Abstract
The frequency of Candida infections is currently rising, and thus adversely impacting global health. The situation is exacerbated by azole resistance developed by fungal pathogens. Candida tropicalis is an opportunistic pathogen that causes candidiasis, for example, in immune-compromised individuals, cancer patients, and those who undergo organ transplantation. It is a member of the non-albicans group of Candida that are known to be azole-resistant, and is frequently seen in individuals being treated for cancers, HIV-infection, and those who underwent bone marrow transplantation. Although the genome of C. tropicalis was sequenced in 2009, the genome annotation has not been supported by experimental validation. In the present study, we have carried out proteomics profiling of C. tropicalis using high-resolution Fourier transform mass spectrometry. We identified 2743 proteins, thus mapping nearly 44% of the computationally predicted protein-coding genes with peptide level evidence. In addition to identifying 2591 proteins in the cell lysate of this yeast, we also analyzed the proteome of the conditioned media of C. tropicalis culture and identified several unique secreted proteins among a total of 780 proteins. By subjecting the mass spectrometry data derived from cell lysate and conditioned media to proteogenomic analysis, we identified 86 novel genes, 12 novel exons, and corrected 49 computationally-predicted gene models. To our knowledge, this is the first high-throughput proteomics study of C. tropicalis validating predicted protein coding genes and refining the current genome annotation. The findings may prove useful in future global health efforts to fight against Candida infections.
Introduction
Candida tropicalis is one of the most common opportunistic human pathogens that causes significant mortality among immune-compromised individuals. It is a diploid, dimorphic fungus, which exists either in the form of yeast-like budding cells or in pseudohyphae (Chai et al., 2010). Studies conducted as early as in the 1930s had demonstrated the pathogenicity of C. tropicalis (Stovall et al., 1933). The incidence of candidiasis caused by C. tropicalis varies across geographical regions. The prevalence of infections is reported to be higher in the Asia-Pacific region and Europe (Negri et al., 2012). Recent studies have reported that C. tropicalis is resistant to fluconazole, which is a broad-spectrum antifungal drug used to treat fungal infections (Pfaller et al., 2009). Secreted proteins are known to play a role in pathogenesis of Candida. However, as opposed to C. albicans, the secreted proteins of C. tropicalis are largely unknown. Identifying secreted proteins that play a role in infection may help in developing better treatment strategies.
The genome of C. tropicalis, strain MYA-3404, was sequenced in 2009 (Butler et al., 2009). The annotated genome assembly consists of 23 supercontigs and 6258 protein-coding genes. However, a large majority of these genes are labeled as either predicted or hypothetical. The usefulness of any genome sequence is largely determined by the quality of annotation, and therefore accurate annotation of protein-coding genes is necessary for sequenced organisms.
Large-scale analyses of proteome using shotgun proteomics have been useful in validating the predicted protein-coding genes in a newly annotated genome. High resolution mass spectrometry can provide valuable data for a proteogenomic approach to refine genome annotation (Renuse et al., 2011). Proteogenomics, briefly, involves the identification of novel peptides (i.e., peptides that are not present in the annotated protein database), using theoretical databases such as 6-frame translated genome database and 3-frame translated RNAseq database. These novel peptides will, in turn, be used to identify novel protein coding regions and corrections in the annotated gene boundaries (Nesvizhskii, 2014).
The methodologies to be followed for proteogenomic studies of micro-organisms have been discussed earlier (Kucharova et al., 2014). Proteogenomics can also be used to predict the translation start site of a protein accurately. Translation start sites play a major role in protein synthesis regulation apart from determining the N-terminus of the nascent protein (Hartmann et al., 2014). In the past, our group has performed genome annotation of various opportunistic yeast pathogens using proteogenomic approaches (Nagarajha Selvan et al., 2014; Prasad et al., 2012).
In this study, we carried out in-depth proteomics analysis of C. tropicalis and validated 2743 predicted protein-coding genes and 329 translation start sites. We also analyzed the conditioned media and report the largest catalogue of secreted proteins of C. tropicalis. Further, we employed proteogenomic strategies to refine the genome annotation and report 86 novel genes, 12 novel exons, and corrected 49 gene models.
Materials and Methods
Strain and growth conditions
Candida tropicalis (MTCC 184) culture was procured from the Microbial Type Culture Collection and Gene Bank, IMTECH, Chandigarh, India. The cells were cultured in Yeast Nitrogen Base (YNB) media (HiMedia Laboratories, Mumbai, India) at 37°C with shaking (300 RPM) for 8–10 hours, until OD600 of culture reached ∼1. Approximately 6 billion cells were pelleted down by centrifuging at 1500 g at 4°C. The pellet was stored at −80°C until further use. The supernatant fraction was also processed further to study secreted fraction of proteome.
Protein isolation from cell pellet and conditioned media
The cell pellets were suspended in cracking buffer (5% SDS, 8 M urea lysis buffer, 0.1 mM EDTA, 40 mM Tris-HCl; pH 6.8) (Printen et al., 1994) and were subjected to disruption with glass beads for 60 min using a cell disruptor (Disruptor Genei SI-D267, Scientific Industries Inc. NY). The samples were centrifuged at 20,000 g for 15 min and the clear supernatant was collected.
