Abstract
Preoperative chemoradiotherapy (CRT) for locally advanced rectal cancer not only improves the postoperative local control rate, but also induces downstaging. However, it has not been established how to individually select patients who receive effective preoperative CRT. The aim of this study was to identify a predictor of response to preoperative CRT for locally advanced rectal cancer. This study is additional to our multicenter phase II study evaluating the safety and efficacy of preoperative CRT using oral fluorouracil (UMIN ID: 03396). From April, 2009 to August, 2011, 26 biopsy specimens obtained prior to CRT were analyzed by cyclopedic microarray analysis. Response to CRT was evaluated according to a histological grading system using surgically resected specimens. To decide on the number of genes for dividing into responder and non-responder groups, we statistically analyzed the data using a dimension reduction method, a principle component analysis. Of the 26 cases, 11 were responders and 15 non-responders. No significant difference was found in clinical background data between the two groups. We determined that the optimal number of genes for the prediction of response was 80 of 40,000 and the functions of these genes were analyzed. When comparing non-responders with responders, genes expressed at a high level functioned in alternative splicing, whereas those expressed at a low level functioned in the septin complex. Thus, an 80-gene expression set that predicts response to preoperative CRT for locally advanced rectal cancer was identified using a novel statistical method.
Keywords: preoperative chemoradiotherapy, histological response, microarray analysis, principle component analysis, rectal cancer
Introduction
Rectal cancer is associated with a high risk of locoregional relapse and remains one of the leading causes of cancer-related mortality. In western countries, preoperative chemoradiotherapy (CRT) followed by total mesorectal excision is the standard treatment for advanced rectal cancer, and it may decrease local recurrence and improve clinical outcome (1–5). To date, a number of clinical trials have demonstrated that 5-fluorouracil (5-FU) is a key chemotherapeutic agent in CRT (6–9). It is also considered that preoperative CRT improves the local control rate in Japan; however, it is still not a standard treatment due to issues regarding its effectiveness and side effects.
Recently, a prospective randomized phase III trial evaluating the survival benefit of adjuvant chemotherapy of an oral fluoropyrimidine (S-1) in patients with advanced rectal cancer demonstrated the superiority of S-1 to tegafur-uracil (10). It was also reported that preoperative CRT using concurrent S-1 and irinotecan achieved excellent long-term survival with acceptable adverse events in patients with rectal cancer (11). We conducted a multi-institutional phase II trial to evaluate the feasibility of preoperative S-1 plus radiotherapy for rectal cancer (UMIN ID: 03396) (12). Our results demonstrated that the treatment completion rate was comparable with that reported by studies conducted in western countries; a high rate of R0 and a low rate of occurrence of adverse side effects were demonstrated (12).
To perform preoperative CRT effectively and avoid adverse side effects, it is crucial to develop a predictive marker of response to CRT. Predictive markers for the therapeutic effects of preoperative CRT have been described in several studies (13–15). Furthermore, it has been reported that peripheral blood lymphocyte subsets were associated with susceptibility to preoperative CRT (16). Additionally, the genes signal transducer and activator of transcription 3, Ras association (RalGDS/AF-6) domain family member 1, docking protein 3 and erbB-2 receptor tyrosine kinase 2, were found to be associated with response to CRT in vitro (17). However, none of these factors have reached the stage where they may be clinically applied. Therefore, a reliable preoperative marker is still required.
Based on our multicenter phase II trial, the aim of the present study was to identify a predictor of response to CRT in patients with locally advanced rectal cancer. We herein attempted to identify a gene expression set for predicting the response to preoperative CRT by principle component analysis (PCA) as a novel statistical approach with microarray analysis using biopsy specimens prior to treatment.
Patients and methods
Patients
From April, 2009 to August, 2011, a total of 37 patients diagnosed with primary rectal cancer were prospectively enrolled in a multicenter phase II study (UMIN ID 03396). The trial design and eligibility criteria have been previously reported (12). Of the 37 patients, 26 whose biopsy specimens were available for microarray analysis were investigated in the present study. The protocol of this study was approved by the local Ethics Committee (Institutional Review Board, Faculty of Medicine, Oita University) and written informed consent was obtained for the therapy and tissue specimen collection from all the patients. Regarding the inclusion criteria, cases with cancer of the upper, middle, or lower third of the rectum, stage T3-4, N0-2 and M0 according to the International Union Against Cancer TNM Classification of Malignant Tumors 7th edition (18), were included in this study.
