Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression in diverse biological process. They act as intracellular mediators that are necessary for various biological processes. MicroRNAs targeting pathways of human disease provide a new and potential powerful candidate for therapeutic intervention against various pathological conditions. Even though, the information about miRNA biology has significantly enriched but we still do not completely understand the mechanism of miRNA gene regulation. Various groups across the globe and pharmaceutical companies are conducting research and developments to explore miRNA based therapy and build a whole new area of miroRNA therapeutics. Consequently, few miRNAs have entered the preclinical and clinical stage and soon might be available in the market for use in humans.
Keywords: Clinical stage, gene expression, microRNA, therapeutics
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs that are approximately 20–25 nucleotides in length. They regulate the expression of multiple target genes through sequence-specific hybridization to the 3′ untranslated region (UTR) of messenger RNAs. These microRNAs block the translation or they can cause direct degradation of their target messenger RNAs. miRNAs do not require perfect complementarity for target recognition, so a single miRNA is responsible for the regulation of multiple messenger RNAs. Although miRNAs exert slight effects on each individual messenger RNA target, the combined effect is significant and produces measurable phenotypic results. miRNAs play integral roles in several biological processes, including immune modulation, metabolic control, neuronal development, cell cycle, muscle differentiation, and stem cell differentiation. Most miRNAs are conserved across multiple animal species, indicating the evolutionary importance of these molecules as modulators of critical biological pathways and processes.
It has been seen that expressions of many miRNAs are altered in different diseases. Some miRNAs are overexpressed whereas some are underexpressed in a particular disease giving rise to signature miRNA pattern. With the discovery of miRNAs and its critical role as regulators in various diseases, it is quiet relevant to explore and understand the possibility of using miRNA as therapeutic agents. Recent studies in animals and humans conducted in different laboratories present data that strengthen the candidature of miRNAs to establish the candidature of miRNAs to establish as a novel class of drugs, i.e., miRNA therapeutics.[1,2] The miRNAs possess a unique characteristic which is very attractive in terms of drug development, i.e.,. they are small, with known sequences and are often conserved among species. Based on antisense technology, very potent oligonucleotide targeted against miRNA known as anti-miR is being developed. An ideal design for an active synthetic miRNA is that it should bind to its respective targeted miRNA with high affinity and specificity.
This review will focus on miRNA and its biogenesis, importance in various diseases, the potential of using miRNAs as a therapeutic modality, and finally an update on the status of clinical trials.
HISTORY
In 1993, first miRNA was discovered during a study on gene Lin-4 of Caenorhabditiselegans.[3] It was seen after the isolation of Lin-4 that the gene encodes a protein that binds to 3′ UTR of Lin-14 mRNA and hence prevents translation of Lin-14. In 2000, second small RNA let-7 was discovered; it repressed Lin-4 mRNA to promote a later developmental transition in C. elegans.[4] Subsequently, it was discovered that lin-4 and let-7 were found in Drosophila and human cells, respectively. Most of the RNAs of this class resembled the Lin-4 and let-7 RNAs, however they had no role in regulating the timing of development thereby suggesting that most of miRNAs were involved in other types of regulatory pathways. Following this, the term “miRNA” to refer to this class of small regulatory RNAs came into existence.
MicroRNAs: Tiny master regulators
The miRNAs are known to be expressed in most organisms from plant to vertebrates (including viruses) and are conserved in all organisms. They regulate gene expression by either degrading or making the targeted mRNAs “silence” rendering their translation into proteins. The miRNAs regulate gene expressions, which affect various biological processes such as cell proliferation, differentiation, survival, and motility. The miRNAs do not require perfect complementarity for target recognition, so a single miRNA is able to regulate multiple mRNAs. On average, a given miRNA can regulate several hundred transcripts whose effector molecules function at various sites within cellular pathways and networks. Consequently, miRNAs are able to switch instantly between cellular programs and are therefore often viewed as tiny master regulators of the human genome.
