Abstract
Background
Anti-Müllerian hormone (amh) or Müllerian-inhibiting substance (mis) is a member of the transforming growth factor-β family of hormones. This gene plays a key role in vertebrate male sex-determination by inhibiting the development of the Müllerian ducts, and has been shown to be the master sex-determinant in the Patagonian pejerrey.
Results
In the lingcod, Ophiodon elongatus, both males and females share one copy of amh, however we have identified a second duplicate copy that appears solely in the male individuals. We have developed a PCR-based assay targeting the TGF-β domain of amh that provides a simple method with which to sex lingcod from a small amount of tissue. An analysis across 57 individuals gave a 100 % success rate in identifying the phenotypic sex.
Conclusions
We present a simple method to sex lingcod through non-lethal tissue sampling. A third, independent, male-specific duplication of amh in a teleost fish has been identified in the lingcod.
Electronic supplementary material
The online version of this article (doi:10.1186/s13104-016-2030-6) contains supplementary material, which is available to authorized users.
Keywords: Sex determination, amh, Anti-Müllerian hormone, Müllerian inhibiting substance, mis, Lingcod, Ophiodon elongatus
Findings
Teleosts do not share a common master sex-determination gene, although certain gene families are consistently implicated for this role. The transforming growth factor-β family contributes two confirmed Master sex determinants (MSD), Gonadal soma-derived factor [1] and Anti-müllerian hormone (amh) (reviewed in [2]). Male-specific duplicated copies of amh (also known as Müllerian-inhibiting substance) have been implicated in controlling sex-determination in the Patagonian pejerrey Odontesthes hatcheri [3] and amh is essential for male sex-determination in the Nile tilapia Oreochromis niloticus [4, 5]. In this work, we show that a male-specific duplicated copy of amh is also strongly linked to sex in the lingcod (Ophiodon elongatus).
The lingcod is a gonochorisitic, bottom-dwelling member of the greenling family (Hexagrammidae) that is found in nearshore marine areas along the west coast of North America [6]. Lingcod form an important component of the commercial, recreational and aboriginal (CRA) groundfish fisheries, with commercial lingcod fisheries forming the ninth most valuable commercial fishery in Pacific Canada in 2013 [7]. This fishery is considered to be sustainably managed yet certain stocks have still become depressed, primarily along the inside passage of Vancouver Island [8]. Given the uncertain nature of the inside stocks and the suggestion of asymmetric migration [9], genetic tools are an important resource that can aid in more comprehensive stock assessment and management.
As mature males have a distinct papilla just forward of the anus it is possible to assign sex of mature lingcod based on size and the presence or absence of this external feature. However, to determine the sex of immature individuals and to confirm the sex of mature lingcod requires euthanization and examination of the gonads. The development of this non-lethal method for the assignment of sex is a valuable tool to support ecological and stock assessment studies of lingcod where examination of the gonads for sex is not possible or desirable.
Samples were collected in 2013 and 2014 by the Government of Canada as part of stock assessment activities; sampling was exempt from requiring an animal use protocol under section 4.1.2.2 of the Canadian Council on Animal Care (CCAC), as determined by Fisheries and Oceans Canada. Sample collection data is summarized in Additional file 1. Genomic DNA was extracted from ethanol-preserved fin clips using DNeasy kits (Qiagen). Samples were sexed by dissection and examination of the gonads by qualified groundfish biologists and technicians.
Initial PCR amplifications were conducted and sequenced using primers designed to conserved portions of sablefish (GenBank:GAJJ01025678 [10] and stickleback (Ensembl:ENSGACT00000016731; ensembl.org) amh transcripts (primers and methods in Additional file 2). Male amh sequences appeared to be highly polymorphic (Fig. 1a), while female amh sequences appeared to be highly conserved (Fig. 1b), which suggested that these primers may amplify two loci in males. The sequences were extended from the original PCRs using genome walking methods, with primers and protocol in Additional file 2 [11]. These primers were used to isolate male-specific (GenBank:KP686073) and shared (GenBank:KP686074) partial amh gene sequences (Additional file 3). Of the seven exons expected based on sablefish and threespine stickleback, the sequences have been generated from the middle of exon 3 through exon 7 and into the 3′-untraslanted region (UTR). Genome walking consistently failed in the intron between exons 2 and 3 due to repetitive sequence, leaving the partial sequences presented.
Fig. 1.