Treatment schedules
The details of the CRT were previously described (12). CRT was administered concurrently with four cycles of chemotherapy. A radiosensitizing dose of S-1 was orally administered at the respective dose on days 1–5, 8–12, 22–26 and 29–33. Radiotherapy comprised 45 Gy of preoperative pelvic radiation in 25 fractions over 5 weeks (1.8 Gy/day on days 1–5, 8–12, 15–19, 22–26 and 29–33).
Pathological evaluation
Histological response was evaluated by grading the post-treatment resection specimens according to the Japanese Classification of Colorectal Carcinoma (19). The absence of residual tumor was defined as grade 3; grade 2 was defined as ≥2/3 degeneration/necrosis area; and grade 0/1a/1b was defined as <2/3 degeneration/necrosis area. Subsequently, patients were classified as ‘responders’ when assigned to regression grade 2 or 3 and as ‘non-responders’ when assigned to grade 0 or 1.
Sample analysis
Biopsy specimens
Biopsy specimens were collected from several enriched cancer lesions in primary tumors prior to preoperative CRT. These specimens were immediately embedded in Tissue-Tek O.C.T Compound (Sakura Finetek Japan Co., Ltd., Tokyo, Japan), frozen in liquid nitrogen and maintained at −80°C until RNA extraction.
Total RNA extraction
Frozen biopsy specimens were stained by hematoxylin and eosin. We examined the specimens microscopically and confirmed that abundant cancer cells were included in each specimen. Subsequently, total RNA was extracted with an RNeasy Mini kit (Qiagen, Valencia, CA, USA). Total RNA (300 ng) was reverse-transcribed to cDNA using murine leukemia virus reverse transcriptase (Invitrogen Corp., Carlsbad, CA, USA).
Microarray analysis
Gene expression was conducted using the Whole Human Genome Oligo DNA Microarray kit (Agilent Technologies, Santa Clara, CA, USA). Labeled cDNA was fragmented and hybridized to an oligonucleotide microarray (Whole Human Genome 4×44 K; G4112F, Agilent Technologies). Fluorescent intensities were determined with an Agilent DNA Microarray scanner and analyzed using G2567AA Feature Extraction Software, version A.7.5.1 (Agilent Technologies), which uses the locally weighted regression curve fit normalization method (20). This microarray study followed the Minimum Information About a Microarray Experiment guidelines issued by the Microarray Gen Expression Data group (21). We performed functional analyses of the expressed genes using GeneSpring version 11.5 (Silicon Genetics, San Carlos, CA, USA). The Database for Annotation, Visualization and Integrated Discovery v6.7 was used to search for each gene and its functions (22,23).
Statistical analysis
Clinical data were statistically analyzed using the χ2 test or Fisher's exact test. All the differences were considered statistically significant if the P-value was <0.05. To evaluate the difference among the expression levels of two groups, i.e., responder and non-responder groups, we used the normalized score of each gene expression. We performed a hypothesis test and assumed as a null hypothesis H that the two groups have equal mean values of a particular expression level. This means that the gene expression in a particular DNA spot on the microarray does not correlate to the type of response. We employed Welch's t-test to perform the hypothesis test for H. This method calculates a P-value for each DNA spot where the P-value is the probability of H. If the P-value is small, it indicates that H should be rejected. Additionally, silhouette distance is a method of validation within clusters of data. This technique provides a precise graphical representation of how well each object lies within its cluster (24). Each DNA spot has a P-value. All DNA spots are sorted in ascending order using the P-value of the existing datasets of 26 subjects. The top-k DNA spots are selected using this order and only the selected DNA spots are used in the next step. To analyze the correlation between gene expression levels, we performed a dimension reduction method called PCA. The k expression levels of the selected k DNA spots were used for each individual. Some of the expression levels were correlated with each other; analysis of this correlation is useful to reduce noise in expression levels and obtain better prediction results.
Results
Comparison of clinical parameters between responders and non-responders to CRT
Of the 26 patients, 11 were classified as responders and 15 as non-responders by histological examination of surgically resected specimens according to the grading system. The characteristics of the 26 patients are summarized in Table I. For various clinical parameters, our results demonstrated that all P-values calculated using the Fisher's exact test were >0.05 in a 2×2 contingency table comparing the responder and non-responder groups. We concluded that there is no significant association between the groups and any of the clinical parameters.
Table I.