The miRNAs are predicted using different approaches such as experimental method, computational approach, expressed sequence tag, and genomic survey sequence analysis. However, only a few predicted miRNAs have undergone validation experimentally. Computational analysis indicates that the total number of miRNAs could be more that 1% of total translated genes and more than 30% of protein-coding genes may be targeted by miRNAs.
MicroRNA biogenesis
The miRNA genes are expressed in the cell nucleus. About 70% of the human miRNAs are transcribed from introns and/or exons. A brief overview of miRNA biogenesis is shown in Figure 1.
Figure 1.
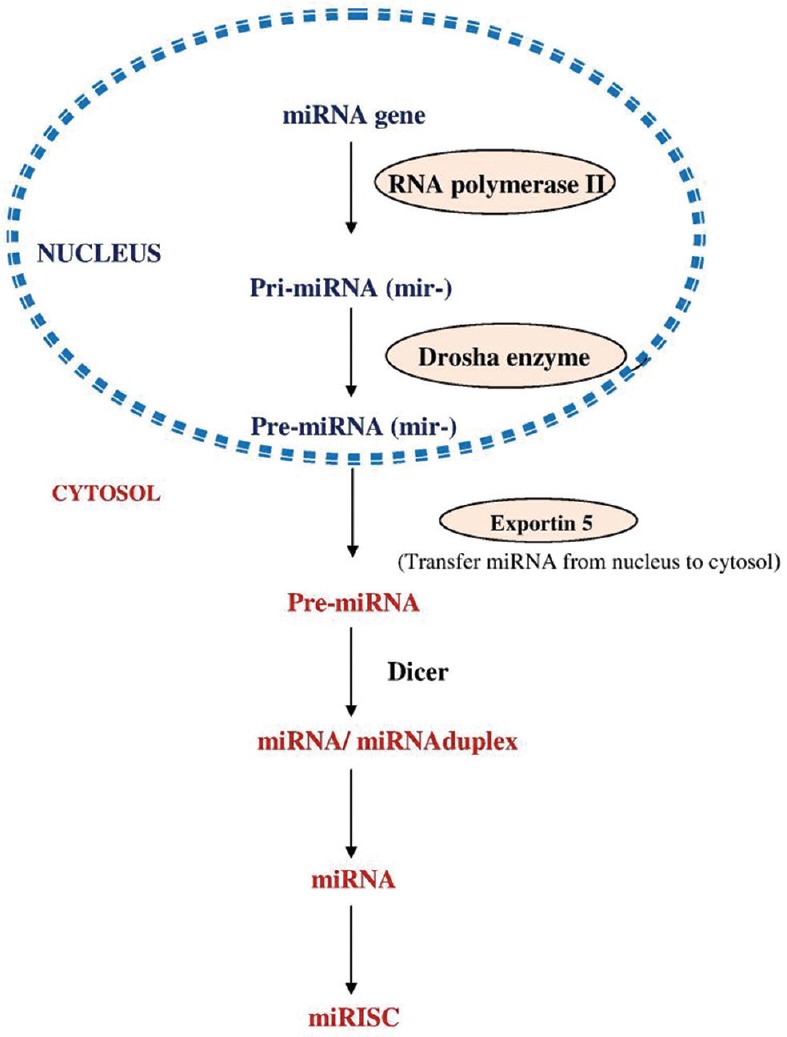
Schematic overview of microRNA biogenesis
Association of microRNA with various diseases
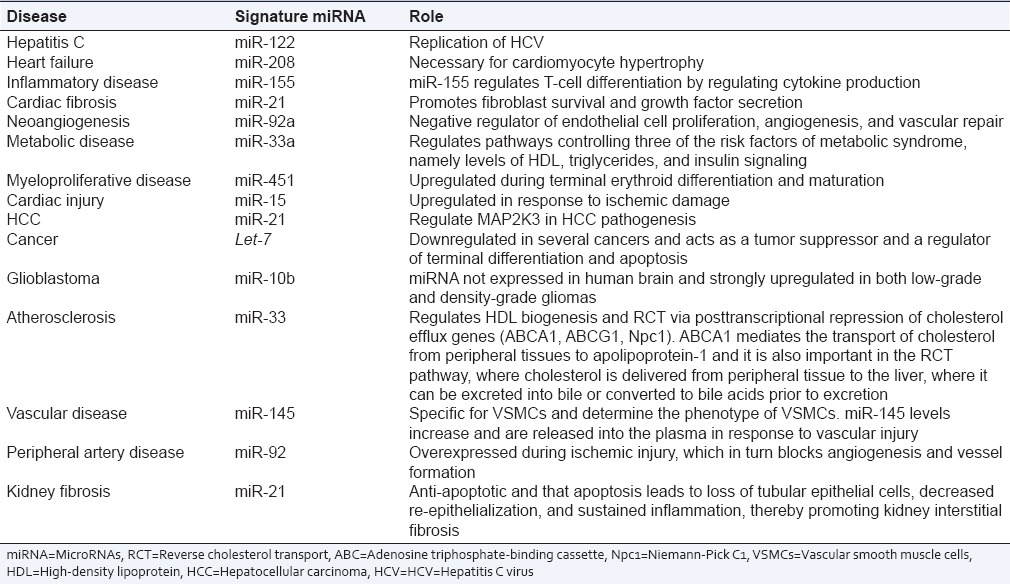
miRNAs are becoming increasingly recognized as their expressions are altered in different diseases such as cancer, hepatitis C infection, myocardial infarction, and metabolic disease. Some miRNAs are overexpressed whereas some are underexpressed in a particular disease giving rise to a signature miRNA pattern. In case of tumors, the miRNAs which are overexpressed may be considered as oncogenes and are called “oncomirs.”[5] They are considered to be involved in tumor development by reciprocally inhibiting tumor suppressor genes (genes that control cell differentiation and apoptosis), for example, miR-17–192 is significantly overexpressed in lung cancer and several other lymphomas. On the other hand, expression of some miRNAs (tumor suppressor) is lower in cancerous cells and usually prevents tumor development by negatively inhibiting oncogenes, for example, let-7 is a tumor suppressor miRNA and aberrant expression of let-7 results in oncogenic loss of differentiation.[6] An overview of the association of signature miRNA with some disease condition is given in Table 1. The signature miRNAs associated with disease are discussed in the following sections.
Table 1.