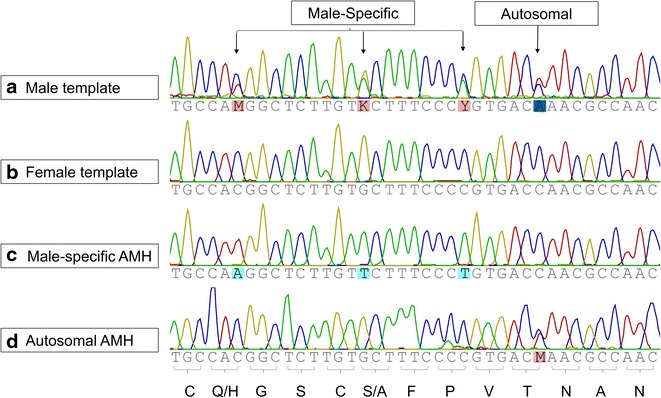
An aligned portion of the TGF-β domain. Primers that amplify both copies of amh were used to amplify a a male template and b a female template. c Male-specific amh primers and d Autosomal amh primers were used on a male template. Male-specific and autosomal variable sites are noted with arrows and the translated consensus is shown along the bottom. Image produced using Geneious v8.0.4 (http://www.geneious.com [12]), sequences are from 1873–1912 bp in KP686073 and 1871–1910 bp in KP686074
From these extended sequences, primers were then designed to target either the male-specific (Fig. 1c) or the shared copy of amh (Fig. 1d) from the male individual used in Fig. 1a. Based on the detection of two alleles (i.e., two copies) at the autosomal polymorphic site (Fig. 1d), it is suggested that the male-specific copy is not a third allele, but a duplicated copy that appears solely on an emerging Y-chromosome. Further evidence is given by the significantly different 3′UTRs in the sequences generated by genome walking (see alignment in Additional file 3). Analysis of the TGF-β domain shows that while coding changes exist (9/93 sites), the seven conserved cysteines (Prosite PDOC00223) required for inter and intra-chain disulfide bonds are conserved in both sequences (Additional file 4). Unlike in tilapia [4], it appears that the TGF-β domain of the male-specific duplicate has remained functional in the lingcod.
We have developed a PCR assay to sex individual lingcod on an agarose gel. Oelo_Sex_For (5′-TYTAGAACAGGGGAGAGCCA-3′) and Oelo_Sex_Rev (5′-TTAGTTTTAGCGGCATCCAC-3′) amplifies both amh copies as a 420 bp band, while Oelo_Male_Rev (5′-GACCCTCGCTGGGCTGTTTGA-3′) with Oelo_Sex_For amplifies only the male version, as a 129 bp band. The 10 μl reactions containing 1X Hot Start PCR Buffer, 0.25 μM of each of the two reverse primers (IDT), 0.5 μM of the forward primer (IDT), 2.0 mM MgCl2, 0.2 mM dNTPs, and 0.5U Maxima Hot Start Taq (Thermo) were incubated for 4 min at 95 °C, then 35 cycles of 30 s at 95 °C, 30 s at 58 °C, 30 s at 72 °C, and a final extension of 10 min at 72 °C. Amplified PCR fragments were then run on a 2.0 % agarose/1XTAE gel stained with EtBr at 100 V for 45 min along with 2.5 μl O’GeneRuler™ 1 kb Plus DNA Ladder (Thermo). These reactions successfully amplified a single band in females and two bands in males (Fig. 2). We positively identified phenotypic sex in 57 individuals (29 males, 28 females) using this PCR and gel based assay. A further seven unsexed museum specimens (Additional file 1) were screened and the male-specific sequence was found in samples from both Washington State and Alaska, confirming the distribution of this sequence across a large geographic range.
Fig. 2.
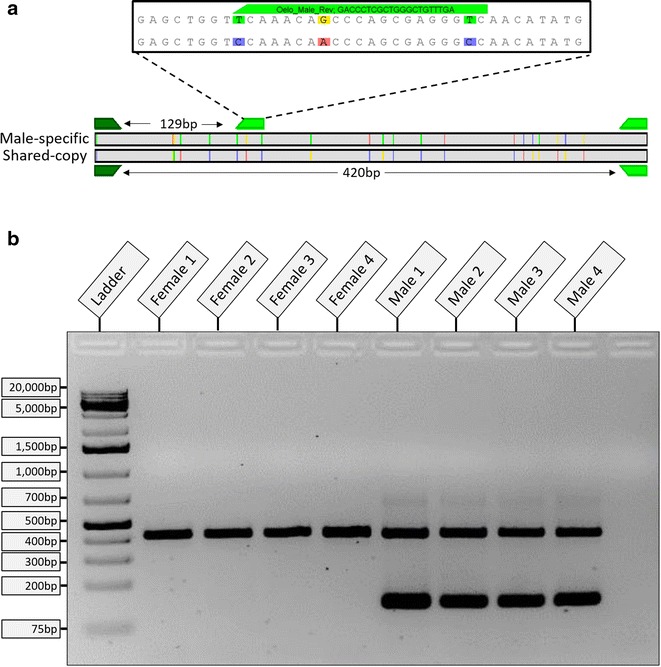
Testing the lingcod PCR assay. The PCR assay strategy is shown in a. The external primers (green polygons) bind both versions while the internal primer binds only in the male due to three internal mismatches. Mismatches are coloured lines, image produced in Geneious v8.0.4 [12]. In b, amplification of four male and four female lingcod templates using the Oelo-Sex three primer PCR assay. Presence of the 129 bp band denotes the presence of the male-specific copy of amh, while the 420 bp band is the shared autosomal copy. Ladder used is 1 kb O’generuler plus (Thermo), 2.0 % agarose gel stained with ethidium bromide
Here, we provide a simple, genetic method to sex lingcod through non-lethal tissue sampling. Although the role of amh as a MSD in lingcod remains to be confirmed, our work shows that a third, independent, male-specific duplication of amh in a teleost fish has occurred, suggesting that multiple evolutions of the amh gene as a novel MSD may have occurred in multiple teleost lineages.