Comparison of various clinical parameters between responders and non-responders to chemoradiotherapy.
| Parameters | Responders (n=11) | Non-responders (n=15) | P-valued |
|---|---|---|---|
| Gender | 0.277 | ||
| Male | 8 | 8 | |
| Female | 3 | 7 | |
| Age (years) | 0.054 | ||
| Mediana | 62.5±8.9 | 54.3±10.8 | |
| Performance status | |||
| 0 | 11 | 15 | |
| ≤1 | 0 | 0 | |
| Tumor location | 0.426 | ||
| Upper rectum | 2 | 4 | |
| Middle-lower rectum | 9 | 11 | |
| Histology | 0.381 | ||
| Well/moderately differentiated | 9 | 14 | |
| Poorly differentiated/mucinous/papillary adenocarcinoma | 2 | 1 | |
| cT stageb | 0.175 | ||
| T3 | 11 | 12 | |
| T4 | 0 | 3 | |
| cN stageb | 0.169 | ||
| Negative | 5 | 3 | |
| Positive | 6 | 12 | |
| cStageb | 0.085 | ||
| II | 5 | 2 | |
| III | 6 | 13 | |
| Surgeryc | 0.384 | ||
| LAR | 6 | 9 | |
| APR | 4 | 6 | |
| Hartmann's procedure | 1 | 0 |
Mean ± standard deviation.
According to the 7th TNM classification.
Includes laparoscopic as well as open procedures.
According to the Chi-square test. LAR, low anterior resection; APR, abdomino-perineal resection.
P-value distribution, silhouette distance validation and processing results
Given a set of 40,000 gene expression levels, we obtained 40,000 P-values for H. The P-value distribution is shown in Fig. 1. If there is no correlation between the DNA spots and the type of response, there will be no obvious peak at a small P-value. However, we observed a peak at the interval between 0.06≤ P-value <0.07. Furthermore, we calculated the silhouette distance of the two sets as Vr and Vn. The prediction results are better with increasing silhouette value. The silhouette analysis is shown in Fig. 2. The silhouette distance increases from k=1 to k=80 and decreases from k=80 to k=1,000. Therefore, k=80 is optimal for prediction. There are 11 two-dimensional vectors Vr for the 11 responders, and 15 two-dimensional vectors Vn for the 15 non-responders. The result of the existing datasets of the 26 subjects is shown in Fig. 3 and the 80-gene expression set correlates with the type of response.
Figure 1.
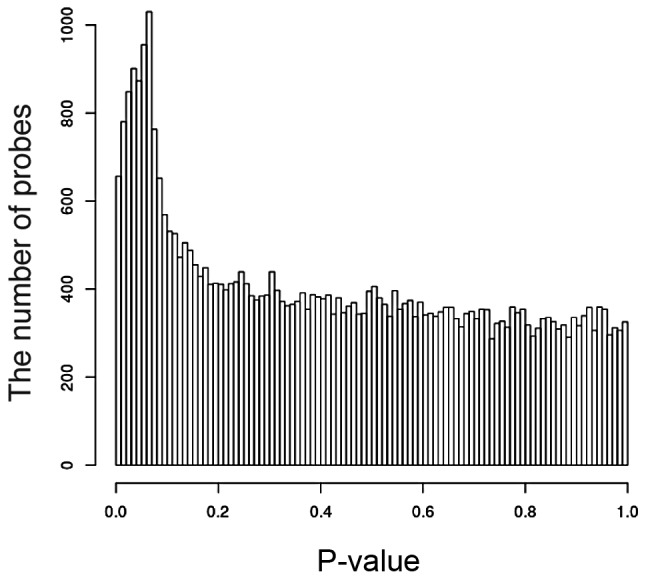
Distribution of P-values. The x-axis represents the P-value. The height of each bin is the number of probes that fall into an interval. If there was no differential expression observed, all the bars would be of approximately the same height.
Figure 2.
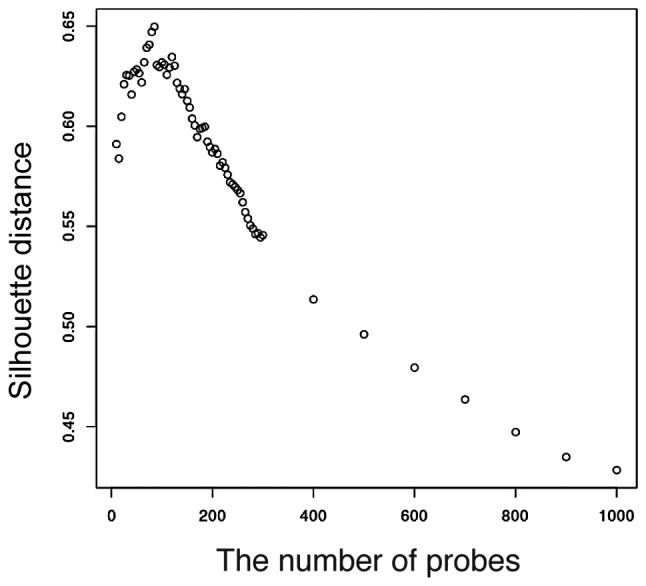
Silhouette distance of the number of probes. The optimal number of probes to identify the sensitivity to chemoradiotherapy is at k=80. This indicates that more than or less than 80 probes produce worse prediction results.
