Abstract
Brief expression of pluripotency-associated factors such as Oct4, Klf4, Sox2 and c-Myc (OKSM), in combination with differentiation-inducing signals, was reported to trigger transdifferentiation of fibroblasts into other cell types. Here we show that OKSM expression in mouse fibroblasts gives rise to both induced pluripotent stem cells (iPSCs) and induced neural stem cells (iNSCs) under conditions previously shown to induce only iNSCs. Fibroblast-derived iNSC colonies silenced retroviral transgenes and reactivated silenced X chromosomes, both hallmarks of pluripotent stem cells. Moreover, lineage tracing with an Oct4-CreER labeling system demonstrated that virtually all iNSC colonies originated from cells transiently expressing Oct4, whereas ablation of Oct4+ cells prevented iNSC formation. Lastly, an alternative transdifferentiation cocktail that lacks Oct4 and was reportedly unable to support induced pluripotency yielded iPSCs and iNSCs carrying the Oct4-CreER-derived lineage label. Together, these data suggest that iNSC generation from fibroblasts using OKSM and other pluripotency-related reprogramming factors requires passage through a transient iPSC state.
Transdifferentiation is defined as the experimentally induced direct conversion of one cell lineage into another. It is typically achieved by ectopic expression of small sets of cell type–specific transcription factors and has been used to convert fibroblasts into a variety of cell types, including myoblasts, cardiomyocytes, hepatocytes and neural cells1. Recently, a more general lineage conversion approach has been developed that uses transient expression of iPSC-inducing transcription factors. For example, transient expression of OKSM in fibroblasts was reported to give rise to iNSCs, pancreatic progenitors, cardiomyocytes, hepatocytes, and endothelial cells when pluripotency-inducing signaling (LIF/Stat3) is removed and cells are cultured in the presence of lineage-specifying extracellular cues2–8. These studies suggested that transient expression of pluripotency-associated transcription factors generates a developmentally plastic state that is amenable to transdifferentiation into other cell types by exposure of cells to different environmental signals9. Several lines of evidence suggested that this process does not involve formation of a transient iPSC state and subsequent differentiation. For example, mouse embryonic fibroblasts (MEFs) subjected to OKSM transdifferentiation protocols failed to show activation of transgenic pluripotency reporters such as Nanog-GFP or Oct4-GFP 2,4,7. Moreover, the kinetics of transdifferentiation was shorter than that of induced pluripotency when using certain culture conditions, suggesting that iPSCs could not have formed2,4,7. Lastly, an alternative transdifferentiation cocktail, Brn4, Klf4, Sox2 and c-Myc (BKSM), also generated iNSCs without apparent production of iPSCs3.
The use of OKSM in both induced pluripotency and transdifferentiation protocols raises the question of how these transcription factors induce such different cell fates. We therefore sought to compare and contrast reprogramming into iPSCs by OKSM and transdifferentiation to iNSCs by either OKSM or BKSM.
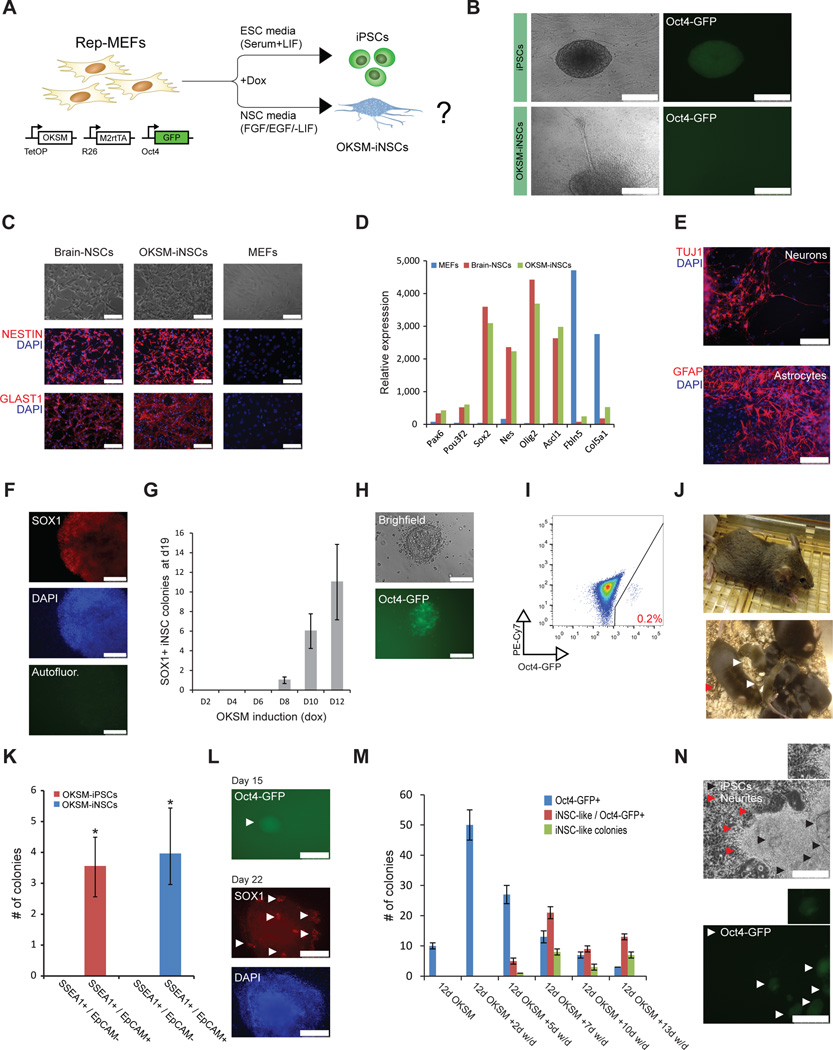
Reprogrammable MEFs (rep-MEFs), which carry the tetOP-OKSM allele in the Col1a1 locus, the M2rtTA transgene in the Rosa26 locus and an EGFP reporter in the endogenous Oct4 locus (Oct4-GFP)10, exposed to doxycycline (dox) gave rise to iPSCs after 1–2 weeks in conventional embryonic stem cell (ESC) medium (serum and LIF)(Fig. 1A, B). In contrast, treatment of rep-MEFs with dox in medium containing the NSC cytokines FGF and EGF but lacking serum/LIF (NSC medium) yielded clusters of cells that morphologically and phenotypically resembled NSC colonies (Fig. 1A, B). For example, cellular projections emanating from iNSC-like colonies suggested formation of neurites, which are typically seen in brain-derived NSC cultures (Fig. 1B). We picked and expanded individual colonies in NSC medium, generating stable clonal cell lines. Immunofluorescence staining and microarray gene expression analysis of these cell lines revealed upregulation of key NSC markers, such as Sox1, Sox2, Nestin and Glast, at levels similar to those of brain-derived NSCs, whereas MEF-related genes, including Col5A1 and Fbln5, were markedly downregulated (Fig. 1C, D and Supplementary Fig. 1A). Unsupervised clustering, correlation and functional annotation analyses of global mRNA expression data further showed that these iNSC-like cells are highly similar to brain-derived NSCs but different from MEFs or iPSCs/ESCs (Supplementary Fig. 1B–D). Cultures expanded from single iNSC-like colonies could differentiate into Tuj1+ neurons and GFAP+ astrocytes, demonstrating multilineage differentiation potential (Fig. 1E). Together, these results show that pulsed OKSM expression with our reprogrammable system in the presence of NSC inductive cytokines produces authentic iNSCs (OKSM-iNSCs) that exhibit multipotency, self-renewal potential and a high similarity to brain-derived NSCs.
Figure 1. Generation of iPSCs and iNSCs under neural culture conditions.