Differentially expressed microRNAs (signature microRNAs) in various diseases
MiR-122 in hepatitis C virus
MiR-122 is the most commonly found miRNA in the adult liver and plays a key role in liver biology that includes development, differentiation, homeostasis, and functions. It controls the metabolism, i.e., cholesterol biosynthesis. miR-122 is involved in the replication of hepatitis C virus (HCV). It binds to the viral genome and enhances viral translation and replication. The binding site in the 5′ end of HCV genome provided the evidence of the direct role of miR-122 into HCV replication.[7,8]
MiR-33a in metabolic disease
miR-33a targets genes involved in cholesterol export such as the adenosine triphosphate-binding cassette (ABC) transporters ABCA1 and ABCG1 and the endolysosomal transport protein Niemann-Pick C1 (Npc1). In agreement with the regulation of ABCA1 by miR-33, modulation of miR-33a levels results in encompassing effects in cholesterol efflux in macrophages thus suggesting that miR-33 may participate in the regulation of high-density lipoprotein (HDL) levels in vivo. Indeed, three independent studies have demonstrated that endogenous inhibition of miR-33 using different strategies leads to a significant increase in hepatic ABCA1 expression and plasma HDL levels, and these findings were later confirmed in the miR-33 knockout mice.[9]
MiR-155 in inflammatory disease
miR-155 is overexpressed in atopic dermatitis and contributes to chronic skin inflammation by increasing the proliferative response of T-helper cells through the downregulation of cytotoxic T-lymphocyte antigen-4.[10] In autoimmune disorders such as rheumatoid arthritis, miR-155 showed higher expression in patients' tissues and synovial fibroblasts.[11] In multiple sclerosis, increased expression of miR-155 has also been measured in peripheral and central nervous system-resident myeloid cells, including circulating blood monocytes and activated microglia. Overexpression of miR-155 leads to chronic inflammatory state in human.