Figure 3.
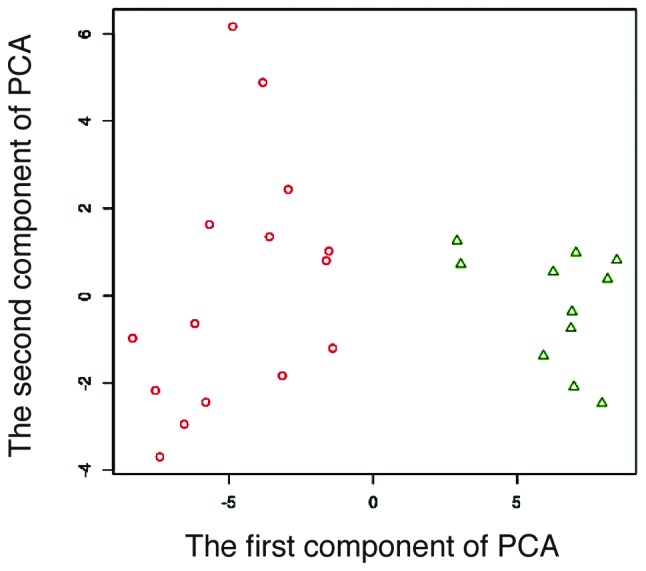
Processing results of the existing datasets of the 26 subjects. Red circles, non-responders (grade 0/1a/1b). Green triangles, responders (grade 2 or 3). When an 80-gene set was selected, the 26 patients were divided into two distinct groups in the two-dimensional graph.
Analysis of the 80-gene expression set
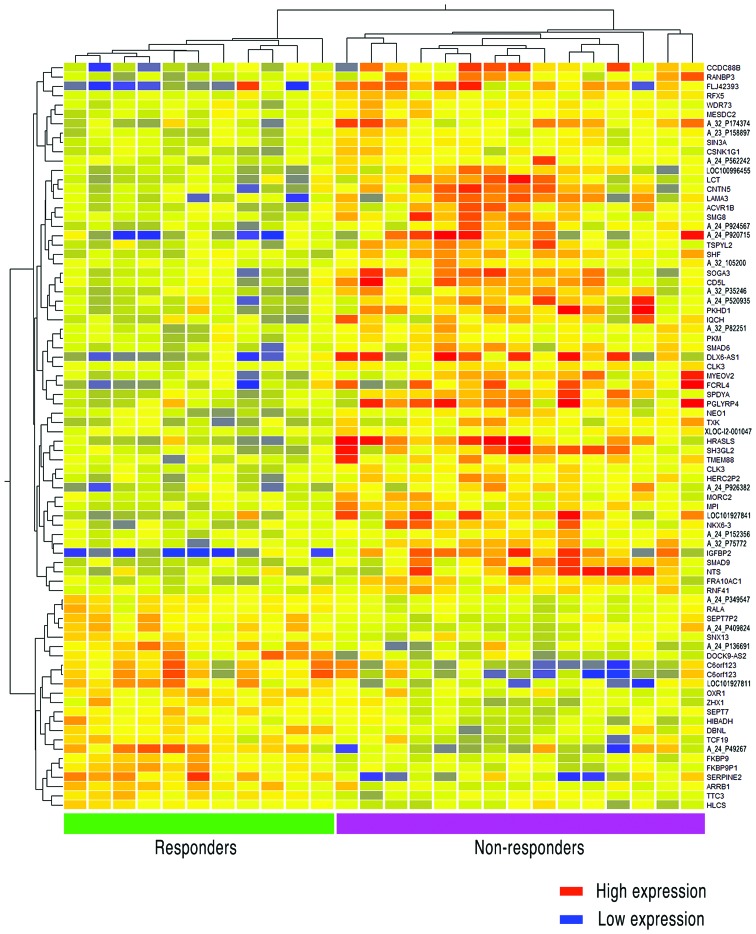
The 80-gene expression set was analyzed using GeneSpring v.11.5 (Agilent Technologies). After having classified the 80 genes according to the response to preoperative CRT, the method of analysis displayed the genes in a hierarchy according to major function. Additionally, the clustering entities were presented in a correlation heatmap (Fig. 4). As 16 of the 80 gene names and their functions were unknown, we described probe ID number as unknown genes according to the database of Agilent Technologies. As shown in Fig. 4, there were 57 genes that were strongly expressed in non-responder for responder, whereas 38 gene names were found to be clear. By contrast, 23 genes strongly expressed in responder for non-responder, and 14 genes of those were found to be clear. The functional annotation results are shown in Table II, which included functions that strongly or weakly emerged in non-responders as compared with responders. In the non-responder group, 38 highly expressed genes were clearly annotated in terms of function. The major function found in 22 of the 38 genes (57.9%) was alternative splicing; the second major function that was found in 21 of the 38 genes (55.3%) was phosphoprotein.
