Abstract
PIWI-associated RNAs (piRNAs or piRs), a new-found class of small non-coding RNAs, which are mainly expressed in germline cells and partly in somatic lines, have a vital role in carcinogenesis by maintaining genomic integrity and regulation of epigenetics. The previous studies have confirmed that the expression of piR-651 is upregulated in several cancer tissue and cell lines, including lung cancer. However, the mechanism of carcinogenesis and piR-651 remains to be elucidated. Therefore, the present study assessed the 95-D high metastasis human lung cancer cell line to detect the effect of piR-651 on carcinogenesis. Firstly, piR-651 promoted the high metastasis characteristic of the 95-D cell lines by Transwell and wound-healing assays. The influence of piR-651 on tumor cell proliferation and apoptosis was detected by the MTT assay and flow cytometry. In conclusion, piR-651 may be an oncogene in lung cancer formation and development. Therefore, piR-651 regulated carcinogenesis by influencing cell proliferation, apoptosis, migration and invasion, and may be a potential tumor marker and therapeutic target.
Keywords: PIWI-associated RNA, small non-coding RNAs, lung cancer, tumor marker, metastasis
Introduction
Cancer is a complex genomic disease with multi-system disorders. In recent years, lung cancer has been one of the highest incidences of cancer, with 1.59 million people succumbing to lung cancer in 2012 worldwide (1,2). Although a great deal of progressive treatment has been applied clinically, including radiotherapy, chemotherapy and biotherapy, locally advanced unresectable lung cancer accounts for ~35% of lung cancer diagnoses, for which treatment remains a challenge. It has been observed that ~90% of patients succumb from metastasis rather than primary tumors (3,4). Therefore, identification of a sensitively predictive tumor marker and understanding of the underlying molecular mechanisms during tumor stage and metastasis have a major importance and may propose an effective therapeutic strategy against non-small cell lung cancer (NSCLC).
Non-coding RNA (ncRNA) regions, which were once thought to be non-functional, account for 98% of the transcriptome of the human genome (5,6); however, it has been found that the ncRNA act as a regulator in flora and fauna biology. Recently, the association of non-coding small regulatory RNAs and tumorigenesis has attracted increasing attention. An increasing number of studies have confirmed that the non-coding small RNA, including the microRNAs (miRNAs), short interfering RNAs (siRNAs) or PIWI-associated RNAs (piRNAs or piRs), have an important role in carcinogenesis (7–9). The research of miRNA and carcinogenesis is the most sufficient among these non-coding small regulatory RNAs, and has identified that the miRNAs may be oncogenes or tumor suppressors in carcinogenesis by regulating their target mRNA (10–12). However, the study regarding piRNA and carcinogenesis remains to be elucidated.
piRNAs are a type of novel non-coding small RNA with a length of 25–33 nucleotides. They have biological roles through the specific combination with the PIWI protein and are produced by the Dicer-independent manner (13–15). The crucial roles of piRNA were involved in maintaining DNA integrity, regulation of epigenetics and germline stem cells, the differentiation of embryonic development, and the occurrence and development of diseases. Several previous studies have identified that analogous with the miRNAs regulating tumorigenesis, the expression levels of piRNAs were also up and downregulated in certain cancer tissue and cell lines, revealing that they may have oncogene or antioncogenic roles in carcinogenesis. For example, the expression levels of piR-49322 (16), piR-823 (17), piR-651 (18), piR-932 (19), piR-4987, piR-20365, piR-20485, piR-20582 (20), piR-DQ594040 (21) and piR-Hep1 (22) were up and downregulated in breast cancer, cervical cancer, gastric cancer, hepatocellular carcinoma, multiple myeloma and others. piR-651, upregulated in several cancer tissues and cell lines including NSCLC, may be an oncogene in carcinogenesis (18). However, the regulatory mechanism of piR-651 and NSCLC carcinogenesis remains to be elucidated. Therefore, the present study focused on the mechanism of piR-651 in regulating carcinogenesis in lung cancer.
Materials and methods
Cell lines and culture conditions
The human high metastasis lung cancer cell line, 95-D, was purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China). 95-D cells were cultured with Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal bovine serum (FBS) (PAA Laboratories, Pasching, Austria). Cells were growth at 37°C in a humidified atmosphere of 5% CO2.