MiR-10b in glioblastoma
The levels of miR-10b are upregulated in human glioblastoma tissues, glioblastoma cell, and stem cell lines as compared to normal human tissues or astrocytes[12] Inhibition of miR-10b inhibits glioblastoma proliferation, reduces cell invasion, and migration in glioblastoma cell and stem cell lines whereas overexpression of miR-10b induced cell migration and invasion.
MiR-33 in atherosclerosis
Plasma HDL levels have a protective role in atherosclerosis. miR-33, an intronic miRNA located within the SREBF2 gene, suppresses the expression of the cholesterol transporter ABC transporter A1 (ABCA1) and lowers HDL levels. Conversely, mechanisms that inhibit miR-33 increase ABCA1 and circulating HDL levels, suggesting that antagonism of miR-33 may be atheroprotective. In a study, mice deficient for the low-density lipoprotein receptor (LDLr–/– mice), with established atherosclerotic plaques treated with anti-miR33 for 4 weeks, showed an increase in circulating HDL levels and enhanced reverse cholesterol transport to the plasma, liver, and feces.[13] Consistent with this, anti-miR33-treated mice showed reductions in plaque size and lipid content, increased markers of plaque stability, and decreased inflammatory gene expression. Notably, in addition to raising ABCA1 levels in the liver, anti-miR33 oligonucleotides directly targeted the plaque macrophages, in which they enhanced ABCA1 expression and cholesterol removal. Thus, raising HDL levels by anti-miR33 oligonucleotide treatment promotes reverse cholesterol transport and atherosclerosis regression.
MiR-21 in hepatocellular carcinoma
Deregulated expressions of several miRNAs were found to correlate with the pathologic and clinical characteristics of hepatocellular carcinoma HCC.[14] The miRNA microarray analysis has revealed that miR-21 was dramatically elevated in HCC tumor cells, with significant reductions of the expressions of several tumor suppressor genes, including PTEN, PDCD4, RECK, and TPM1 (PTEN).[15] MAP2K3 has also been identified as a novel direct target of miR-21. The study of loss-of-function of miR-21 by transduction of miR-21 sponge in HepG2 cells indicated that miR-21 might regulate cell proliferation, apoptosis, and invasiveness partially by targeting MAP2K3.
MicroRNA as therapeutic target and tool
The expression of miRNAs is altered in various diseases and it is now feasible to manipulate miRNA expression by injecting miRNAs similar to the use of antisense mRNAs and RNAi (widely used techniques for investigating gene function and in gene therapy).[16] As the activation of oncogenes could cause cancer, artificial antisense miRNAs could be synthesized and employed to block their targeted oncomirs to prevent the formation of cancer. However, various critical prerequisite data must be available, for example, identification of signature miRNAs, their mechanism of action, applicability by RNAi, delivery of miRNAs, and their active form in vivo. Once this information is available, miRNA will have a bright future and become a novel therapeutic tool. The process of building miRNA therapeutics is similar to drug discovery and development. Following steps are involved in the discovery and development of miRNA therapeutics Figure 2:
Figure 2.
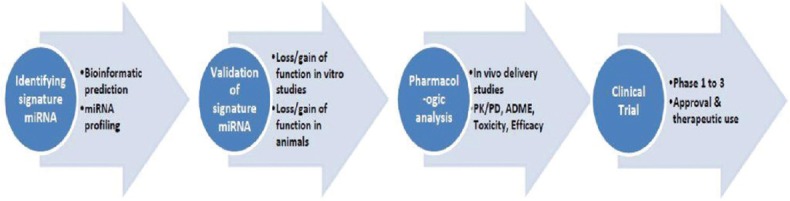
Process of microRNA discovery and development
Identification of signature miRNA (done by miRNA profiling in disease)
Validation of signature miRNA (loss/gain of function studies in vitro and in animal models)
Pharmacological analysis (in vivo miRNA delivery studies, pharmacokinetics/pharmacodynamics, (absorption, distribution, metabolism, excretion, and toxicity studies)
Clinical trials (studies on the evaluation of efficacy and safety).