(A) Experimental design to generate iPSCs and iNSCs in different culture conditions using reprogrammable MEFs (rep-MEFs) harboring the Col1a1-tetOP-OKSM and Rosa26-M2rtTA alleles and an Oct4-GFP reporter. (B) Representative image of Oct4-GFP+ iPSC colony generated in ESC medium (serum + LIF) (upper panel) and Oct4-GFP iNSC-like colony generated in NSC medium (−LIF/+FGF, EGF) showing typical spheroid morphology with neurite-like structures projecting from the center (bottom panel). Scale bar is 250µM. (C) Representative immunofluorescence images showing staining for indicated NSC markers in brain-derived NSCs and OKSM-iNSCs. MEFs served as a negative control. Scale bar is 100µM. (D) Expression of NSC or MEF-associated markers in the indicated cell lines based on microarray gene expression analysis. (E) Differentiation potential of OKSM-iNSCs into Tuj1+ neurons and GFAP+ astrocytes. Scale bar is 100µM. (F) Representative immunofluorescence image of a Sox1+ iNSC colony obtained after 10 days of OKSM expression, followed by 9 days of dox-independent growth. Scale bar is 100µM. Autofluor., autofluorescence control. (G) Graph showing the minimal number of days required to generate stable Sox1+ iNSC colonies using conventional NSC medium. Doxycycline was applied for indicated lengths of time before scoring for iNSC colonies at day 19 to capture stable, transgene-independent colonies. For each replicate, 3x104 cells were used (n=3 independent replicates for each time point; error bars, standard error of the mean (s.e.m.) for 3 independent experiments). (H) Detection of a rare Oct4-GFP+ iPSC-like colony under transdifferentiation conditions in NSC medium. Scale bar is 100µM. (I) Flow cytometric analysis for Oct4-GFP expression in bulk rep-MEF cultures subjected to the NSC transdifferentiation protocol. The PE-Cy7 channel was used to detect autofluorescence. (J) Contribution of NSC medium–derived iPSCs to chimeras as indicated by agouti coat color (upper image). Germline offspring (white arrowheads) obtained from a male chimera (red arrow) (lower image). (K) Potential of sorted SSEA1+/EpCAM+ and SSEA1+/EpCAM intermediates after 6 days of OKSM expression to produce iPSCs in ESC medium or iNSCs in conventional NSC medium, respectively. For each replicate, 10x105 cells were plated (n=3 independent replicates; error bars, s.e.m for 3 independent experiments, *p<0.05). (L) Representative images of an Oct4-GFP+ colony (top image) and a Sox1+ iNSC colony (bottom image) obtained from sorted SSEA1+/EpCAM+ intermediates (day 6) in NSC medium. White arrowheads indicate an Oct4-GFP colony (top) or clusters of Sox1+ expressing cells (bottom). Scale bar is 500µM (Oct4-GFP) or 250µM (Sox1). (M) Quantification of Oct4-GFP+ colonies, Oct4-GFP+/iNSC hybrid colonies and Oct4-GFP/iNSC colonies at the indicated time points after dox withdrawal (w/d). For each replicate, 3x104 cells were used (n=3 independent replicates for each time point; error bars, s.e.m. for 3 independent experiments). (N) Representative image of iPSC/iNSC hybrid colonies detected during the transdifferentiation protocol. Note the dome-shaped iPSC-like colonies in the center and emanating neurites (indicated by black and red arrowheads, respectively). Bottom images shows patches of Oct4-GFP reporter expression (white arrowheads) within the same colony as shown above. Scale bar is 250µM. Insets show magnification of neurites (top) and a representative Oct4-GFP colony (bottom).
We previously showed that the formation of stable iPSCs requires a minimum of 8–10 days of OKSM expression under conventional culture conditions11. To determine the minimal time needed to generate iNSCs, we induced rep-MEFs with dox in NSC medium for different lengths of time, followed by dox withdrawal before counting iNSC colonies at day 19. OKSM expression was required for at least 8 days to detect iNSC colonies, as determined by staining for Sox1 (Fig. 1F, G). The apparent similarity in temporal factor requirement between iNSC and iPSC generation prompted us to ask whether iPSC colonies could form in NSC medium, a condition that is incompatible with long-term culture of iPSCs. Notably, forced expression of OKSM in rep-MEFs consistently gave rise to rare Oct4-GFP+ iPSC-like cells (0.2% of total cell population) in this differentiation-stimulating condition (Fig. 1H, I). Unlike brain-derived NSCs or OKSM-iNSCs, these colonies co-expressed Oct4 and Nanog, ruling out the possibility that they were primitive neurectodermal cells that continued to express Oct4 upon exit from pluripotency12 (Supplementary Figs. 1E, 2A, B). The iPSC-like colonies could be stably propagated in ESC medium in the absence of exogenous transgene expression (i.e., after dox withdrawal) and showed expression of the pluripotency-associated markers alkaline phosphatase, PECAM1, EpCAM and Nanog at levels that were comparable to ESCs, indicating self-renewal capacity and faithful molecular reprogramming to pluripotency (Supplementary Fig. 2C–E). Notably, the expanded iPSC-like clones contributed to coat-color chimeric mice and to their offspring upon injection into blastocyst embryos—a rigorous measure of pluripotency (Fig. 1J). We conclude that forced OKSM expression overrides the requirement for serum and LIF/Stat3 signaling and counteracts differentiation-inducing signals (EGF, FGF), generating authentic, germline-competent iPSCs under adverse growth conditions.
These data raise the possibility that certain previously published transdifferentiation protocols using OKSM overexpression might have in fact generated transient iPSC cells that then differentiated into other cell lineages in response to extracellular cues. To test this hypothesis, we first examined the order of events in transdifferentiation by tracking cell surface markers by flow cytometry. During reprogramming toward iPSCs, MEFs first downregulate the fibroblast-associated marker Thy1 and then upregulate the pluripotency-associated marker SSEA1 at day 3, followed by EpCAM expression by day 611,13. We observed a similar downregulation of Thy1 and an upregulation of EpCAM in a fraction of cells (∼5%) at day 6 of OKSM expression under iNSC transdifferentiation conditions, although established iNSCs and brain-derived NSCs do not express EpCAM (Supplementary Fig. 2F, G). Analysis of SSEA1 expression was not informative because it is also expressed by NSCs14. These results show that OKSM-induced lineage conversion generates similar phenotypic intermediates as OKSM-induced pluripotency, despite very different extracellular stimuli.
Sorted EpCAM+ intermediates obtained after 6 days of OKSM expression in ESC medium give rise to rare dox-independent iPSC colonies, whereas EpCAM− intermediates do not13. To prospectively determine whether these subpopulations can give rise to iNSCs, we sorted day 6 SSEA1−/EpCAM−, SSEA1+/EpCAM− and SSEA1+/EpCAM+ intermediates generated in ESC medium and cultured them in either ESC or NSC medium without dox (Fig. 1K, L). Only SSEA1+/EpCAM+ intermediates could produce transgene-independent iPSCs or iNSCs (Fig. 1K, L and data not shown). These findings show that iPSCs and iNSCs originate from the same rare subpopulation of SSEA1+/EpCAM+ cells that emerge shortly after OKSM expression.
Considering that EpCAM+ intermediates typically give rise to Oct4-GFP+ iPSCs under ESC culture conditions13 and that rare Oct4-GFP+ cells emerge under NSC transdifferentiation conditions (Fig. 1H, I), we next determined the fraction of nascent iNSC colonies that expressed the pluripotency gene Oct4 in situ over time using the same Oct4-GFP knock-in reporter. The majority of colonies forming at early time-points of OKSM expression (days 10–12) in NSC medium indeed contained small clusters of Oct4-GFP+ cells, whereas colonies at intermediate time points (days 17–22) consisted of Oct4-GFP+ clusters, iNSC-like clusters or a combination of both (Fig. 1M, N). Oct4-GFP expression was entirely silenced upon picking and expansion of these iNSC-like colonies in NSC medium without dox (data not shown).