Figure 4.
Cluster analysis of differentially expressed genes in 26 patients with a functional annotation clustering analysis using a hierarchical method. Clustering analysis in 26 patients (columns) and gene probes/names (rows) is shown. Red indicates high expression, while blue indicates low expression. Eleven responders are clustered on the left side and 15 non-responders are clustered on the right side. As 16 of the 80 gene names were unknown, we described probe ID number as unknown genes according to the database of Agilent Technologies. For the notation, ‘A_ (number) _P (number)’ was used for an unknown gene name.
Table II.
Molecular functions of genes with high expression and low expression levels in non-responders as compared with those in responders.
| Genes | Molecular functions | Counta | %b | P-valuec |
|---|---|---|---|---|
| Genes exhibiting high expression | Alternative splicing | 22 | 57.9 | 0.0058 |
| Phosphoprotein | 21 | 55.3 | 0.01 | |
| Regulation of phosphorylation | 5 | 13.2 | 0.011 | |
| Regulation of phosphorus metabolic process | 5 | 13.2 | 0.013 | |
| Regulation of phosphate metabolic process | 5 | 13.2 | 0.013 | |
| Dwarfin | 2 | 5.3 | 0.015 | |
| MAD homology, MH1 | 2 | 5.3 | 0.015 | |
| Transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 7.9 | 0.017 | |
| DWB | 2 | 5.3 | 0.017 | |
| Transforming growth factor β receptor, cytoplasmic mediator activity | 2 | 5.3 | 0.018 | |
| TGF-β signaling pathway | 3 | 7.9 | 0.02 | |
| Signal | 12 | 31.6 | 0.02 | |
| MAD homology 1, Dwarfin-type | 2 | 5.3 | 0.022 | |
| DWA | 2 | 5.3 | 0.026 | |
| SMAD domain-like | 2 | 5.3 | 0.028 | |
| Regulation of protein kinase activity | 4 | 10.5 | 0.028 | |
| Regulation of kinase activity | 4 | 10.5 | 0.03 | |
| Kinase | 5 | 13.2 | 0.032 | |
| Regulation of transferase activity | 4 | 10.5 | 0.034 | |
| Phosphotransferase | 3 | 7.9 | 0.049 | |
| Genes exhibiting low expression | Septin complex | 2 | 14.3 | 0.0084 |
| Septin cytoskeleton | 2 | 14.3 | 0.0084 | |
| Cell cortex | 3 | 21.4 | 0.0044 | |
| GTP binding | 3 | 21.4 | 0.038 | |
| Guanyl nucleotide binding | 3 | 21.4 | 0.04 | |
| Guanyl ribonucleotide binding | 3 | 21.4 | 0.04 |
Molecular functions were analyzed by using the Database for Annotation, Visualization and Integrated Discovery.
Number of expressed genes in each function.
Rate of expressed genes in the high or low expressed functions.
P-values for the genes loaded onto the specified functional cluster. DWA, domain A in dwarfin protein; DWB, domain B in dwarfin proteins.
