondy ML. Molecular epidemiology of primary brain tumors. Neurotherapeutics. 2009;6:427–435. doi: 10.1016/j.nurt.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Shete S, Hosking FJ, Robertson LB, Bondy ML, Houlston RS. New insights into susceptibility to glioma. Arch Neurol. 2010;67:275–278. doi: 10.1001/archneurol.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W, Rossin A, Clifford JL, Gronemeyer H. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene. 2006;25:3735–3744. doi: 10.1038/sj.onc.1209410. [DOI] [PubMed] [Google Scholar]
- 21.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 23.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu LX, Ho SC, Wong TK. Effectiveness of health education programs on exercise behavior among patients with heart disease: A systematic review and meta-analysis. J Evid Based Med. 2013;6:265–301. doi: 10.1111/jebm.12063. [DOI] [PubMed] [Google Scholar]