The conditioned media was obtained by processing the supernatant after pelleting out C. tropicalis cells from the culture. The supernatant was filtered using a 0.22 μm filter (Millipore Corporation, Billerica, MA) and then concentrated using a 3000 Da molecular mass cut-off spin filter (Millipore Corporation). Protein estimation for both cell lysate and conditioned media was performed using bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Rockford, IL). The assay involved construction of a standard curve with known concentrations of Bovine Serum Albumin (BSA) and thus estimating protein concentration in the sample.
In-gel digestion
For both total proteome and secreted fraction, 300 μg of protein was resolved on a 10% SDS-PAGE gel and stained with colloidal Coomassie blue. The lane of the cell lysate proteome was divided into 23 bands. As the proteome of the conditioned media was not complex, the lane was divided into eight bands. In-gel digestion was performed for these protein gel bands as described earlier (Balakrishnan et al., 2014). Briefly, the gel bands were incubated in 5 mM dithiothreitol (DTT) at 60°C for 45 min to reduce the disulfide bonds of the proteins. The gel bands were later incubated in 20 mM iodoacetamide (IAA) at room temperature for 10 min in dark to alkylate the reduced bonds. Trypsin (modified sequencing grade; Promega, Madison, WI) was added and digestion was carried out at 37°C for 16 h. 100 μL of 1% formic acid was added to quench the reaction. Peptides were extracted using 40% ACN and dried. They were then reconstituted in 0.1% formic acid and desalted using C18 STAGE tips (Rappsilber et al., 2003) and vacuum dried. The dried peptides were stored at −80°C till LC-MS/MS analysis.
In-solution trypsin digestion
Two mg of the protein lysate from the cell pellet and ∼900 μg of protein from the conditioned media were separately subjected to buffer exchange with 50 mM triethylammonium bicarbonate (TEAB buffer) using 30,000 Da molecular mass cut-off spin filter (Millipore Corporation). The proteins were then reduced by incubating the lysate in 5 mM dithiothreitol (DTT) at 60°C for 60 min. Alkylation was carried out by incubating the lysate in 20 mM iodoacetamide (IAA) at room temperature for 10 min in dark. L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK) treated trypsin (Worthington Biochemical Corporation, Lakewood, NJ) was added at 1:10 (w/w) final concentration and digested for 16 h at 37°C. The reaction was quenched by acidification with formic acid. The peptides were lyophilized and stored at −80°C until further use.
Basic pH RPLC
Lyophilized samples of in-solution digested peptides were reconstituted in bRPLC solvent A (10 mM TEABC buffer, pH ∼8.5) and then loaded onto a Waters XBridge column (Waters Corporation, Milford, MA; 130 Å, 5 μm, 250 × 9.4 mm) using an Agilent 1200 series HPLC system, with a flow rate of 1 mL/min for 4.5 min. Separation of peptides was performed using a 50 min gradient, at a flow rate of 1 mL/min of solvent A (10 mM TEABC buffer, pH ∼8.5) and B (10 mM TEABC buffer, 90% acetonitrile, pH ∼8.5). The fractionation program was 95% solvent A for 5 min, continued by a short (30 sec) gradient of 5%–8% of solvent B, followed by a gradient of 8%–40% of B in 35 min, and a 40%–100% gradient for 1 min. Fractionation was completed by a gradient of 100% B for 4 min and then equilibrated in 95% of solvent A for 5 min. Flow through fractions (95 in total) were collected into a 96-well plate from 4.5–50 min. The fractions were concatenated into 24 in cell lysate and 12 in conditioned media, respectively. Pooled samples were lyophilized and stored at −80°C till they were subjected to LC-MS/MS analysis.
LC-MS/MS analysis
The peptide fractions were analyzed on LTQ-Orbitrap Velos (Thermo Scientific, Bremen, Germany) interfaced with an Easy-nLC (Thermo Scientific). The LC system consisted of an enrichment column (75 μm × 2 cm, C18 material 5 μm, 100 Å) and an analytical column (75 μm × 10 cm, C18 material 5 μm, 100 Å). Both the columns were packed in-house. A flow rate of 350 nL/min and a linear gradient of 7% to 30% solvent B (0.1% formic acid, 90% ACN) was used to separate the peptides on the analytical column. The spray voltage was 2.0 kV, and the heated capillary temperature was 225°C. Mass spectrometry data were acquired in a data-dependent mode with a resolution of 60,000 at 400 m/z for MS scans and 7500 for MS/MS scans. The 20 most intense precursor ions from each survey scan were selected for fragmentation. Peptide fragmentation was performed by higher-energy collision dissociation (HCD) with normalized collision energy of 39%. The automatic gain control and maximum ion injection time for FT MS was 0.5 million ions and 100 ms respectively; while it was 0.1 million ions and 200 ms for FT MS/MS.
Database searches for peptide and protein identification
The MS/MS data acquired was searched against two databases: i) C. tropicalis protein database (6,258 entries; 3,034,039 amino acids), and ii) six-frame translated genome database (992,921 entries; 25,161,168 amino acids). The whole genome and protein databases were downloaded from the resources of Broad Institute (http://www.broadinstitute.org/annotation/genome/candida_group/MultiDownloads.htmL). The six-frame translated genome database was created using in-house Python scripts generated. Six-frame translation of the genomic sequence was carried out from stop codon to stop codon and the sequences that were not unique and/or shorter than 7 amino acids in length were removed.