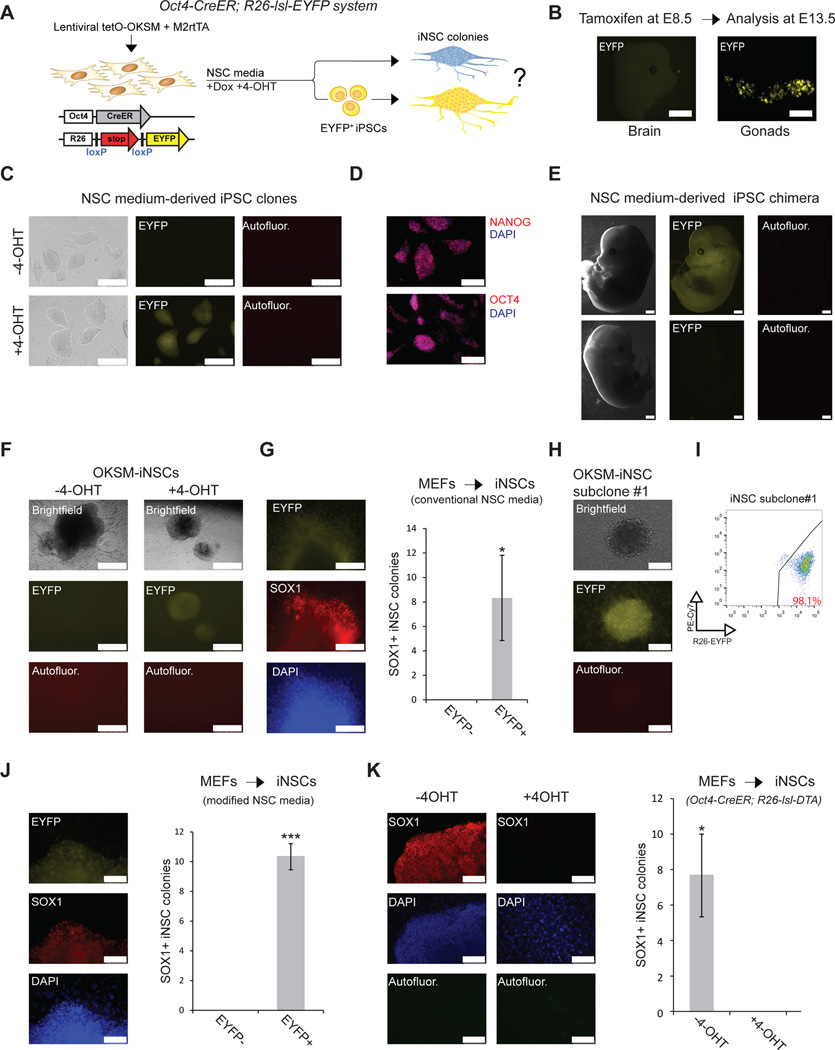
To accurately determine how many colonies expressed Oct4 before becoming iNSCs, we used cells carrying a 4-hydroxytamoxifen (4-OHT)-inducible Oct4-CreER allele in combination with a Rosa26-loxSTOPlox-EYFP (R26-lsl-EYFP) reporter cassette15 (Fig. 2A). We first determined the specificity and sensitivity of this lineage tracing system for labeling pluripotent cells. To demonstrate that primitive Oct4+ neurectodermal cells12 are not labeled with this system in vivo, we treated pregnant females harboring Oct4-CreER; R26-lsl-EYFP bitransgenic embryos with tamoxifen at E8.5 and examined resultant fetuses at E13.5. We did not detect EYFP expression in the brains of five E13.5 fetuses or in derivative NSC cultures (Fig. 2B and Supplementary Fig. 3A, B). However, we noticed strong EYFP signal in primordial germ cells in the genital ridge, which are known to express Oct4 at E8.5 (Fig. 2B). To ensure that culture of brain-derived NSCs does not spuriously activate the Oct4-CreER; R26-lsl-EYFP reporter, we derived NSCs from the brains of E13.5 fetuses with this genotype and exposed cells to 4-OHT. These NSC lines remained EYFP− regardless of whether 4-OHT was added to the culture (Supplementary Fig. 3C). Next, to confirm that short-term expression of OKSM does not directly activate the endogenous Oct4 locus, we transduced Oct4-CreER; R26-lsl-EYFP MEFs with lentiviral vectors co-expressing tetOP-OKSM and M2-rtTA for 2 or 4 days before measuring EYFP expression by flow cytometry. No notable EYFP reporter signal was detected at these early time points (Supplementary Fig. 4A). We also failed to detect Oct4-GFP reporter expression after 2 and 4 days of OKSM expression (Supplementary Fig. 4B). In contrast, long-term induction of OKSM in Oct4-CreER; R26-lsl-EYFP MEFs in ESC or NSC media generated EYFP+ iPSCs in a 4-OHT-dependent manner (Fig. 2C–E, Supplementary Fig. 5A and data not shown). EYFP+ iPSCs derived under NSC conditions expressed Nanog and Oct4, and they contributed to midgestation chimeras upon injection into blastocysts, demonstrating pluripotency (Fig. 2C–E and Supplementary Fig. 5B, C). Based on these controls, we feel confident that this lineage tracing system and our culture conditions have high specificity and sensitivity, allowing us to determine whether nascent iNSCs go through a transient Oct4+ state. Similar to EYFP-labeled iPSCs produced in NSC medium (Fig. 2C), all recovered Sox1+ iNSC colonies derived from OKSM-expressing Oct4-CreER; R26-lsl-EYFP MEFs were EYFP+ in the presence of 4-OHT, whereas no EYFP+ iNSC colonies were detected in the absence of 4-OHT (Fig. 2F, G). EYFP+ iNSC clusters could be single-cell subcloned and propagated for over ten passages while maintaining NSC marker expression, indicating self-renewal capacity (Fig. 2H, I and Supplementary Fig. 6A–C). These results show that all examined iNSCs generated under NSC conditions passed through a transient Oct4+ state.
Figure 2. Oct4 lineage tracing and ablation during iNSC induction.
(A) Schematic showing lineage tracing approach to test whether iNSCs pass through an Oct4+ state. (B) Head and gonads from an Oct4-CreER; R26-lsl-EYFP embryo that was induced in utero with 4-OHT (E8.5) and recovered at E13.5. Note that the brain is EYFP negative, while the gonads are EYFP positive, indicating specificity of the lineage-tracing allele in-vivo. (C) Generation of EYFP+ iPSCs from Oct4-CreER; R26-lsl-EYFP MEFs generated in NSC medium. Note that iPSCs are EYFP+ only when 4-OHT is administered to the reprogramming medium. Scale bar is 250µM. Autofluor., autofluorescence control. (D) Representative immunofluorescence images for the indicated pluripotency markers in NSC medium–derived EYFP+ iPSCs. Scale bar is 100µM. (E) EYFP+ iPSCs, derived in NSC medium, contribute to an E13.5 midgestation chimera (upper images); non-chimeric littermate lacks EYFP signal (bottom images). (F) Images of EYFP+ OKSM-iNSC colonies with typical neurites protruding from the center. OKSM-iNSC colonies remain EYFP when 4-OHT is not applied. Scale bar is 500µM. (G) Quantification of Sox1+ EYFP+ and Sox1+ EYFP iNSC colonies generated in conventional NSC medium after 8–12 days of OKSM induction, followed by 14–18 days of dox-independent growth. For each replicate, 3x104 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, *p<0.05). A representative EYFP+ iNSC colony with patches of Sox1+ expressing cells is shown to the left. Scale bar is 100µM. (H) Immunofluorescence for EYFP of OKSM-iNSC subclone #1 at passage 20, demonstrating continuous and uniform EYFP expression. Scale bar is 100µM. (I) Flow cytometric analysis for EYFP expression of the same OKSM-iNSC subclone #1. The PE-Cy7 channel was used to control for autofluorescence. (J) Quantification of Sox1+ EYFP+ and Sox1+ EYFP iNSC colonies, generated in modified NSC medium after 4–5 days of OKSM induction, followed by 2–4 weeks of dox-independent growth. For each replicate, 5x105 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, ***p<0.0005). A representative EYFP+/Sox1+ iNSC colony is shown to the left. Scale bar is 100µM. (K) Quantification of Sox1+ iNSC colonies obtained from Oct4-CreER; R26-lsl-DTA (Diphtheria Toxin fragment A) MEFs after 5 days of OKSM expression, followed by 3 weeks of dox-independent growth in modified NSC medium in the absence or presence of 4-OHT. For each replicate, 5x105 cells were used (n=3 independent replicates; error bars, s.e.m. for 3 independent experiments, *p<0.05). Scale bar is 100µM. Autofluor., autofluorescence. Representative images are shown to the left.