Cell transfection
The piR-651 inhibitor (Shanghai GenePharma Co., Shanghai, China) was transfected into 95-D cells to decrease the expression of endogenous piR-651 and detect the effect of piR-651 on carcinogenesis. The specific 2′-O-methyl oligo RNA oligonucleotides were synthesized and purified by Shanghai GenePharma Co. Their sequences were as follows: 5′-GACGCUUUCCAAGGCACGGGCCCCUCUCU-3′ (piR-651 inhibitor) and 5′-CAGUACUUUUGUGUAGUACAA-3′ [non-specific sequences for the negative control (NC)]. In brief, the cells were seeded in 6-well plates 1 day before transfection. When the cells reached 50–60% confluence, 100 pmol of the piR-651 inhibitor or 100 pmol of NC was transfected using the Lipofectamine 2000 reagent (Invitrogen). The transfecting rate was determined by fluorescence microscope and flow cytometry (BD LSR II; Becton-Dickinson, Franklin Lakes, NJ, USA).
Transwell invasion and migration assays
A Transwell chamber with 8-mm membrane pores (Corning Costar, Inc., Corning, NY, USA) was used to test the effect of piR-651 on the migration and invasion in 95-D cells. For migration, 3–4×104 transfected cells per well in serum-free medium were placed into the upper chamber with the non-coated membrane. For invasion, 5–6×104 cells in serum-free medium were seeded in the upper chamber, which was pre-coated with Matrigel (BD Biosciences, Bedford, MA, USA). The assays were conducted three independent times.
Wound-healing assays
The 95-D cells were seeded in 6-well plates to near confluence. A linear wound was carefully generated by moving a 10-µl sterile pipette tip across the confluent cell monolayer, and the cell fragments were removed by washing with phosphate-buffered saline (PBS) and incubated with DMEM without FBS. Subsequently, images of the wounded monolayers were captured 48 h after wounding.
Proliferation assay
To observe the effect of piR-651 on cell proliferation, the MTT assay was used. Each group was performed in six repeated wells in this assay. 95-D cell line was cultured in 96-well plates for 24 h and transfected with the piR-651 inhibitor and NC, respectively. After transfection at 12, 24, 48 and 72 h, the optical density (OD) values of each dose were determined. In total, 20 µl MTT (Sigma-Aldrich, St. Louis, MO, USA) solution was added into each of the 96-wells. After the cells were incubated at 37°C for 4 h in the dark, the supernatant was removed and 150 µl of dimethylsulphoxide (Thermo Fisher Scientific, Loughborough, UK) was added to dissolve the crystalization. The 96-well plate was mildly agitated on the shaking table for ≥10 min. The OD values were measured at 490 nm using a Wellscan reader (Labsystems, Santa Fe, NM, USA). Growth proliferation was calculated as a percentage as follows: (OD experiment/OD control) × 100%. Three independent experiments were performed.
Cell apoptosis analysis
In the cell apoptosis assays, cells were collected and washed with cold PBS twice after 24 h of transfection. Subsequently, 5×105 cells/ml were stained with Annexin V-propidium iodide in the dark, according to the manufacturer's protocol (Biosea Biotechnology, Co., Beijing, China). The cells were suspended in 300 µl binding buffer and assessed using flow cytometry (BD Biosciences, San Jose, CA, USA) in 1 h. All the assays were performed in triplicate.
Western blotting
To quantify the expression levels of apoptosis-related proteins following downregulation of piR-651, western blotting was used. In summary, the cells were collected 48 h after transfection and were lysed with radioimmunoprecipitation assay buffer (Beyotime Biotechnology, Jiangsu, China), and subsequently the lysates underwent centrifugation at 12,000 × g for 10 min at 4°C. The supernatants were collected, boiled and subsequently separated by sodium dodecyl sulfate-polyacrylamidegels, blotted onto polyvinylidene fluoride membrane (Millipore Corporation, Bedford, MA, USA) and finally detected with appropriate antibodies for bax (mouse anti-human monoclonal antibody; cat. no. sc-7480)/B-cell lymphoma-2 (bcl-2; mouse anti-human monoclonal antibody; cat. no. sc-7382), β-actin (mouse anti-human monoclonal antibody; cat. no. sc-58679; all from Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or from for caspase-p17 (rabbit anti-human polyclonal antibody; cat. no. 25546-1-AP; Proteintech Group, Inc., Wuhan, China). Two or three independent experiments were performed for this analysis.