Sequences of commonly encountered protein contaminants such as BSA, trypsin, and keratins were added to both databases. The searches were performed using SEQUEST and MASCOT through Proteome Discoverer (Version 1.4) software suite (Thermo Scientific). The search parameters included trypsin as the proteolytic enzyme with two missed cleavages and semi-tryptic identifications allowed.
Oxidation of methionine, deamidation of asparagine and glutamine, carbamylation of the peptide N-terminus and lysine, and acetylation of the protein N-terminus were set as dynamic modifications, while carbamidomethylation at cysteine was set as a static modification. Precursor and fragment mass tolerance were set to 10 ppm and 0.05 Da, respectively. Only those peptide-spectrum matches (PSMs) that qualified a 1% false discovery rate (FDR) were considered as authentic identifications. The FDR was calculated using target-decoy database searches.
Workflow for proteogenomic analysis
The peptides identified uniquely in the six-frame translated genome database were used for proteogenomic analysis. These peptides, referred to as Genome Search Specific Peptides (GSSPs), were considered for analysis only if they qualified the 1% FDR threshold and matched uniquely with six frame translated genome database. The GSSPs in the intergenic region were analysed to identify novel exons and novel protein coding regions while the GSSPs that were in close proximity with the existing genome annotation were used to refine the existing gene models. The quality of MS/MS spectra of these GSSPs was manually verified to ensure correct peptide assignments. The genome of C. tropicalis, annotated genes, peptides identified from known proteins, GSSPs and gene models derived from publicly available RNA-Seq data (www.ncbi.nlm.nih.gov/sra/SRX470927) were overlaid in the Integrated Genome Viewer (IGV) Version 2.3 (Thorvaldsdottir et al., 2013) for analysis.
Results
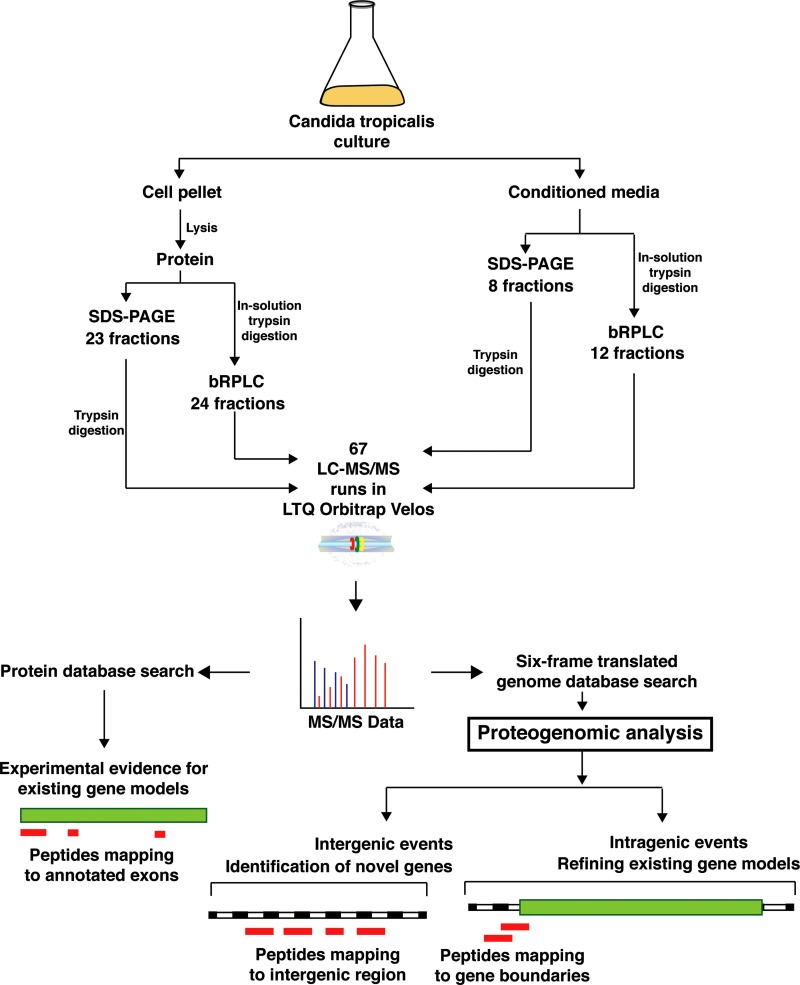
The genome of C. tropicalis was sequenced in 2009, and 6258 protein coding genes were annotated (Butler et al., 2009). In this study, we aimed to carry out in-depth proteomic profiling of this opportunistic pathogen by performing an unbiased global proteomic analysis. We also intended to identify novel genes and refinements in the genome architecture using a proteogenomic approach. To achieve this, we took a multipronged strategy with different fractionation techniques. A total of 67 fractions (23 in-gel and 24 bRPLC fractions from the cell lysate; 8 in-gel and 12 bRPLC fractions from the conditioned media) were subjected to LC-MS/MS analysis that resulted in a total of 357,343 MS/MS spectra. These data were searched against two different databases as described in the Methods section. A schematic of the workflow followed in this study is presented in Figure. 1.
FIG. 1.