We next applied the Oct4-CreER lineage tracing system to an alternative transdifferentiation protocol7 that gave rise to iNSCs but not iPSCs7 after only 4–5 days of Oct4 expression, coupled with constitutive expression of Sox2, Klf4 and c-Myc7. We were able to derive Sox1+ iNSC lines after only 4–5 days of OKSM expression, followed by 14–21 days of OKSM-independent growth, similar to a previous report7 (Fig. 2J and Supplementary Fig. 7A). All examined Sox1+ iNSC colonies were also EYFP+ (Fig. 2J and Supplementary Fig. 7A). This alternative transdifferentiation protocol uses a modified NSC medium containing KnockOut Serum Replacement (KOSR) and low amounts of serum, which are not included in conventional NSC medium7. We surmise that the presence of ascorbic acid in KOSR accelerates iPSC formation, as was shown previously16. thus explaining the faster appearance of iPSCs and derivative NSCs.
We used the modified transdifferentiation protocol in most subsequent experiments because of the higher recovery rate of iNSCs.
To rule out the possibility that our visual quantification of EYFP-marked cells missed single iNSCs or small iNSC colonies that underwent direct transdifferentiation, we repeated the experiment with MEFs harboring the Oct4-CreER allele in combination with a R26-lsl-DTA (Diphtheria Toxin fragment A) allele (Supplementary Fig. 7B). Expression of the DTA gene in 4-OHT-induced cells undergoing transdifferentiation or reprogramming is expected to ablate nearly all Oct4-expressing cells (Supplementary Fig. 7B–D). Using this system, we could not detect any Sox1+ iNSC colonies or Sox1+ single cells in 4-OHT-treated cultures compared to untreated cultures (Fig. 2K), confirming that virtually all iNSCs pass through an Oct4+ state.
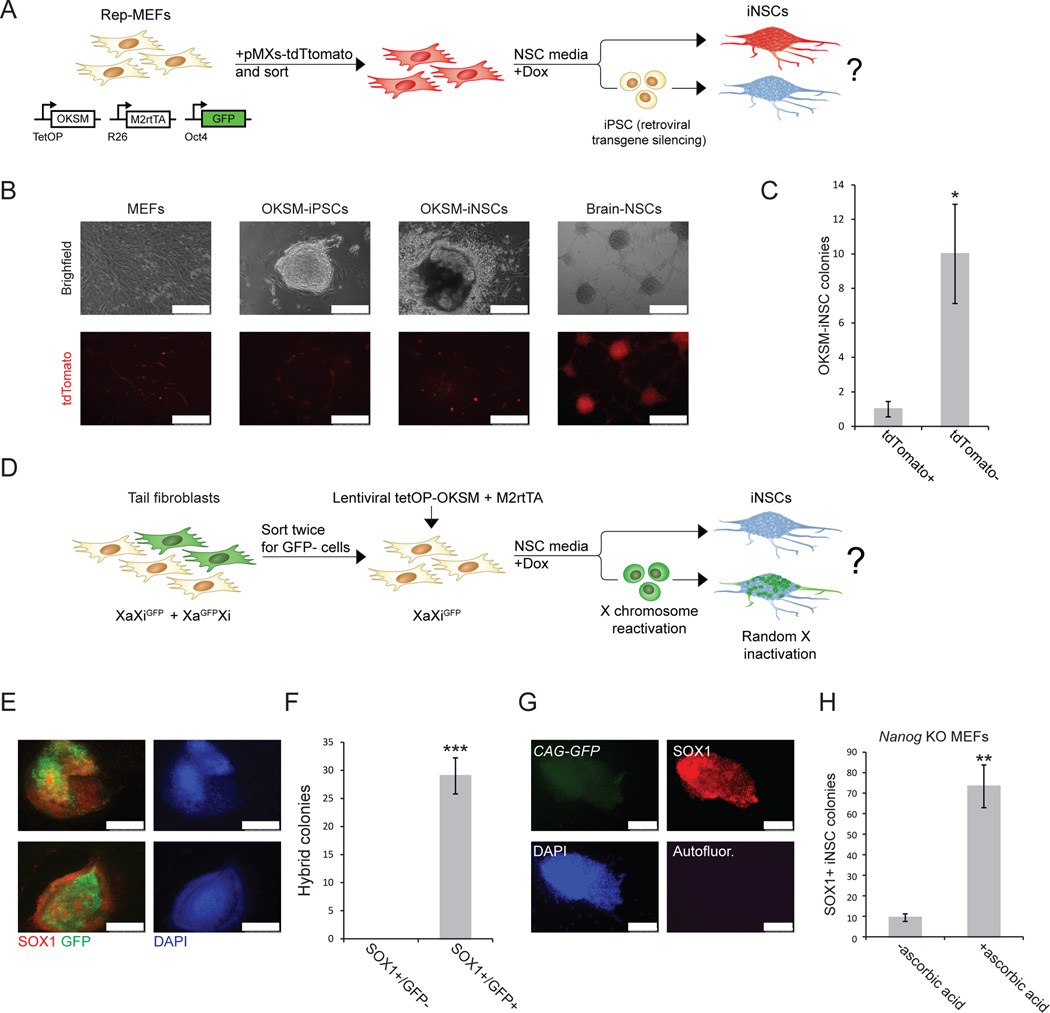
To complement our Oct4 lineage labeling and ablation experiments with independent assays of pluripotency, we first examined retroviral silencing in iNSC cultures. Unlike most somatic cells, including NSCs, iPSCs efficiently silence retroviral transgenes due to the activity of de novo DNA methyltransferases11. We infected rep-MEFs with a retroviral vector constitutively expressing tdTomato (Fig. 3A), purified tdTomato+ Rep-MEFs by FACS, and exposed them to dox and NSC medium (Fig. 3A, B). Approximately 90% of iNSC colonies were tdTomato−, and ∼10% were tdTomato+ (Fig. 3C). We believe that the latter colonies represent nascent hybrid iPSC–iNSC colonies that had not yet undergone complete retroviral silencing (see also Fig. 1M). Notably, culture of sorted tdTomato+ MEFs in NSC medium without dox did not elicit silencing of retroviral transgenes (Supplementary Fig. 8). We previously showed that reprogramming of female fibroblasts to iPSCs reactivates the silenced X chromosome, and subsequent differentiation of the iPSCs results in random X inactivation17. To determine whether X chromosome reactivation occurs during iNSC induction, we sorted GFP− cells from a mixed population of GFP+ and GFP− female tail tip fibroblasts (TTFs) carrying an X-linked GFP reporter in a heterozygous configuration (i.e., mix of XaGFPXi and XiGFPXa cells). Thus, all sorted GFP− TTFs carried the GFP transgene on the inactive X chromosome (XiGFPXa) (Fig. 3D). Infection of these TTFs with lentiviruses expressing tetOP-OKSM (STEMCCA)18 and M2rtTA gave rise to iNSC colonies after 4–5 days of dox induction, followed by 2–4 weeks of dox-independent growth in the modified NSC medium. All Sox1+ iNSC colonies reactivated the silent X chromosome as judged by GFP expression in a fraction of cells (Fig. 3E, F). GFP expression declined in some clones with continuous passaging of these iNSCs, possibly owing to silencing of the transgene or loss of the transgenic X chromosome (Supplementary Fig. 9A). However, we did not detect reactivation of GFP in sorted GFP− TTFs after 14 days of culture in NSC medium in the absence of OKSM expression, indicating that the silenced X chromosome is not spuriously activated by culture in NSC medium (Supplementary Fig. 9B). Because the percentage of GFP+ cells varied among established iNSC clones, we confirmed reactivation of the silenced X chromosome in 7 clonally derived iNSC cultures by quantitative RT-PCR for an X-linked marker that distinguishes the maternal (mus musculus) and paternal (mus castaneous) X chromosome, respectively (Supplementary Fig. 9C). These data reinforce the interpretation that iNSCs transition through a pluripotent intermediate state with concomitant X reactivation before undergoing differentiation.
Figure 3. Silencing of retroviral transgenes and X chromosome reactivation during iNSCs induction.