Statistical analysis
Statistical analysis was performed using the Statistical Program for Social Sciences (SPSS) version 17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Decrease in the expression of piR-651 decreases the cell proliferation rate in 95-D cells
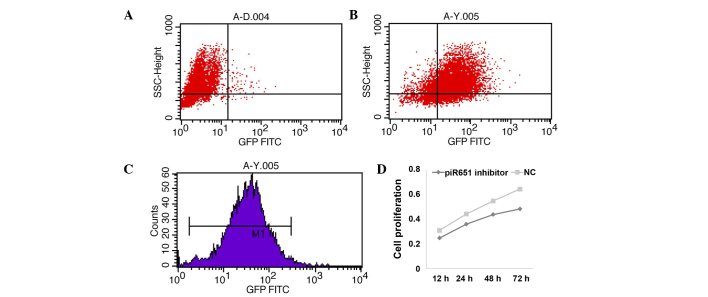
Numerous studies have identified that piR-651 was upregulated in lung cancer tissues and cell lines by deep sequencing analysis. Therefore, the expression of endogenous piR-651 was decreased by transfecting with its inhibitor. The transfecting rate was detected using the FAM genes. The results showed that 82.5% of all cells had fluorescent tags, and the mean fluorescence intensity was 31.87% (Fig. 1A–C). Cell proliferation was suppressed following transfection of the piR-651 inhibitor and the proliferation rate was decreased in a time-dependent manner in the 95-D cells (Fig. 1D) (P<0.001).
Figure 1.
piR-651 inhibitor suppresses cell proliferation in a time-dependent manner. (A-C) The transfection rate of the piR-651 inhibitor was measured by flow cytometry. (D) In the 95-D cells, the cell proliferation rate was decreased following transfection with the piR-651 inhibitor in the 95-D cells in a time-dependent manner compared with the NC (P<0.001). NC, negative control; GFP FITC, green fluorescent protein fluorescein isothiocyanate.
Decrease in the expression of piR-651 induces cell apoptosis and regulates the apoptosis-related protein expression level in 95-D cells
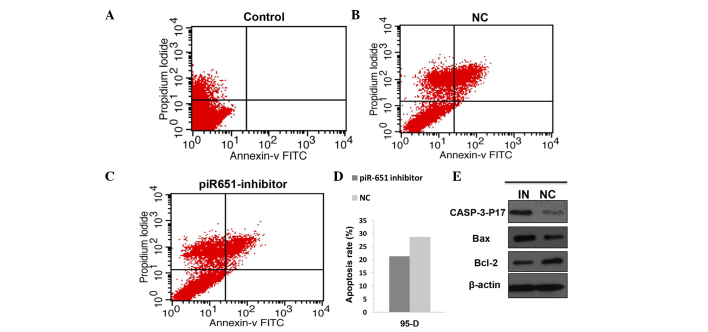
Numerous previous studies have confirmed that apoptosis has an important role in the maintenance of the internal environment and this balance is lost in a number of tumor patients. In the present study, the cell apoptosis rate in the piR-651 inhibitor cells was evidently higher compared to the NC in the 95-D cell line (P<0.001). The expression level of the apoptosis-related protein was assessed by western blotting. The expression of piR-651 was reduced by transfecting the piR-651 inhibitor and the transfection rate was assessed by observing the fluorescence. The expression levels of caspase-p17 (P=0.022) and bax (P<0.001) were upregulated and bcl-2 (P<0.001) was downregulated (Fig. 2).
Figure 2.
piR-651 inhibitor induces cell apoptosis and regulates the expression of the apoptosis-related protein compared with the NC. (A-D) The cell apoptosis rate of the piR-651 inhibitor was higher compared to the NC (P<0.001). (E) The expression of caspase-p17 and bax was upregulated and bcl-2 was downregulated in the piR-651 inhibitor. IN, piR-651 inhibitor; NC, negative control; FITC, fluorescein isothiocyanate; bcl-2, B-cell lymphoma-2.