Workflow of the study: C. tropicalis was cultured in YNB media, and the cells and supernatant were collected. Protein extract from cells and conditioned media were subjected to in-gel and in-solution digestion. Peptides obtained from in-solution digestion were separated by basic pH reversed-phase liquid chromatography. The peptide fractions obtained were analyzed using Fourier transform mass spectrometry, and the data obtained were searched against the known protein database of C. tropicalis to confirm the predicted genes. The data were also searched against six-frame translated genome database to identify novel gene models and refinements to the current annotation of the genome.
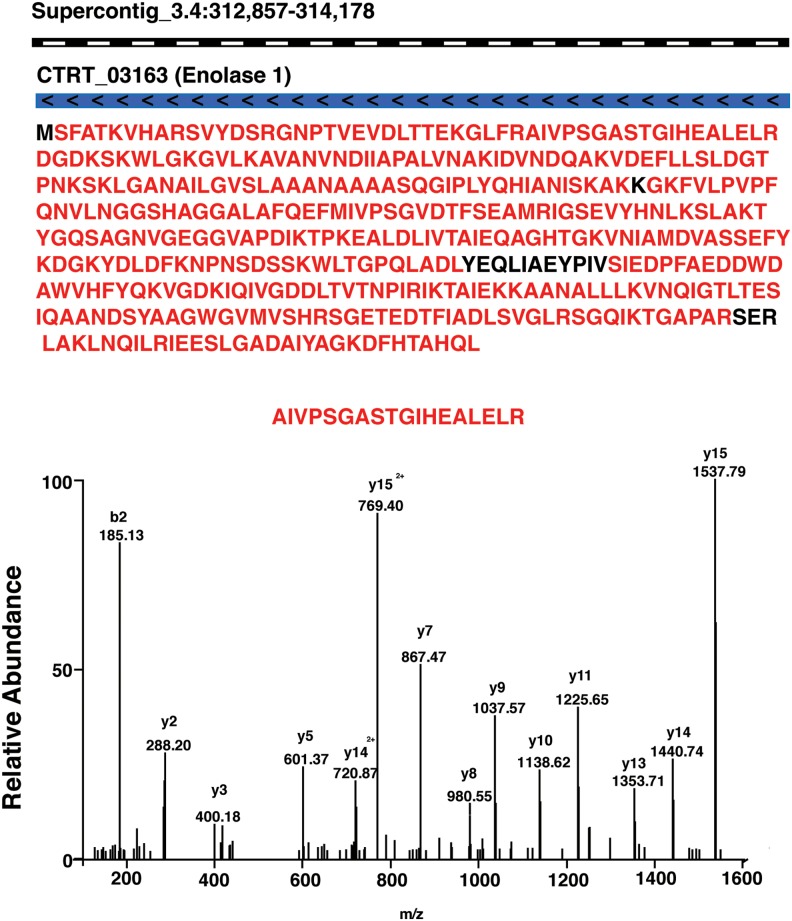
The database searches of MS/MS spectra against C. tropicalis protein database using MASCOT and SEQUEST resulted in 23,173 unique peptides supported with 170,364 peptide-spectrum matches (PSMs) with 1% FDR cut-off, which mapped to 2,743 protein groups accounting for ∼44% of the C. tropicalis predicted proteome. A large majority (∼66%) of the detected proteins were identified based on two or more unique peptides. For example, Figure 2 shows confirmation of a 440 amino acid protein, enolase 1 (CTRT_03163) from C. tropicalis. 203 peptides, of which 184 were unique, were mapped to this protein covering 96% of the protein sequence. To our knowledge, this study provides the largest dataset for the proteome of C. tropicalis.
FIG. 2.
Peptide evidence for predicted gene Enolase 1: It is encoded by the CTRT_03163 (blue bar) gene located on Supercontig 3.4. It has a 441 amino acid sequence. The sequences in red font indicate the peptides identified in this study. A representative MS/MS spectra of the peptide AIVPSGASTGIHEALELR is shown in the lower panel.
Proteomic analysis of cell lysate
The proteins extracted from the cell pellet was subjected to LC-MS/MS analysis after fractionation by two methods: (i) the proteins were separated on SDS-PAGE and in-gel digestion was performed for the 23 bands that were excised, and (ii) the proteins were digested using trypsin and the peptides were fractionated using bRPLC and pooled into 24 fractions. The proteomic analysis of the cell lysate resulted in the identification of 2591 proteins out of which 384 were uniquely identified from SDS-PAGE fractions, while 965 were uniquely identified from bRPLC fractions. Multiple fractionation strategies have been followed in a number of proteogenomic investigations in order to increase the number of protein identifications. The list of proteins that were identified in the cell lysate and the peptides that lead to these identifications are provided in Supplementary Table S1 and Supplementary Table S2, respectively (supplementary material is available online at ftp.liebertpub.com/omi).
Analysis of conditioned media proteome
Secreted proteins are known to play a major role in virulence and cellular stability in Candida species (Sorgo et al., 2013). To identify the secreted proteins of C. tropicalis, we carried out proteomic analysis of the conditioned media using two different fractionation strategies (SDS-PAGE and bRPLC). We identified 780 proteins (36 unique proteins from SDS-PAGE fractions and 615 unique proteins from bRPLC fractions) in the conditioned media. Out of the 780 proteins identified, 152 proteins were detected exclusively in the conditioned media, and not from the cell lysate proteome in this study. All the proteins that were identified in the conditioned media and the peptides that led to these identifications are listed in Supplementary Table S3 and Supplementary Table S4, respectively.