(A) Experimental design to assess retroviral silencing during iNSC formation. (B) Representative images comparing tdTomato expression in MEFs, iPSCs, OKSM-iNSCs and brain-derived NSCs. Note that tdTomato+ non-reprogrammed MEFs surround the iPSC and iNSC colonies. Scale bar is 250µM. (C) Quantification of iNSC colonies expressing tdTomato+. For each replicate, 3x104 cells were used (n=5 independent replicates; error bars, s.e.m. for 5 independent experiments, *p<0.05). (D) Experimental design to follow X chromosome inactivation status during iNSC formation. Tail tip fibroblasts (TTFs) from a female mouse heterozygous for an X-linked CMV-GFP reporter (mix of XiGFPXa and XiXaGFP cells) were sorted twice for cells that carried the GFP transgene on the inactive X chromosome (XiGFPXa). These cells were reprogramed for 4–5 days in modified NSC medium, after which dox was withdrawn and GFP expression was assessed in emerging Sox1+ iNSC colonies 2–4 weeks later. (E) Representative immunofluorescence images of Sox1+ iNSC colonies that show reactivation of the silent XiGFP reporter. Scale bar is 250µM. (F) Quantification of Sox1+ iNSC colonies that reactivated the silenced X chromosome as determined by GFP reporter reactivation. All examined Sox1+ iNSC colonies showed patchy GFP expression. For each replicate, 3x104 cells were used (n=3 independent replicates; error bars, s.e.m. for 3 independent experiments, ***p<0.0005). (G) Representative immunofluorescence images showing Nanog knockout Sox1+ iNSC colony generated in the presence of ascorbate. Nanog-deficient iNSCs express the constitutive CAG-GFP marker, confirming their origin from Nanog-mutant MEFs. Scale bar is 100µM. Autofluor., autofluorescence control. (H) Quantification of Sox1+ iNSC colonies generated from Nanog-deficient MEFs with and without ascorbate. For each replicate, 3x104 cells were used (n=3 independent replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005).
Next, we investigated whether interfering with genes involved in iPSC reprogramming affects the generation of iNSCs. Nanog is expressed in pluripotent cells but not in iNSCs (Supplementary Fig. 1E), and Nanog deficiency leads to a severe reduction in iPSC formation that can be rescued by exposure of cells to ascorbic acid19. Nanog knockout MEFs gave rise to 6–8 times more Sox1+ iNSC colonies in the presence of ascorbic acid compared to control cultures lacking ascorbate (Fig. 3G, H). Ascorbate acts synergistically with the GSK3β inhibitor CHIR-99021 to increase iPSC formation efficiency20. Formation of iNSCs from wild-type MEFs was enhanced by exposure to both molecules compared to ascorbate alone, further demonstrating that iNSC generation responds to the same signaling pathways as iPSC formation (Supplementary Fig. 10A, B).
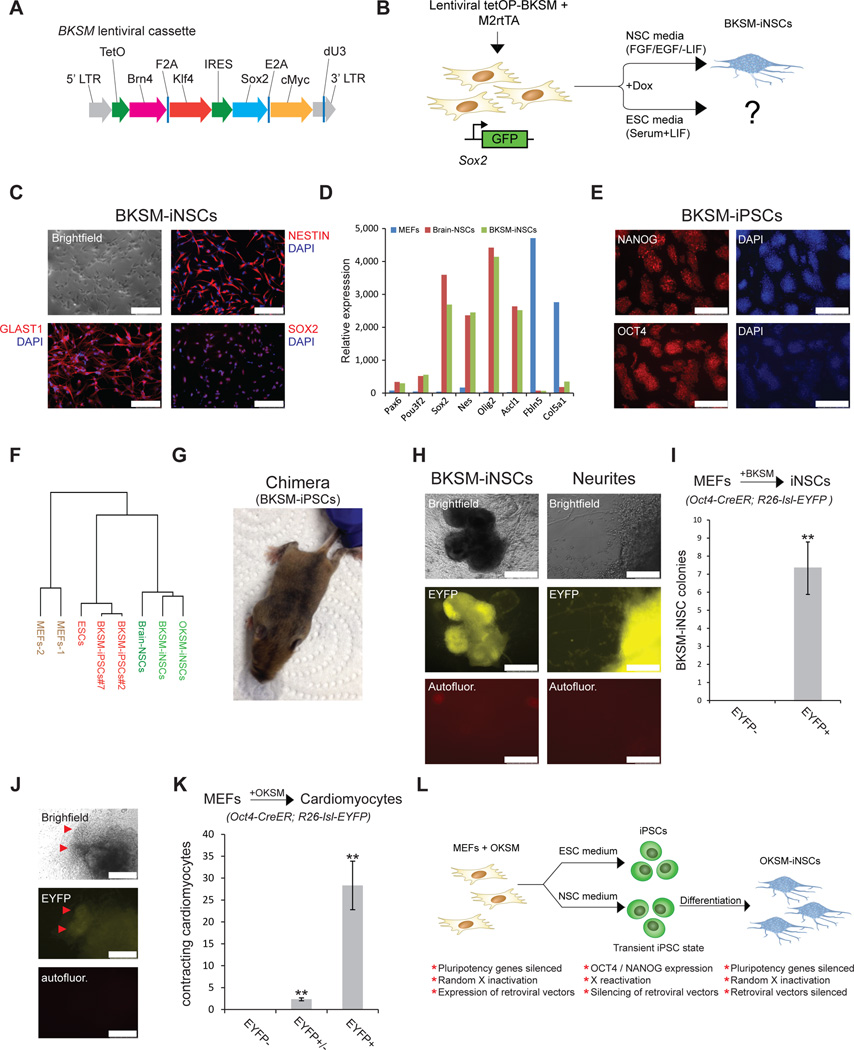
Finally, we studied a different iNSC transdifferentiation gene cocktail that did not yield iPSCs in ESC or NSC medium, namely Brn4, Klf4, Sox2 and c-Myc (BKSM)3. To ensure homogeneous expression of all four factors, we engineered a polycistronic lentiviral vector encompassing the coding regions for Brn4, Sox2, Klf4 and c-Myc under the control of a dox-inducible promoter, akin to the OKSM/STEMCCA system18 (Fig. 4A, B and Supplementary Fig. 11A–C). Infection of Sox2-GFP MEFs with lentiviral constructs expressing tetOP-BKSM and M2rtTA gave rise to Sox2-GFP+ iNSC-like colonies in the presence of dox and neural-inducing cytokines, in agreement with ref. 6 (Supplementary Fig. 12A). Of note, formation of iNSC clusters in conventional NSC medium required at least 12 days of BKSM expression, rather than 8 days with the OKSM cocktail, and we typically obtained ∼10 times fewer colonies (data not shown). BKSM-iNSCs could be propagated without dox, demonstrating independence from exogenous factor expression and self-renewal potential (Fig. 4C, Supplementary Fig. 12A and data not shown). Accordingly, BKSM-iNSCs activated NSC markers, suggesting faithful molecular reprogramming towards an NSC-like state (Fig. 4C, D and Supplementary Fig. 12B, C). Global gene expression analysis documented a high degree of similarity between OKSM-iNSCs, BKSM-iNSCs and brain-derived NSCs regardless of their origin from either fibroblasts or fetal brains, , whereas these samples were quite different from the starting MEF population (Fig. 4F and Supplementary Fig. 12B, C).
Figure 4. An alternative transdifferentiation cocktail generates iNSCs and iPSCs via an Oct4+ intermediate stage.