Discussion
In the present study, we demonstrated that an 80-gene expression set may predict response to preoperative CRT with an oral fluorouracil agent (S-1) for locally advanced rectal cancer. The 80 genes identified in this study were selected from 40,000 genes by a novel statistical technique, i.e., the dimension reduction method referred to as PCA. Several investigators previously reported that one gene or a few genes were associated with response to preoperative CRT for rectal cancer (13–15,17,25). However, it was considered that the prediction of CRT response using a few genes as biomarkers may be difficult, as this comprehensive treatment has various mechanisms associated with antitumor effects. In this study, we focused on a new statistically analytical system of gene expression by microarray analysis, in order to identify a set of genes predicting response to CRT using biopsy samples of tumors prior to CRT treatment. To the best of our knowledge, this is the first study to investigate a gene set as a biomarker of response to radiation with various chemotherapies using a novel statistical technique.
We obtained the expression levels of 40,000 genes from DNA spots in a microarray for each patient. One of the main aims of this study was to automatically identify and predict the type of response to preoperative CRT and classify it into either responders or non-responders based on the expression levels. We were then certain that there must be a particular number of genes correlated with susceptibility. The basic idea was to use the existing datasets of 26 subjects for prediction. Herein, we proposed a new method of prediction. First, the existing datasets of 26 subjects were processed to prepare an automatic predictor, as explained in Materials and methods. Our goal was to estimate the optimal number k of DNA spots that were obtained in the optimal predication results. To evaluate the prediction, we calculated the silhouette distance of the two sets Vr and Vn. Our results demonstrated that these 80 genes were correlated with the type of response. If the number of genes were > or <80 DNA spots, the prediction would be incorrect.
It was previously reported that alternative splicing was significantly associated with cancer development (26). In rectal cancers of the same pathological differentiation, alternative splicing of a gene may lead to different resistances to various treatments, including CRT. Haley et al and Cutillas et al reported that phosphoproteomics enabled tumor cells to survive against chemotherapeutics (27,28). However, the septin complex was the major annotated function among genes expressed at a low level, accounting for 4 out of 14 genes (28.6%). It has also been reported that septin is associated with the cytoskeleton and cell division (29), and that the septin family facilitates differentiation between patients without cancer and those with cancer (30). These reports support the idea that our functional cluster analysis of the gene expression set is a possible predictive marker for CRT response in rectal cancer. Although Nishino et al reported that the organic anion transporter 2 gene was associated with the major uptake transporters of 5-FU, this gene was not found in our 80-gene set (25). In the future, it is necessary to elucidate the functions of unknown genes and to evaluate the interaction of the expression of these genes with response to preoperative CRT.
Our study had several limitations. First, the heterogeneity of cancer tissue is a factor that makes anticancer treatment difficult; thus, it was necessary to consider heterogeneity when the tissue samples were obtained. Therefore, we decided to sample from the pretreatment biopsy site, which was located in an elevated part of the tumor nearest to the anus. Second, it was necessary to evaluate whether prediction using this 80-gene set is possible for radiation therapy with other types of chemotherapy. Furthermore, the accuracy of the 80-gene expression set could not be validated, which will be addressed in a future multi-institutional study to evaluate the utility of this 80-gene expression set for CRT prior to surgery for locally advanced rectal cancer (UMIN ID 03398). The results of this validation trial will demonstrate whether this 80-gene expression set is reliable as a predictive biomarker of response to preoperative CRT.
In conclusion, the 80-gene expression set identified in our study may be a predictor of response to preoperative CRT for locally advanced rectal cancer. We demonstrated that the application of PCA is useful for the identification of a gene set in this context. To the best of our knowledge, this is the first study to develop a prediction gene set based on microarray analysis for response to CRT using pretreatment biopsy specimens.
Acknowledgments
The following are acknowledged for their contribution to this study: Dr Toshifumi Matsumoto and Dr Teijiro Hirashita, National Hospital Organization Beppu Medical Center; Dr Koichiro Tahara and Dr Satoshi Sugita, National Hospital Organization Oita Medical Center; Dr Akio Morimoto, Dr Atsushi Sasaki, Dr Yu Takeuchi and Dr Fumitaka Yoshizumi, Japan Community Health Care Organization Nankai Medical Center; Dr Toshio Bando and Dr Yasuhiro Hirabayashi, Oita Prefectural Hospital; Dr Akihiko Kuwahara, Dr Koichi Ishikawa, Dr Kazuya Sonoda, Tsukumi Central Hospital; Dr Akio Shiromizu, Oita Red Cross Hospital; Dr Kyuzo Fujii and Dr Toru Kusano, Nakatsu Municipal Hospital; Dr Nobuhiro Kubo, Dr Kohei Shibata and Dr Takuya Noguchi, JA Oita Koseiren Tsurumi Hospital; Dr Koshi Mimori and Dr Hidetoshi Eguchi, Kyushu University Beppu Hospital; Dr Tadahiko Kinoshita, Dr Yuji Morii, Dr Masaaki Tajima and Dr Yohei Kono, Bungoono City Hospital; Dr Katsuhiro Shimoda, Dr Tadashi Ogawa, Dr Kosuke Suzuki, Dr Yoko Komori, Usuki Cosmos Hospital; Dr Kazuaki Hiroishi and Dr Shigeo Ninomiya, Arita Gastrointestinal Hospital; Dr Toshiya Abe, Dr Masahiro Fukano, Dr Seitaro Hirano, Nakatsu Gastrointestinal Hospital; Dr Masanori Aramaki and Dr Hiroshi Sato, Oita Oka Hospital; Dr Kenji Zeze, Zeze Hospital; Dr Toshihiro Suematsu, Oita Tobu Hospital. We would like to express our gratitude to Ms. Mami Kimoto, Ms. Mayumi Takeda and Ms. Yuiko Aso for the technical assistance.