To elucidate the secretory potential of these proteins, bioinformatics analyses were performed on these 780 proteins using SignalP 4.0 (Petersen et al., 2011) and TMHMM (Krogh et al., 2001). These analyses led to the finding that 102 proteins have an N-terminal signal peptide and 23 out of the 102 have one or more transmembrane domains. Therefore, it can be concluded that 79 proteins that were identified in the conditioned media possess the potential to be secreted.
The large majority of proteins identified in the conditioned media of C. tropicalis are known to play biological roles in cell wall assembly and maintenance and virulence. C. tropicalis protein EPD2 (CTRT_03942) was identified with six unique peptides and was exclusively detected from the conditioned media. PHR1, the C. albicans ortholog of this protein, is known to be expressed to adapt to a neutral pH of the environment, such as blood (Saporito-Irwin et al., 1995). C. tropicalis candidapepsin-7 (CTRT_04491) was identified with nine unique peptides and was also exclusive to the conditioned media. Secreted aspartyl proteinase 7 (SAP7), the C. albicans ortholog is a known virulence factor and is reported to be expressed during murine vaginal and intravenous infection (Ran et al., 2013). Orthologs of a large number of proteins that are known to play a role in pathogenesis of C. albicans were identified in this study (Table 1). To our knowledge, this is the largest catalogue of secreted proteins in C. tropicalis.
Table 1.
A Partial List of Secreted Proteins Identified in This Study
| Candida tropicalis ID | Candida albicans gene symbol | Protein description/function |
|---|---|---|
| CTRG_04491 | SAP7 (orf19.756) | Secreted aspartyl proteinase |
| CTRG_01870 | ABG1 (orf19.1597) | Altered budding growth pattern |
| CTRG_02140 | UTR2 (orf19.1671) | Cell-surface factor |
| CTRG_05456 | CHT2 (orf19.3895) | Chitinase |
| CTRG_05827 | CHT33 (orf19.7586) | Chitinase |
| CTRG_03988 | COI1 (orf19.5063) | Ciclopirox olamine-induced |
| CTRG_04579 | ENG1 (orf19.3066) | Endoglucanase |
| CTRG_00476 | ECE1 (orf19.3374) | Extent of cell elongation |
| CTRG_02093 | MP65 (orf19.1779) | Mannoprotein of 65 kDa |
| CTRG_03942 | PHR1 (orf19.3829) | PH responsive |
| CTRG_06145 | PGA4 (orf19.4035) | Predicted GPI-anchored |
| CTRG_03687 | RBE1 (orf19.7218) | Repressed by Efg1p |
| CTRG_04180 | RBT4 (orf19.6202) | Repressed by TUP1 |
| CTRG_00109 | RBT5 (orf19.5636) | Repressed by TUP1 |
| CTRG_00299 | RHD3 (orf19.5305) | Repressed during hyphae development |
| CTRG_02113 | SAP1 (orf19.5714) | Secreted aspartyl proteinase |
| CTRG_01856 | YWP1 (orf19.3618) | Yeast-form wall protein |
| CTRG_04359 | ASM3 (orf19.6037) | Putative secreted acid sphingomyelin phosphodiesterase |
| CTRG_00169 | BGL2 (orf19.4565) | Cell wall 1,3-beta-glucosyltransferase |
| CTRG_00046 | DAG7 (orf19.4688) | Secretory protein |
| IOB_CT_NG_63 | Ecm33 (orf19.3010.1) | GPI-anchored cell wall protein |
| CTRG_04664 | EXG2 (orf19.2952) | GPI-anchored cell wall protein |
| CTRG_01272 | MSB2 (orf19.1490) | Mucin family adhesin-like protein |
| CTRG_04132 | PHO100 (orf19.4424) | Putative inducible acid phosphatase |
| CTRG_04296 | PHR2 (orf19.6081) | Glycosidase |
| CTRG_01230 | PLB4.5 (orf19.1442) | Phospholipase B |
| CTRG_05458 | SCW11 (orf19.3893) | Cell wall protein |
| CTRG_03668 | SIM1 (orf19.5032) | Adhesin-like protein |
| CTRG_01132 | SOD4 (orf19.2062) | Cu-containing superoxide dismutase |
| CTRG_02944 | SUN41 (orf19.3642) | Cell wall glycosidase |
| CTRG_02154 | Tos1 (orf19.1690) | Protein similar to alpha agglutinin anchor subunit |
| CTRG_04334 | XOG1 (orf19.2990) | Exo-1,3-beta-glucanase; 5 glycosyl hydrolase family member |
Identification of translational start sites
The N-termini of most eukaryotic proteins are acetylated after the cleavage of the initiator methionine. Translational start sites of a protein can be correctly annotated by the identification of N-terminal acetylated peptides. In C. tropicalis, we identified a total of 379 unique acetylated peptides corresponding to 329 proteins. Out of the 379, 61 peptides had an acetylated methionine residue, while 253 and 39 peptides had an acetylated serine and alanine residue, respectively. This is the most comprehensive dataset of acetylation for C. tropicalis and validates the initiation site of 329 predicted proteins. The list of acetylated peptides that were identified in this study is provided in Supplementary Table S5.