(A) Replacement of the Oct4 gene within the Oct4-Klf4-Sox2-cMyc (OKSM cassette) by the neuronal transcription factor Brn4 (BKSM cassette). (B) Experimental design to generate iPSCs and iNSCs using the BKSM cassette from MEFs harboring a Sox2-GFP knock-in allele in ESC or NSC culture conditions. (C) Immunofluorescence of typical NSC markers in an expanded BKSM-iNSC clone. Scale bar is 100µM. (D) Expression of selected NSC-associated genes in indicated cell lines based on microarray data. Microarray analysis was performed on expanded clonal iNSC cultures. (E) Immunofluorescence for indicated pluripotency markers in expanded BKSM-iPSC clone. Scale bar is 250µM. (F) Dendrogram based on microarray analysis of indicated samples. Note that BKSM-iNSCs and OKSM-iNSCs cluster with brain-derived NSCs while BKSM-iPSCs cluster with mouse ESCs. (G) Contribution of BKSM-iPSCs to an adult chimera as shown by agouti coat color. (H) Representative images of EYFP+ BKSM-iNSC colonies with emanating neurites generated from MEFs in conventional NSC medium. Scale bar is 500µM for iNSC colony (left) and 100µM for neurites (right). Autofluor., autofluorescence control. (I) Quantification of EYFP+ BKSM-iNSC colonies, demonstrating that all of the examined BKSM-iNSC colonies transitioned through an Oct4+ state. For each replicate, 5x105 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005). (J) Representative images showing EYFP+ cells in a cluster of beating cardiomyocytes (red arrowheads). (K) Quantification of EYFP+, EYFP and EYFP+/EYFP contracting induced cardiomyocyte colonies derived from Oct4-CreER; R26-lsl-EYFP MEFs upon short OKSM expression in cardiac transdifferentiation conditions. For each replicate, 5x105 cells were used (n=3 biological replicates; error bars, s.e.m. for 3 independent experiments, **p<0.005). (L) Summary.
In contrast to previous observations3, the BKSM combination generated iPSC-like colonies in both ESC and NSC media (Supplementary Fig. 13A and data not shown). As with OKSM-iPSCs derived in NSC medium, BKSM-iPSC-like clones activated key endogenous pluripotency genes, including Nanog, Oct4 and Zfp42, when replated in ESC medium without dox, indicating transgene-independent self-renewal (Fig. 4E and Supplementary Fig. 13B). The global transcriptional profile of BKSM-iPSC-like cells was highly similar to those of ESCs and OKSM-derived iPSCs (Fig. 4F and Supplementary Fig. 13B, C). Lastly, BKSM-iPSCs supported the formation of teratomas and the generation of fetal and adult chimeras upon blastocyst injection, demonstrating pluripotency (Fig. 4G and Supplementary Fig. 13D, E). Like BKSM-iNSCs, BKSM-iPSCs were recovered at a lower frequency and with delayed kinetics compared to OKSM-mediated iPSCs (data not shown), further supporting the notion of a linked process. Notably, infection of Oct4-CreER; R26-lsl-EYFP MEFs with lentiviral constructs expressing M2rtTA and tetOP-BKSM yielded exclusively EYFP+ iNSC colonies in the presence of 4-OHT, dox, EGF and FGF (Fig. 4H, I). These BKSM-iNSC colonies continued to grow without dox and could be converted into iPSCs upon re-addition of dox (Supplementary Fig. 14A–C). Together, these results show that transdifferentiation into iNSCs using our polycistronic BKSM cassette inevitably passes through a transient Oct4+ state.
A key question is whether our observations extend to other types of lineage conversion using either pluripotency or lineage-specific factors. We examined activation of the Oct4-CreER; R26-lsl-EYFP reporter during OKSM-induced cardiomyocyte generation2 and Ascl1/Brn2/Myt1l-driven induced neuron (iN) formation21 from MEFs. OKSM-induced cardiomyocyte clusters exhibited rhythmic contractions and expression of cardiac troponin and activated the Oct4-CreER; R26-lsl-EYFP reporter in nearly all cases (Fig. 4J, K and Supplementary Fig. 15A, B). By contrast, transdifferentiation of MEFs into iN-like cells did not activate the EYFP reporter, suggesting that passage through an Oct4+ state may be specific to the use of pluripotency-related transcription factors (Supplementary Fig. 15C–E).
Here we show that pulsed expression of OKSM or BKSM, two gene cocktails, previously used for direct lineage conversion of fibroblasts into iNSCs, causes cells to pass through a transient Oct4+ state that silences retroviral transgenes and reactivates silenced X chromosomal genes in female somatic cells. We further demonstrate that forced OKSM and BKSM expression produces bona fide iPSCs under culture conditions that are incompatible with long-term culture of established pluripotent cell lines. These results imply that transdifferentiation protocols using pluripotency-associated factors often, if not always, involve reprogramming into short-lived iPSCs that rapidly differentiate into various lineages as determined by the extracellular environment (Fig. 4L). Our results are consistent with previous findings that retroviral transgenes used to convert fibroblasts into iNSCs by brief OKSM expression are predominately silenced3,7, whereas induced neural progenitor cells generated by forced expression of lineage-specific factors22 remain transgene-dependent. Our data might also explain the recent observation that cells generated through transdifferentiation with pluripotency factors exhibit chromosomal aberrations typically associated with pluripotent stem cell lines23.
Transient iPSC formation was ruled out in previous transdifferentiation studies owing to (i) the lack of Nanog-GFP or Oct4-GFP signal, (ii) the inability to derive iPSCs in culture conditions lacking LIF and containing differentiation-associated cytokines, and (iii) the different kinetics of direct lineage conversion and iPSC formation. We believe that the Nanog-GFP reporter may not be sufficiently sensitive to detect rare reprogramming events in heterogeneous bulk cultures. Consistent with this notion, previous studies recorded transient Nanog mRNA upregulation between days 6–8 of transdifferentiation by qPCR analysis even though no reporter signal was detected2,4. Moreover, transdifferentiation experiments have often been performed under optimized culture conditions compared to control iPSC experiments, which may inadvertently facilitate fast yet transient iPSC formation. Indeed, several transdifferentiation supplements (e.g., CHIR99021, BMP4, bFGF, KOSR/ascorbic acid) strongly enhance and accelerate iPSC formation16,20,24. Accordingly, we could reduce the minimum number of days of OKSM expression required to derive stable iNSCs from 8 to 4 days by modifying the culture medium.
Our results show that the absence of LIF and the presence of differentiation-inducing cytokines (e.g., FGF, EGF) are not sufficient to block iPSC formation. Thus, forced transcription factor expression can effectively compensate for the lack of growth factors important for iPSC self-renewal and counteract culture conditions that normally trigger ESC/iPSC differentiation. This finding is reminiscent of the observation that forced expression of Stat3 or Nanog bypasses the requirement for LIF signaling during ESC derivation and iPSC formation25,26. We believe that our single-copy transgenic reprogramming system allowed us to increase the efficiency reprogramming through homogeneous expression of the four factors, facilitating mechanistic dissection of critical intermediate stages. By contrast, iNSC generation in some previous studies was extremely inefficient (1–3 iNSC colonies per infection with BKSM; 11 iNSC colonies per 130,000 infected cells or 0.008% with OKSM) and lengthy (3 weeks with OKSM, 4–6 weeks with BKSM expression)3,7 , which was likely due to inefficient retroviral infection of fibroblasts or an inadequate expression stoichiometry of all four transgenes. The low efficiency of iNSC formation may also explain why iPSCs were not detected upon BKSM overexpression, whereas our use of a polycistronic, dox-inducible expression vector gave rise to iPSCs in ESC or NSC conditions, albeit at much lower efficiency than OKSM-mediated reprogramming.
Pluripotency factor–driven transdifferentiation has been proposed as a safer and more efficient alternative to induced pluripotency for therapeutic applications as it does not require lengthy iPSC differentiation protocols and avoids potential teratoma formation from rare undifferentiated cells. Our study highlights the practical importance of dissecting the mechanisms underlying transdifferentiation to determine whether cells transit through a pluripotent state. While transient passage through a pluripotent state is not expected to impact in vitro applications of this technology, it will be important to exclude the presence of residual iPSCs in potential clinical settings.
Materials and methods
Cell culture
Mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 1% Glutamax, 1% non-essential amino acids (NEAA), 1% Penicillin-Streptomycin, 0.5% β-mercaptoethanol (all from Life Technologies) and 10% FBS (Gibco). Mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) were grown on irradiated MEFs (GlobalStem) in KO-DMEM medium (Life Technologies) supplemented with 15% FBS, 1% Glutamax, 1% NEAA, 1% Penicillin-Streptomycin, 1% β-mercaptoethanol, and 1000 Unit/ml LIF (purified in-house). Neural stem cells (NSCs) were cultured in DMEM/F12 (Life technologies) supplemented with 1XB27 and 1XN2 cell-supplements (Life Technologies), 0.01% Fraction V BSA 7.5% solution (Life Technologies), 1% Penicillin-Streptomycin and 10ng/ml bFGF (R&D) and 20ng/ml mouse EGF (Life Technologies) (“conventional NSC medium”).