Strategies for microRNA manipulation
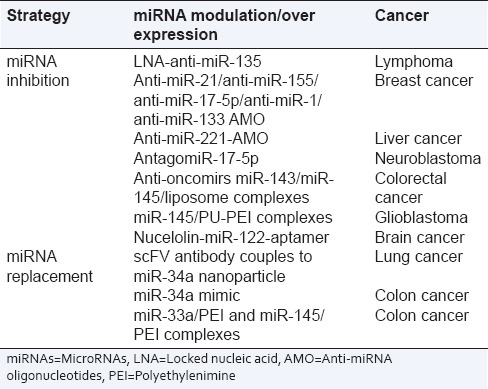
There are two main strategies to manipulate miRNAs which are dependent on whether the targeted miRNA expression needs to be downregulated or to re-introduce miRNAs function to restore loss of function. The strategies for miRNA manipulation are summarized in Table 2.
Table 2.
Various microRNAs-based therapeutic strategies investigated in cancer
Antisense inhibition of mature microRNA (inhibiting oncomirs)
The miRNA antagonists are oligonucleotides containing the complementary sequences of endogenous miRNAs. Antisense oligonucleotide (AMO), also called antagomir, is the most commonly used anti-miRNA antisense oligomer.[2] The locked nucleic acid possess stronger affinity to targeted miRNA, more resistant to nucleases, and have lower toxicity. The peptide nucleic acid (PNA) is an artificial synthesized peptide-structured polymer and is similar to DNA and RNA. The PNA binds the targeted nucleotide more tightly than the nucleotide/nucleotide binding, are relatively stable, have low toxicity, and could be administered systematically.[17] The latest new strategy available is miRNA sponge and miRNA masking. The miRNA sponge downregulates the targeted miRNA and possesses multiple complementary sites to the targeted miRNA,[18] whereas miRNA masking have complimentary miRNA binding site in the 3′ UTR of the target mRNA to inhibit competitively and decreases the activity of endogenous miRNA. The various antisense inhibitors employed for downregulating various miRNAs are depicted in Table 3.
Table 3.
Various methods available for the delivery of microRNAs
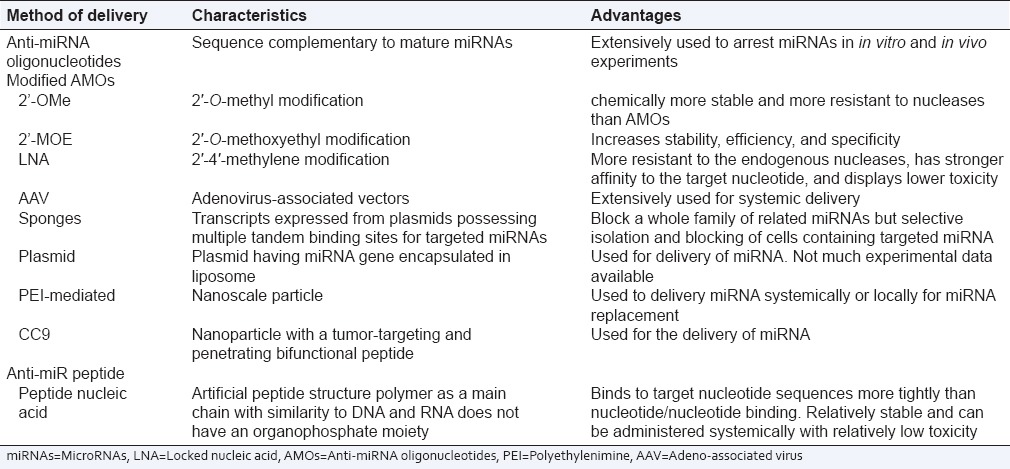
Replacement of microRNAs
In diseases such as cancer, expression levels of downregulated miRNAs or altered miRNA could be done using vector, overexpressing the targeted miRNA or by the transfection of double-stranded miRNAs. Therefore, studies have been conducted by introducing artificial double-stranded miRNA (mimic of targeted downregulated miRNA). For example, overexpression of downregulated miR-26a in hepatocellular carcinoma (HCC) in mouse liver resulted in inhibition of cancer proliferation and initiation of apoptosis. Subsequently in another study, the downregulated miR-34a level was increased by delivering artificial miR-34a with NOV340 liposome in an orthotopic model of HCC.[19] This resulted in significant tumor reduction, prolonged survival, and disease protection in animals.