Enhancement of genomic information through proteomic data
Utilizing a conceptually derived six-frame translated genome database of C. tropicalis for MS/MS data analysis, we identified 627 peptides (Genome Search Specific Peptides—GSSPs), which do not correspond to any of the known proteins annotated in the C. tropicalis genome. GSSPs were identified using FDR of 1% as cut-off for our analysis. These GSSPs, when mapped on to the annotated genome, led to the identification of novel protein coding regions and gene structure correction events in the C. tropicalis genome. Using a comparative genomics approach, we investigated conservation of these novel protein-coding genes and revised gene models in closely related species. The succeeding sections discuss our findings through proteogenomic analysis in greater depth.
GSSPs reveal 86 novel genes in C. tropicalis genome
Our study provides experimental evidence for 86 novel protein-coding regions in the C. tropicalis genome, which were missed out in the current genome annotation. Of these 86, 63 novel genes were identified with at least 2 GSSPs. Irrespective of the number of GSSPs, the sequence of the translated ORF was analyzed for homology with related Candida species such as C. albicans and C. glabrata. Section A of Supplementary Table S6 provides the complete list of novel ORFs identified in this study, details of GSSPs, and genome coordinates.
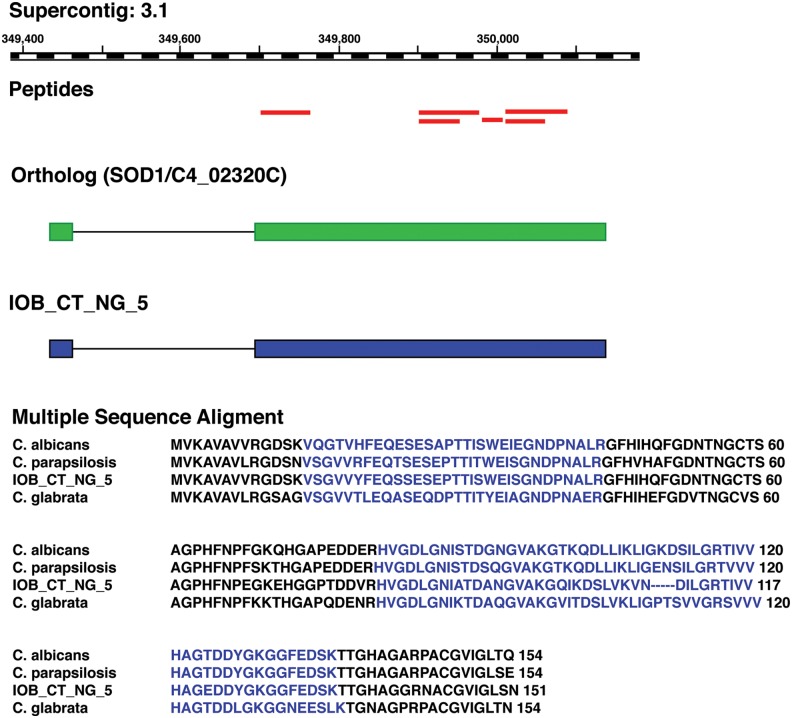
We discovered a novel protein coding gene IOB_CT_NG_5 (Fig. 3), supported by the identification of 6 GSSPs clustering in the intergenic region between the genes CTRT_00158 and CTRT_00160. ORF analysis of IOB_CT_NG_5 revealed a 151 amino acid coding potential for this genomic region with 55.6% coverage by GSSPs. Homology-based conservation analysis for this novel protein identified the corresponding ortholog in Candida albicans (SOD1/C4_02320C), Candida glabrata (SOD1/CAGL0C04741g), and Candida parapsilosis (CPAR2_500390), among others, indicating conservation of this protein-coding gene among closely related species. Further domain analysis using SMART predicted a copper-zinc superoxide dismutase domain for the translated product indicating its role in synthesis of superoxide radicals.
FIG. 3.
Illustration of a novel protein coding gene: Six GSSPs (red bars) were identified in the positive strand of Supercontig 3.1 where no gene has been annotated. Ortholog evidences from different species of Candida suggest the existence of a protein coding gene in this region (Multiple Sequence Alignment). Based on the combined evidences, we propose the gene model for IOB_CT_NG_5.
GSSPs substantiate refinement of current gene models
Predicted gene models are often affected by shortcomings of ab initio algorithms, such as improper annotation of start and stop sites (Mathe et al., 2002). Our study identifies 61 amendments of annotated gene boundaries based on novel peptides as evidence. We have GSSPs mapping upstream to 35 annotated gene models extending their N-terminus. We also observed GSSPs that mapped to downstream regions of 8 genes indicating the C-terminal extension of such gene models. Further, we propose the presence of 12 novel exons based on novel peptides identified and their orthologs in other species. Additionally, we identified 6 examples, wherein we provide evidence of merging of two genes into one. The details of all the gene boundary corrections, along with the corresponding GSSPs evidence are provided in Supplementary Table S5.