Generation of iNSCs using conventional NSC medium
Conventional NSC medium contained DMEM/F12 (Life Technologies) supplemented with 1XB27 and 1XN2 cell-supplements (Life Technologies), 0.01% Fraction V BSA 7.5 % solution (Life Technologies), 1% Penicillin-Streptomycin and 10ng/ml bFGF (R&D) and 20ng/ml mouse EGF (Life Technologies); this media formulation was previously used to generate iNSCs3. We termed this medium “conventional NSC medium” because it does not contain LIF, serum or serum replacement components. For the generation of OKSM-iNSCs, reprogrammable MEFs (rep-MEFs) from mice carrying a tetOP-OKSM allele in the Col1a1 locus and an M2rtTA transgene in the Rosa26 locus10 as well as an EGFP transgene knocked into one allele of the Oct4 locus were used for some of the experiments10. Alternatively, MEFs or tail tips fibroblasts (TTFs) were reprogrammed into iNSCs by infection with lenti-viral vectors expressing M2rtTA in combination with either tetOP-OKSM or tetOP-BKSM cassettes. In conventional NSC medium, at least 8 days (OKSM) or 12 days (BKSM) of dox exposure, followed by 7–21 days (OKSM) and 12–24 days (BKSM) of dox-independent growth, were usually required to generate iNSCs. For most experiments, either 3x104 cells were plated per well of a 6 well plate or 5x105 cells per 10cm plate in MEF medium a day before dox/NSC medium were administered. Of note, low-passage MEFs reprogrammed more efficiently and yielded iNSC colonies faster than high-passage MEFs. Individual iNSC colonies were picked onto Poly-L-Ornithine (P4957, Sigma) coated plates and propagated in conventional NSC medium. For the generation of Nanog knockout (KO) iNSCs, Nanog KO MEFs were infected with lentiviral vectors expressing M2rtTA and tetOP-OKSM as previously described19. For each replicate, 3x104 cells were plated per well a day before dox administration. Cells were reprogrammed in conventional NSC medium for 10 days, and then switched to the same NSC medium without dox for an additional 7 days before scoring for Sox1+/CAG-GFP+ transgene-independent iNSC colonies. For the generation of iNSCs from rep-MEFs using small molecules, a concentration of 50 µg/ml ascorbic acid (Sigma) and 3µM GSK3 inhibitor CHIR-99021 (Tocris) were used in conventional NSC medium. For each replicate, 3x104 cells were plated on 6 well plates the day before dox administration. Serum was added for the first 3 days of transdifferentiation to reduce cell death. Small molecules were withdrawn at the same time as dox and as previously described20. A concentration of 2 µg/ml doxycycline (dox) (Sigma) was used for all transdifferentiation and reprogramming experiments.
Generation of iNSCs using modified NSC medium
Modified NSC medium contained DMEM/F12, 2% FBS, 8% KOSR (Gibco), 1xN2, 1% Glutamax, 1% NEAA and 0.5% β-mercaptoethanol (all purchased from Life Technologies) and 1000 U/ml LIF (purified in-house). This medium was previously used to generate iNSCs7. We termed this NSC medium “modified” because it contains low levels of serum, KOSR and LIF. To achieve iNSC induction with this medium, MEFs or TTFs were transduced with lentiviral vectors expressing M2rtTA and tetOP-OKSM. For each replicate, 5x105 cells were plated in a 10cm dish a day before dox administration. OKSM expression was induced by adding dox for 4–5 days in modified NSC medium, followed by 2–4 weeks of dox-independent growth to select for transgene-independent iNSCs. The generation of iNSCs and iPSCs with modified NSC medium required a shorter duration of OKSM expression compared to conventional NSC medium (4 vs. 8 days). Moreover, low-passage MEFs (passage 3 and below) reprogrammed more efficiently and faster than high-passage MEFs. A concentration of 2 µg/ml doxycycline (dox) (Sigma) was used for all transdifferentiation and reprogramming experiments.
Generation of iPSCs in NSC medium
Reprogrammable MEFs or MEFs infected with lentiviral vectors expressing M2rtTA and tetOP-OKSM were reprogrammed in conventional NSC medium supplemented with dox for 8–12 days to initiate OKSM expression, before switching to ESC medium without dox. Oct4-GFP+ colonies were first observed in NSC medium at around day 10–12; however, these cells tended to differentiate into iNSCs after a few days without dox when kept in NSC medium. Switching to ESC medium (serum/LIF) prevented this rapid differentiation and allowed propagation of dox-independent iPSC colonies.
Generation of Oct4-CreER; Rosa26-loxSTOPlox-EYFP MEFs
Oct4-CreER mice, termed B6(SJL)-Pou5f1tm1.1(Cre/Esr1*)Yseg/J, were purchased from Jackson laboratory (Stock number 016829)15. Oct4-CreER mice were crossed with Rosa26-lox-STOP-lox-EYFP mice27 to produce bitransgenic reporter MEFs and NSCs27. Genotyping of the Oct4-CreER allele was performed as previously described15. Cells with the correct genotype were infected with lentiviral vectors harboring M2rtTA and tetOP-OKSM alleles and reprogrammed in either modified or conventional NSC medium. MEFs carrying the same genotype were used to derive iPSCs in conventional NSC medium (as shown in Fig. 2). 4-hydroxytamoxifen (4-OHT) was used at a concentration of 100nM for all experiments and was continuously added to the culture medium during and after dox treatment to ensure labeling of Oct4+ cells at all stages of transdifferentiation. For in vivo induction, 1mg tamoxifen (Sigma) was diluted in corn oil (10mg/ml) and injected into the peritoneum of a pregnant mouse at embryonic day E8.5 The brains and gonads of recovered embryos were dissected and analyzed at E13.5.
Derivation of NSCs from fetuses
Brain-derived NSCs were derived from the heads of E13.5 fetuses as previously described28. We used conventional NSC medium for expansion and culture of NSCs.
Scoring for X reactivation during iNSC formation
Tail tip fibroblasts (TTFs) obtained from female mice carrying a CMV-GFP transgene on one X chromosome29 were infected with lenti-viral vectors carrying the M2rtTA and tetO-OKSM constructs without dox. Infected cells were sorted twice on a FACSAriaII machine (BD Biosciences) to select for the GFP-negative fraction (sort purity 99.9–100% after 2 sorting rounds). For each replicate, 3x104 cells were sorted a day before dox administration and treated in a 6 well plate. iNSC induction was performed in modified NSC medium in the presence of dox for 4–5 days, followed by 2–4 weeks of dox-independent growth before scoring for the fraction of GFP+ Sox1+ iNSC colonies.
Retroviral silencing during iNSC induction
Rep-MEFs were infected with retroviral vectors carrying the pMXs-tdTomato construct. Cells were then sorted on a FACSAriaII machine (BD Biosciences) to isolate the tdTomato+ cell-fraction. For each replicate, 3x104 cells were sorted per well of a 6 well plate and cultured in ESC or NSC media supplemented with dox for 12 days. Scoring of iPSC or iNSC colonies was performed 7–10 days after dox withdrawal. iNSC induction was performed in the presence of serum for the first 3 days of OKSM expression to minimize cellular senescence. This modification is in accordance with other reports that used low serum during iNSC induction4,7. Fetal brain-derived NSCs were infected with pMXs-tdTomato retroviral vectors, sorted for the tdTomato+ fraction and cultured for an additional 4 weeks in conventional NSC medium before measuring the fraction of tdTomato+ NSC spheres.