Availability of supporting data
The data supporting the results of this article are included within the article and its additional files, or uploaded to NCBI, accessions GenBank: KP686073-KP686074.
Authors’ contributions
EBR and BFK conceived the experiment, EBR and CVL performed the experimental work and analysis, SCJ collected and determined the gender of the lingcod, EBR and SCJ drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Malcolm Wyeth, Kathryn Temple and the other members of the Pacific Biological Station Groundfish Survey team for their work in sampling the tissues required for analysis. We would also like to thank the Burke Museum at the University of Washington and in particular Katherine Maslenikov for providing additional tissue samples. This work was funded by the University of Victoria and the Natural Sciences and Engineering Research Council.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- Amh
Anti-Müllerian hormone
- Mis
Müllerian inhibiting substance
- MSD
master-sex determinant
- TGF-β
transforming growth factor beta
- UTR
untranslated region
Additional files
10.1186/s13104-016-2030-6 Sampling information. Additional information on the collection of lingcod tissues used in the study.
10.1186/s13104-016-2030-6 Primers and protocol. Primers and protocol used to amplify lingcod amh KP686073 and KP686074.
10.1186/s13104-016-2030-6 Partial amh gene sequences. Sequences of male-specific (KP686073) and shared (KP686074) partial amh gene sequences and corresponding alignment (Geneious v8.1.7 default alignment settings).
10.1186/s13104-016-2030-6 TGF-β domain. Translation of TGF-β domain and identification of conserved cysteines between amh and amhy.
Contributor Information
Eric B. Rondeau, Email: erondeau@uvic.ca
Cassandra V. Laurie, Email: cvl@uvic.ca
Stewart C. Johnson, Email: stewart.johnson@dfo-mpo.gc.ca
Ben F. Koop, Phone: 1-250-472-4071, Email: bkoop@uvic.ca
References
- 1.Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, Hamaguchi S, Sakaizumi M. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 2012;191(1):163–170. doi: 10.1534/genetics.111.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heule C, Salzburger W, Böhne A. Genetics of sexual development: an evolutionary playground for fish. Genetics. 2014;196(3):579–591. doi: 10.1534/genetics.114.161158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, Sakamoto T, Fernandino JI, Somoza GM, Yokota M, Strüssmann CA. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA. 2012;109(8):2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eshel O, Shirak A, Dor L, Band M, Zak T, Markovich-Gordon M, Chalifa-Caspi V, Feldmesser E, Weller J, Seroussi E, Hulata G, Ron M. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus) BMC Genom. 2014;15(1):774. doi: 10.1186/1471-2164-15-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K, Jiang D, Zhou L, Sun L, Tao W, Nagahama Y, Kocher TD, Wang D. A tandem duplicate of Anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015;11(11):e1005678. doi: 10.1371/journal.pgen.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cass AJ, Beamish RJ, McFarlane GA. Lingcod (Ophiodon elongatus) Canadian Spec Publ Fisheries Aquat Sci. 1990;109:1–40. [Google Scholar]
- 7.Preliminary Summary Commercial Statistics. 2013. [http://www.pac.dfo-mpo.gc.ca/stats/comm/summ-somm/annsumm-sommann/2013/ANNUAL13_USER_three_party_groups-eng.htm].
- 8.DFO. Strait of Georgia lingcod (Ophiodon elongatus) assessment and advice for fishery management. DFO Can Sci Advis Sec Sci Advis Rep. 2005. 2005/042.
- 9.Marko P, Rogers-Bennett L, Dennis A. MtDNA population structure and gene flow in lingcod (Ophiodon elongatus): limited connectivity despite long-lived pelagic larvae. Mar Biol. 2007;150(6):1301–1311. doi: 10.1007/s00227-006-0395-0. [DOI] [Google Scholar]
- 10.Rondeau EB, Messmer AM, Sanderson DS, Jantzen SG, von Schalburg KR, Minkley DR, Leong JS, Macdonald GM, Davidsen AE, Parker WA, Mazzola RSA, Campbell B, Koop BF. Genomics of Sablefish (Anoplopoma fimbria): expressed genes, mitochondrial phylogeny, linkage map and identification of genetic sex markers. BMC Genom. 2013;14:452. doi: 10.1186/1471-2164-14-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23(6):1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]