Decrease in the expression level of piR-651 restrains the cell ability of migration and invasion
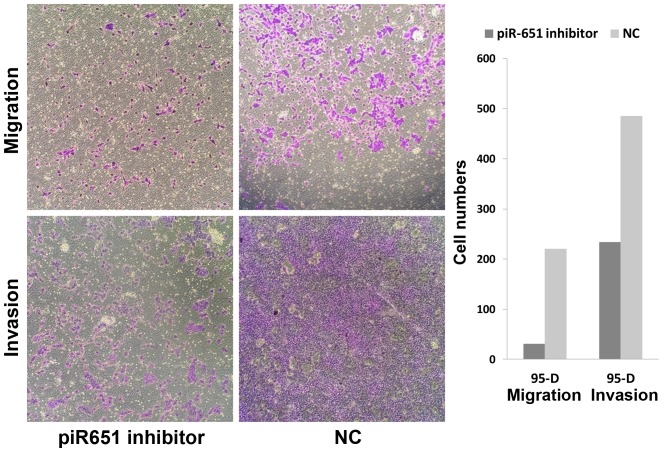
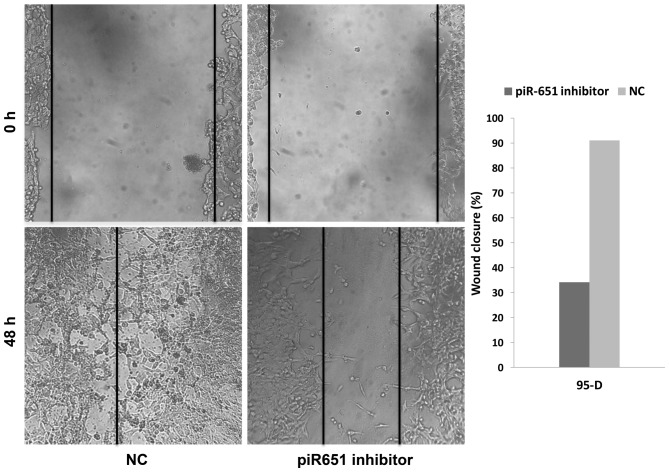
Numerous previous studies have shown that the majority of patients succumb from metastasis rather than primary tumors. Metastasis and invasion are some of the most important features of tumor cells. In the present study, the number of migrating cells in the piR-651 inhibitor cells was evidently less compared to the NC in 95-D (Fig. 3) (P<0.001). As shown in Fig. 3, downregulation of piR-651 impeded the invasion of 95-D cells compared with that in the control cells (P<0.001). In the wound-healing assays, the wound closure (%) in the piR-651 inhibitor was larger compared to the NC, as shown in Fig. 4 (P=0.001). These experiments illustrated that piR-651 may have an important role in regulating the ability of migration and invasion.
Figure 3.
Reduction of the cell ability for invasion and migration following transfection with the piR-651 inhibitor compared with the NC. The cell number in the NC was higher compared to the NC in the migration (P<0.001) and invasion assays (P<0.001). NC, negative control.
Figure 4.
Comparison of the wound-healing assays between the piR-651 inhibitor and NC cells. The inhibitory effect of the piR-651 inhibitor on cell migration was confirmed (P<0.001). NC, negative control.
Discussion
In recent years, the research of non-coding small RNA, including miRNA, siRNA and piRNA has become a significant area of interest in the field of biological gene research. Increasing evidence has demonstrated that the abnormal non-coding small RNA expression was detected in numerous human cancers and cell lines, including lung cancer, demonstrating their various functions by regulating several biological characteristics (23). Different from the miRNAs, by collectively regulating a substantial fraction of the transcriptome, the piRNAs mainly protect the genome from transposons (24). A previous study confirmed that the piRNAs are mainly expressed in germline cells, but several studies also demonstrated that piRNAs are expressed in certain somatic cells and cancer cells (25,26). Therefore, with the development of deep sequencing, several studies have demonstrated that the piRNAs were dysregulated in a number of cancer tissue and cell lines (17–22).