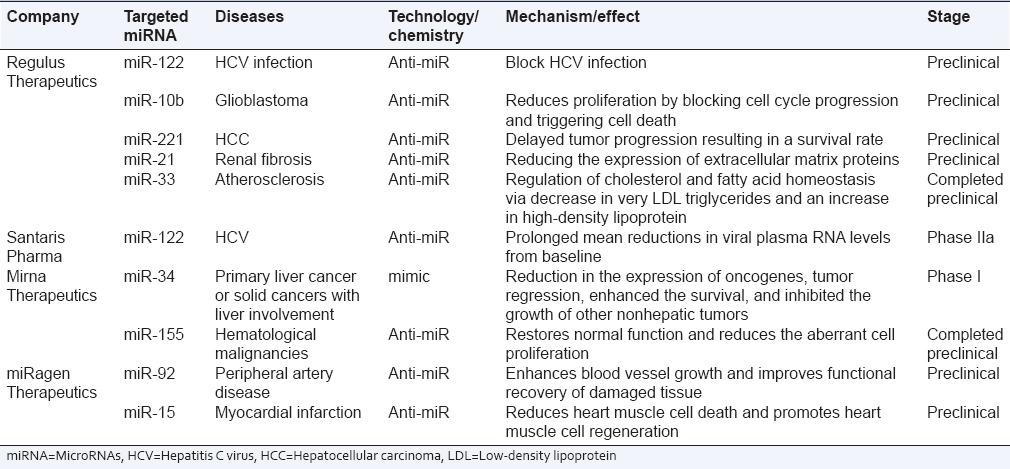
Patent and clinical trial status of microRNA therapeutics
There is a great excitement regarding miRNA use as therapeutic entities. In terms of scientific perspective, miRNA represents a novel and an attractive target which could manipulate the body functions. Consequently, there has been considerable increase in the number of patent applications filed over the decade. The annual number of US and European published patent applications and issued patents related to miRNAs is close to 500. A snapshot of the current development status of miRNA-based therapeutics is summarized in Table 4.
Table 4.
Overview of microRNAs therapeutics development status
Future perspectives of microRNA therapeutics
The functionality of miRNA in controlling diverse gene expression in cancer and various other important diseases makes miRNA an ideal candidate for therapeutic applications. Recent data demonstrate that the miRNA expression is altered in various human diseases and its selective modulation through antisense inhibition or replacement could significantly affect the prognosis of a disease. With the technological advancement and ease of administration of miRNA through local or parenteral injection routes and its sufficient uptake in the tissue without the need of developing formulation gives miRNA therapeutics an extra edge. We hope that with increasing research and development on miRNA, increase in the filed patents and increased attention and interest of the biotechnology companies in miRNA-based therapeutics would follow suit. This promises the availability of miRNA therapeutics in the market in the coming years for the treatment of various diseases.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. 2006;354:1194–5. doi: 10.1056/NEJMcibr060065. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Jopling CL, Schütz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 9.Marquart TJ, Allen RM, Ory DS, Baldán A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–32. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Tian F, Wang F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-a and IL-1ß in PBMCs. Int J Mol Sci. 2013;14:23910–21. doi: 10.3390/ijms141223910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–63. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–31. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravalli RN. Development of MicroRNA Therapeutics for hepatocellular carcinoma. Diagnostics (Basel) 2013;3:170–91. doi: 10.3390/diagnostics3010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 16.Vidal L, Blagden S, Attard G, de Bono J. Making sense of antisense. Eur J Cancer. 2005;41:2812–8. doi: 10.1016/j.ejca.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 17.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–75. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11:316–22. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bader AG. miR-34 – A microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]