Evidence of exon extension and existence of novel exon
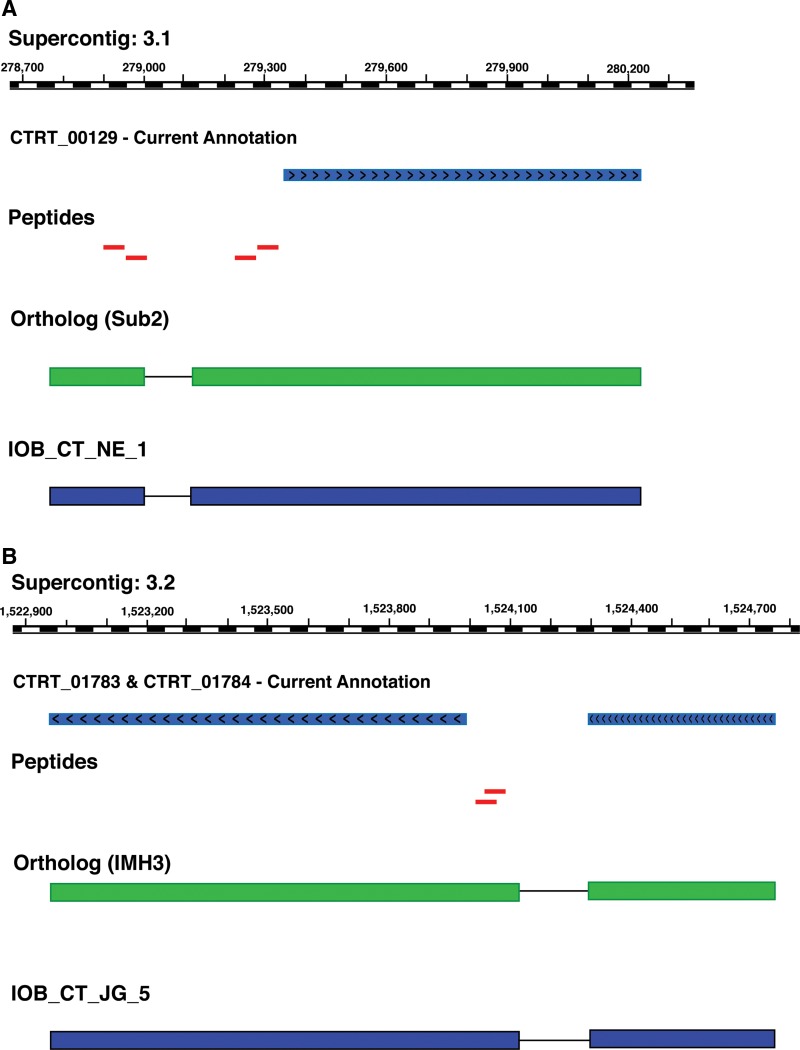
We identified four GSSPs that were found to map to the intergenic region upstream to the gene ATP-dependent RNA helicase SUB2 (CTRT_00129) in C. tropicalis (Fig. 4A). Further analysis of two of these GSSPSs (TAVFVLSTLQQLDPVPGEISTLVICHTR and ELAYQIRNEYAR) confirmed N-terminal extension of CTRT_00129 gene. Interestingly, the remaining two GSSPs (GSYVGIHATGFR and DFLLKPELLR) were observed to map ∼400 nucleotides upstream of the annotated CTRT_00129 gene, but, in a different frame. Orthologous evidence from C. albicans (Sub2), C. glabrata (CAGL0L06908g), and C. dubliniensis (CD36_40290), among others, suggest that a 73 amino acid coding region/exon of CTRT_00129 was not annotated by prediction algorithms. Based on our analysis, we propose the presence of an upstream novel exon and an exon extension event at the N-terminus and to the annotated CTRT_00129.
FIG. 4.
Proteogenomic analysis: (A) N terminal extension and novel gene. Four GSSPs (red bars) were identified upstream of the annotated gene CTRT_00129. Ortholog evidences suggest N terminal extension of the annotated gene and also the presence of an upstream exon. (B) Joining of genes. Two GSSPs (red bars) were identified in the intergenic region of the annotated genes CTRT_01783 and CTRT_01784. Ortholog evidences suggest that it is a single gene with two exons.
GSSPs bridge the gap between two predicted genes
Prediction programs may erroneously annotate a single gene as many split genes or merge two different genes as one (Mathe et al., 2002). In our study, we identified TWO GSSPs (LVGIITSR and TGGKLVGIITSR) that mapped to the intergenic region between CTRT_01783 and CTRT_01784 in the C. tropicalis genome (Fig. 4B). In the absence of a proper ORF in this region, presence of a novel protein-coding region was ruled out. An orthology based approach to investigate these GSSPs revealed that CTRT_01783 and CTRT_01784 are indeed a single gene. We found ortholog evidence from C. albicans (IMH3), C. parapsilosis (CPAR2_104580), and C. dubliniensis (CD36_20770), which proposed the merged gene to be inosine-5'-monophosphate dehydrogenase. SMART domain analysis of the merged translated product of these two genes predicts the existence of a monophosphate dehydrogenase domain.
Public accessibility of mass spectrometry-derived data
The mass spectrometry proteomics data have been deposited to the Proteome Xchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001071.
Discussion
The genome of C. tropicalis was sequenced as a part of a study that reported the genomes of eight Candida species. Genomic sequencing reiterated the fact that C. tropicalis is a strong pathogen and is very closely related to C. albicans (Butler et al., 2009). Although numerous studies are being undertaken for the better understanding of C. albicans, studies reporting molecular details of C. tropicalis are almost nil. Further, the 6258 protein coding genes predicted on the genome of C. tropicalis do not have any experimental evidence. Here, we report a high-resolution mass spectrometry based proteomic study of this pathogen. Using both gel-based and gel-free approaches, we validate ∼44% of the predicted proteome of this medically important yeast. Though a subset of proteins have been identified with a single unique peptide, they remain confident identifications as they are based on high statistical significance of 1% false discovery rate (FDR). In addition, utilizing an N-terminomics approach, we validate the translational start sites of 329 proteins.