Glossary
Abbreviations
- CRT
chemoradiotherapy
- 5-FU
5-fluorouracil
- S-1
oral fluoropyrimidine
- PCA
principle component analysis
References
- 1.Garcia-Aguilar J, Smith DD, Avila K, Bergsland EK, Chu P, Krieg RM. Timing of Rectal Cancer Response to Chemoradiation Consortium: Optimal timing of surgery after chemoradiation for advanced rectal cancer: Preliminary results of a multicenter, nonrandomized phase II prospective trial. Ann Surg. 2011;254:97–102. doi: 10.1097/SLA.0b013e3182196e1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Påhlman L, Glimelius B, van de Velde CJ. Dutch Colorectal Cancer Group: Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet. 2011;12:575–582. doi: 10.1016/S1470-2045(11)70097-3. [DOI] [PubMed] [Google Scholar]
- 3.Sauer R, Becker H, Hohenberger W, Rödel C, Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 4.Bosset JF, Collette L, Calais G, Mineur L, Maingon P, Radosevic-Jelic L, Daban A, Bardet E, Beny A, Ollier JC. EORTC Radiotherapy Group Trial 22921: Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 5.Martling A, Holm T, Johansson H, Rutqvist LE, Cedermark B. Stockholm Colorectal Cancer Study Group: The Stockholm II trial on preoperative radiotherapy in rectal carcinoma: Long-term follow-up of a population-based study. Cancer. 2001;92:896–902. doi: 10.1002/1097-0142(20010815)92:4<896::AID-CNCR1398>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 6.Bosset JF, Calais G, Mineur L, Maingon P, Stojanovic-Rundic S, Bensadoun RJ, Bardet E, Beny A, Ollier JC, Bolla M, et al. Fluorouracil-based adjuvant chemotherapy after preoperative chemoradiotherapy in rectal cancer: Long-term results of the EORTC 22921 randomised study. Lancet Oncol. 2014;15:184–190. doi: 10.1016/S1470-2045(13)70599-0. [DOI] [PubMed] [Google Scholar]
- 7.Gérard JP, Conroy T, Bonnetain F, Bouché O, Chapet O, Closon-Dejardin MT, Untereiner M, Leduc B, Francois E, Maurel J, et al. Preoperative radiotherapy with or without concurrent fluorouracil and leucovorin in T3-4 rectal cancers: Results of FFCD 9203. J Clin Oncol. 2006;24:4620–4625. doi: 10.1200/JCO.2006.06.7629. [DOI] [PubMed] [Google Scholar]
- 8.Sebag-Montefiore D, Stephens RJ, Steele R, Monson J, Grieve R, Khanna S, Quirke P, Couture J, de Metz C, Myint AS, et al. Preoperative radiotherapy versus selective postoperative chemoradiotherapy in patients with rectal cancer (MRC CR07 and NCIC-CTG C016): A multicenter, randomised trial. Lancet. 2009;373:811–820. doi: 10.1016/S0140-6736(09)60484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschele C, Cionini L, Lonardi S, Pinto C, Cordio S, Rosati G, Artale S, Tagliagambe A, Ambrosini G, Rosetti P, et al. Primary tumor response to preoperative chemoradiation with or without oxaliplatin in locally advanced rectal cancer: Pathologic results of the STAR-01 randomized phase III trial. J Clin Oncol. 2011;29:2773–2780. doi: 10.1200/JCO.2010.34.4911. [DOI] [PubMed] [Google Scholar]
- 10.Murata A, Yoshida K, Maeda K, et al. A randomized phase III trial comparing S-1 versus UFT as adjuvant chemotherapy for stage II/III rectal cancer (JFMC35-C1: ACTS-RC) ASCO abstract. 2015;#3515 doi: 10.1093/annonc/mdw162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T, Yamashita K, Sato T, Ema A, Naito M, Watanabe M. Neoadjuvant chemoradiation therapy using concurrent S-1 and irinotecan in rectal cancer: Impact on long-term clinical outcomes and prognostic factors. Int J Radiat Oncol Biol Phys. 2014;89:547–555. doi: 10.1016/j.ijrobp.