A previous study confirmed that piR-651 was upregulated in gastric, colon, lung and breast cancer tissues and cell lines compared with the paired non-cancerous tissues (18). Therefore, we speculated that piR-651 had a role in tumorigenesis as an oncogene. However, the mechanism of piR-651 in regulating carcinogenesis remains to be elucidated. In the present study, the cell proliferation rate was evidently decreased in a time-dependent manner following transfection with the piR-651 inhibitor in 95-D cells. The flow cytometry assays confirmed that the piR-651 inhibitor induced cell apoptosis, and therefore we deduced that the biological function of piR-651 was closely associated with proliferation and apoptosis of tumor cells. Subsequently, up and downregulation of the apoptosis-related protein following a change in the expression of endogenous piR-651 confirmed this deduction. A previous study identified that the caspase family had a vital role in regulating cell apoptosis, and caspase-3 was the main executor of this process (27). Bax is a member of the bcl-2 family, which is involved in the process of cell apoptosis. When cell apoptosis was induced, bcl-2 was downregulated and bax was overexpressed with the removal from cytochylema to mitochondria and karyotheca (28). In the present study, the level of expression of cleaved caspase-3 and bax was upregulated and bcl-2 was downregulated following a decrease in the expression of endogenous piR-651. Therefore, we can infer that piR-651 regulated tumorigenesis by inhibiting apoptosis and altering the expression level of the apoptosis-related protein.
Invasion and migration, which are usually recognized as the main reason for the high recurrence and fatality rates of lung cancer, restrict the efficacy of surgery and other therapies (29). In the present study, the ability of invasion and migration was reduced following a decrease in the expression of endogenous piR-651 in 95-D cells. Therefore, we can infer that piR-651 has an important role in regulating invasion and migration.
In conclusion, the present study demonstrated that piR-651 has an important role in human NSCLC pathogenesis. Reduced endogenous piR-651 expression may restrain the tumor progression by inhibiting cell proliferation, migration and invasion, and inducing cell apoptosis. These results indicate that targeting endogenous piR-651 may constitute a potential treatment modality for lung cancer.
References
- 1.WHO International Agency for Research on Cancer (IARC): Globocan 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. WHO; Geneva: 2012. [Google Scholar]
- 2.Chen B, Zhang B, Xia L, Zhang J, Chen Y, Hu Q, Zhu C. Knockdown of eukaryotic translation initiation factor 4E suppresses cell growth and invasion, and induces apoptosis and cell cycle arrest in a human lung adenocarcinoma cell line. Mol Med Rep. 2015;12:7971–7978. doi: 10.3892/mmr.2015.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Ai X, Shen S, Lu S. NF-κB-mediated miR-124 suppresses metastasis of non-small-cell lung cancer by targeting MYO10. Oncotarget. 2015;2:1–11. doi: 10.18632/oncotarget.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. International Human Genome Sequencing Consortium: Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Ohno S. So much ‘junk’ DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 7.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 9.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Dieckmann KP, Spiekermann M, Balks T, Flor I, Löning T, Bullerdiek J, Belge G. MicroRNAs miR-371-3 in serum as diagnostic tools in the management of testicular germ cell tumours. Br J Cancer. 2012;107:1754–1760. doi: 10.1038/bjc.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, Boland CR, Goel A. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7:e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 14.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 15.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y, Zhang S, Ma Y. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010;43:635–641. doi: 10.5483/BMBRep.2010.43.9.635. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22:217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 21.Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, Zhong D, Ma L, Tong N, Qin C, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2 Pt B):561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Yan S, Pei C, Cui Y. Decreased microRNA-132 and its function in human non-small cell lung cancer. Mol Med Rep. 2015;11:3601–3608. doi: 10.3892/mmr.2015.3222. [DOI] [PubMed] [Google Scholar]
- 24.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meister G. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113:373–380. doi: 10.1002/jcb.23363. [DOI] [PubMed] [Google Scholar]
- 27.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 28.Huang F, Huang M, Zhang H, Zhang C, Zhang D, Zhou G. Changes in apoptotic factors and caspase activation pathways during the postmortem aging of beef muscle. Food Chem. 2016;190:110–114. doi: 10.1016/j.foodchem.2015.05.056. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Mayo MW, Xiao A, Hall EH, Amin EB, Kadota K, Adusumilli PS, Jones DR. Loss of BRMS1 promotes a mesenchymal phenotype through NF-κB-dependent regulation of Twist1. Mol Cell Biol. 2015;35:303–317. doi: 10.1128/MCB.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]