Knowledge of proteins that a human pathogen secretes is valuable. It gives insights into host–pathogen interactions and may lead to identification of vaccine candidates and diagnostic markers. In this study, we have cultured C. tropicalis in a medium devoid of proteins and analyzed the conditioned media (or spent medium) for the presence of secretory proteins. We report the largest catalogue of secreted proteins in C. tropicalis that includes a number of molecules related to virulence.
By subjecting the mass spectrometry-derived data to proteogenomic analysis, we identified 86 novel protein coding genes and 12 novel exons. In addition, we could refine the gene models of 49 computationally predicted genes.
Conclusions
The frequency of Candida infections is on the rise globally. The situation is exacerbated by the azole resistance being developed by several fungal species. High-resolution mass spectrometry-based proteomic studies of various opportunistic pathogens will provide a platform for the identification of proteins that are involved in virulence and pathogenesis through comparative proteomic analyses. In this study, we carried out an in-depth proteomic profiling of Candida tropicalis, one of the most frequent causative agent of opportunistic infections of the Candida-non-albicans group. We have mapped ∼44% of C. tropicalis proteome using high-resolution mass spectrometry. We have also identified a large number of novel protein-coding genes and refinements to the current gene structures. Similar studies under different biological conditions such as pseudohyphae and biofilms would provide differential protein expression data which will facilitate better understanding of the molecules involved in virulence and pathogenesis.
Supplementary Material
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics. We thank the Infosys Foundation for research support to the Institute of Bioinformatics. Keshava K. Datta and Gourav Dey are recipients of Senior Research Fellowships from the University Grants Commission (UGC), Government of India. Anil K. Madugundu is a recipient of a BINC-Senior Research Fellowship from DBT. Harsha Gowda is a Wellcome Trust-DBT India Alliance Early Career Fellow.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Balakrishnan L, Nirujogi RS, Ahmad S, et al. (2014). Proteomic analysis of human osteoarthritis synovial fluid. Clin Proteomics 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai LY, Denning DW, and Warn P. (2010). Candida tropicalis in human disease. Critical reviews in microbiology 36, 282–298 [DOI] [PubMed] [Google Scholar]
- Hartmann EM, and Armengaud J. (2014). N-terminomics and proteogenomics, getting off to a good start. Proteomics 14, 2637–2646 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, and Sonnhammer EL. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305, 567–580 [DOI] [PubMed] [Google Scholar]
- Kucharova V, and Wiker HG. (2014). Proteogenomics in microbiology: Taking the right turn at the junction of genomics and proteomics. Proteomics 14, 2360–2675 [DOI] [PubMed] [Google Scholar]
- Mathe C, Sagot MF, Schiex T, and Rouze P. (2002). Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res 30, 4103–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajha Selvan LD, Kaviyil JE, Nirujogi RS, et al. (2014). Proteogenomic analysis of pathogenic yeast Cryptococcus neoformans using high resolution mass spectrometry. Clin Proteomics 11, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri M, Silva S, Henriques M, and Oliveira R. (2012). Insights into Candida tropicalis nosocomial infections and virulence factors. Eur J Clin Microbiol Infect Dis 31, 1399–1412 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI. (2014). Proteogenomics: Concepts, applications and computational strategies. Nature Methods 11, 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, and Nielsen H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Boyken L, Hollis RJ, et al. (2009). Comparison of results of fluconazole and voriconazole disk diffusion testing for Candida spp. with results from a central reference laboratory in the ARTEMIS DISK Global Antifungal Surveillance Program. Diagn Microbiol Infect Dis 65, 27–34 [DOI] [PubMed] [Google Scholar]
- Prasad TS, Harsha HC, Keerthikumar S, et al. (2012). Proteogenomic analysis of Candida glabrata using high resolution mass spectrometry. J Proteome Res 11, 247–260 [DOI] [PubMed] [Google Scholar]
- Printen JA, and Sprague GF., Jr (1994). Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y, Iwabuchi K, Yamazaki M, Tsuboi R, and Ogawa H. (2013). Secreted aspartic proteinase from Candida albicans acts as a chemoattractant for peripheral neutrophils. J Dermatol Sci 72, 191–193 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, and Mann M. (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Analyt Chem 75, 663–670 [DOI] [PubMed] [Google Scholar]
- Renuse S, Chaerkady R, and Pandey A. (2011). Proteogenomics. Proteomics 11, 620–630 [DOI] [PubMed] [Google Scholar]
- Saporito-Irwin SM, Birse CE, Sypherd PS, and Fonzi WA. (1995). PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol 15, 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgo AG, Heilmann CJ, Brul S, de Koster CG, and Klis FM. (2013). Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett 338, 10–17 [DOI] [PubMed] [Google Scholar]
- Stovall WD, and Pessin SB. (1933). Classification and pathogenicity of certain monilias. Am J Clin Pathol 3, 347–365 [Google Scholar]
- Thorvaldsdottir H, Robinson JT, and Mesirov JP. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinformatics 14, 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.