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Inomata M, Akagi T, Nakajima K, Etoh T, Shiraishi N, Tahara K, Matsumoto T, Kinoshita T, Fujii K, Shiromizu A, et al. A prospective feasibility study to evaluate neoadjuvant-synchronous S-1 with radiotherapy for locally advanced rectal cancer: A multicentre phase II trial. Mol Clin Oncol. 2016;4:510–514. doi: 10.3892/mco.2016.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Aguilar J, Chen Z, Smith DD, Li W, Madoff RD, Cataldo P, Marcet J, Pastor C. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg. 2011;254:486–493. doi: 10.1097/SLA.0b013e31822b8cfa. discussion 492–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, Iinuma H, Konishi T, Horie H, Ikeuchi H, et al. Gene expression signature and response to the use of leucovorin, fluorouracil and oxaliplatin in colorectal cancer patients. Clin Transl Oncol. 2011;13:419–425. doi: 10.1007/s12094-011-0676-z. [DOI] [PubMed] [Google Scholar]
- 15.Nishioka M, Shimada M, Kurita N, Iwata T, Morimoto S, Yoshikawa K, Higashijima J, Miyatani T. Gene expression profile can predict pathological response to preoperative chemoradiotherapy in rectal cancer. Cancer Genomics Proteomics. 2011;8:87–92. [PubMed] [Google Scholar]
- 16.Tada N, Kawai K, Tsuno NH, Ishihara S, Yamaguchi H, Sunami E, Kitayama J, Oba K, Watanabe T. Prediction of the preoperative chemoradiotherapy response for rectal cancer by peripheral blood lymphocyte subsets. World J Surg Oncol. 2015;13:30. doi: 10.1186/s12957-014-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spitzner M, Emons G, Kramer F, Gaedcke J, Rave-Fränk M, Scharf JG, Burfeind P, Becker H, Beissbarth T, Ghadimi BM, et al. A gene expression signature for chemoradiosensitivity of colorectal cancer cells. Int J Radiat Oncol Biol Phys. 2010;78:1184–1192. doi: 10.1016/j.ijrobp.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobin LH, Gospodarowicz MK, Wittekind C, editors. TNM Classification of Malignant Tumours. 7th. Wiley-Blackwell; Hoboken: 2009. [Google Scholar]
- 19.Japanese Society for Cancer of the Colon and Rectum: Japanese Classification of Colorectal Carcinoma. 2nd English. Kanehara Shuppan Co. Ltd.; Japan: 2009. [Google Scholar]
- 20.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 21.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 22.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Comput Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 25.Nishino S, Itoh A, Matsuoka H, Maeda K, Kamoshida S. Immunohistochemical analysis of organic anion transporter 2 and reduced folate carrier 1 in colorectal cancer: Significance as a predictor of response to oral uracil/ftorafur plus leucovorin chemotherapy. Mol Clin Oncol. 2013;1:661–667. doi: 10.3892/mco.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal S, Gupta R, Davuluri RV. Alternative transcription and alternative splicing in cancer. Pharmacol Ther. 2012;136:283–294. doi: 10.1016/j.pharmthera.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Haley J, White FM. Adaptive protein and phosphoprotein networks which promote therapeutic sensitivity or acquired resistance. Biochem Soc Trans. 2014;42:758–764. doi: 10.1042/BST20140038. [DOI] [PubMed] [Google Scholar]
- 28.Cutillas PR. Role of phosphoproteomics in the development of personalized cancer therapies. Proteomics Clin Appl. 2015;9:383–395. doi: 10.1002/prca.201400104. [DOI] [PubMed] [Google Scholar]
- 29.Mostowy S, Cossart P. Septins: The fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 30.Connolly D, Abdesselam I, Verdier-Pinard P, Montagna C. Septin roles in tumorigenesis. Biol Chem. 2011;392:725–738. doi: 10.1515/BC.2011.073. [DOI] [PubMed] [Google Scholar]