Generation of Oct4-CreER; Rosa26-loxSTOPlox-DTA MEFs
Oct4-CreER mice were crossed with Rosa26-loxSTOPlox-DTA animals30. MEFs were isolated from bitransgenic fetuses at E13.5. Generation of iNSCs was performed in modified NSC medium. For each replicate, 5x105 cells infected with lenti-viral vectors carrying the M2rtTA and tetO-OKSM constructs were plated on a 10cm plate a day before dox treatment. Dox was administered for 4–5 days, followed by 2–3 weeks of dox-independent growth. 4-OHT was added throughout the transdifferentiation protocol at a concentration of 100nM for all experiments.
Generation of induced neuronal (iN)-like cells from MEFs
Oct4-CreER; R26-lsl-EYFP MEFs were infected with tetOP-Brn2, tetOP-Ascl1, tetOP-Myt1l and M2rtTA and reprogrammed as described previously21. Dox was added to the medium for 12 days, followed by 7 days of dox-independent growth before measuring Tuj1+ iN cells. 4-OHT was used at a concentration of 100nM for all experiments.
Subcloning of iNSC clones
OKSM-iNSCs (EYFP+) derived from Oct4-CreER; R26-lsl-EYFP MEFs were plated at 50 cells per 6 well plate using a FACSAriaII sorter (BD Biosciences). Clonal colonies were detected 7–10 days after plating. Subclones were picked onto poly-ornithine coated plates and propagated until passage 25.
Generation of induced cardiomyocytes
Oct4-CreER; R26-lsl-EYFP MEFs were infected with lentiviral vectors expressing M26rtTA and tetOP-OKSM and treated precisely as reported previously to generate induced cardiomyocytes2. For each replicate, 5x105 cells were plated on a 10cm plate the day before dox exposure to initiate transdifferentiation. Briefly, cells were first cultured in KO-DMEM (Life Technologies) supplemented with 15% FBS, 5% KOSR, 1% Glutamax, 1% non-essential amino acids, 1% penicillin-streptomycin and 1% β-mercaptoethanol. After 6 days of growth in this culture condition, cells were switched to medium containing KO-DMEM supplemented with 14% KOSR, 1% FBS, 1% Glutamax , 1% non-essential amino acids, 1% penicillin-streptomycin, 1% β-mercaptoethanol. During these 9 days, JAK inhibitor (EMD) was added to the medium at a concentration of 0.5µM. At day 9, cells were switched to “CDM medium”2 containing RPMI-1640 (Life Technologies), 0.5 XN2, 1XB27, 0.05% BSA fraction V, 1% Glutamax and 0.5% β-mercaptoethanol. 20ng/ml BMP4 (Stemgent) was added for the first 5 days of culture in CDM medium as previously described2. Dox was administered for 6–9 days for all experiments. iPSC-like colonies appeared 3–4 days after dox withdrawal whereas contracting induced cardiomyocytes were first detected 7–8 days after dox withdrawal. Beating colonies could be detected as long as 3–4 weeks after dox withdrawal. Scoring of induced cardiomyocytes was performed by counting the fraction of EYFP+ colonies that showed rhythmic contractions or by co-immunostaining with antibodies against Cardiac Troponin (cTnT) and EYFP 1–2 weeks after dox withdrawal. 4-OHT was used at a concentration of 100nM for all experiments.
Viral vector production
For retroviral generation, approximately 2x106 T-293 cells were transfected with 4.5 µg PCL-ECO (ecotropic envelope) and 4.5 µg pMXs-tdTomato vectors, mixed in transfection solution consisting of 300µL Opti-MEM (Gibco) and 27µl of TransIT®-LT1 (Mirus). Transfection was performed in regular MEF medium without Penicillin-Streptomycin. 24 hours post transfection, medium was replaced, and 48 and 72 hours post transfection the supernatant was collected, filtered through a 0.45µM filter (Westnet), supplemented with 4–8 µg/ml Polybrene (Sigma) and added freshly to the cells. For lentiviral generation, ∼6x106 T-293 cells were transfected in a 10cm culture dish with a solution consisting of 770µL Opti-MEM and 50 µl of TransIT®-LT1, Δ8.9 (8.5 µg), VSV-G (5.5 µg) and 11µg of the target plasmid (either M2rtTA11, tetOP-OKSM11, tetOP-BKSM, tetOP-Ascl121, tetOP-Brn221 and tetOP-Myt1l21). Viral supernatant was harvested 48 and 72 hours post transfection, filtered and added freshly to the cells. Similar ratios of M2rtTA and tetOP-OKSM or tetOP-BKSM (1:1) were used for maximal infection efficiency.
Construction of the BKSM polycistronic expression vector
To achieve co-expression of Brn4, Klf4, Sox2 and cMyc, a novel lentiviral construct (“BKSM”) was generated by reengineering the STEMCCA reprogramming vector18. First, a DNA fragment consisting of the Oct4 (Pou5f1) gene followed by an F2A peptide and part of the Klf4 gene was excised from STEMCCA (OKSM) using NotI and BstZ17I. Next, we used AccuPrimeTM Pfx DNA Polymerase (Life Technologies) to generate a fragment corresponding to Brn4-F2A-Klf4 by overlapping PCR. To accomplish this, two separate reactions were performed using the primer pairs Brn4 5’ NotI/Brn4-F2A 3’ and F2A-Klf4 5’/Klf4 3’ BstZ17I (see below) under the following cycling conditions: initial denaturation at 95°C for 2 minutes followed by 35 cycles of 15 seconds at 95°C and 1 minute at 68°C. Aliquots of the two purified amplicons were then mixed in a 1:1 ratio and used in a third PCR with primers Brn4 5’ NotI and Klf4 3’ BstZ17I under the following conditions: initial denaturation at 95°C for 2 minutes; 5 cycles of 15 seconds at 95°C, 30 seconds at 58°C, and 2 minutes at 68°C; and 30 cycles of 15 seconds at 95°C, 30 seconds at 63°C, and 2 minutes at 68°C. The resulting fragment (Brn4-F2A-Klf4) was gel-purified, digested and inserted by directional cloning into the NotI- and BstZ17I-digested STEMCCA lentiviral vector. We confirmed the identity of the BKSM polycistronic vector by restriction analysis; DNA sequencing and immunostaining (see supplementary data). The primer sequences used were:
Brn4 5’ NotI: ATAAGAATGCGGCCGCCATGGCCACAGCTGCCTCGAATCCCTAC
Brn4-F2A 3’: CTTGAGAAGGTCAAAATTCAAAGTCTGTTTCACGCCACTTCCGAG ATCGTGGCA GGACGCGTCTGTTTTCACC
F2A-Klf4 5’: AAACAGACTTTGAATTTTGACCTTCTCAAGTTGGCGGGAGACGTGGAGTCCAACCCAGGGCCCATGGCTAGCGACGCTCTGCTCCC
Klf4 3’ BstZ17I: TCTTTCGTATACTGGGTCCAACTCCGGCCGCAGGAGC
Differentiation potential of iNSCs
To generate neurons, OKSM-iNSCs were grown in NSC medium following removal of the growth factors bFGF and EGF for 5–10 days; neuronal cells were identified by immunostaining with rabbit anti-Tuj1 antibody (Covance, IgG1, 1:200). Astrocytes were generated using NSC medium supplemented with 5% FBS for a duration of 10 days, after which they were stained with rabbit anti-GFAP antibody (Dako Z0334, IgG, 1:200). The secondary antibody used was donkey anti-rabbit IgG (A10040, Alexa Fluor 546) at a concentration of 1:250. See “Immunocytochemistry” section for a detailed protocol.
Immunocytochemistry
Cells were washed twice with 1xPBS, cross-linked in 4% PFA solution for 5–10 minutes, washed with 1xPBS and blocked for ½ hour at room temperature (RT) with blocking solution (2% BSA dissolved in PBS and 0.1% Triton-X-100). Primary antibody staining was performed at RT (1 hour). Appropriate secondary antibodies were added after two washes with 1xPBS, followed by incubation for 1 hour at RT. Primary and secondary antibodies were diluted in blocking solution. DAPI was used for nuclear counterstaining. The following antibodies were used in this study: rabbit anti-Nanog (AB80892 Abcam, IgG, 1:200), goat anti-Oct4 (Santa Cruz sc-8628, IgG 1:200), goat anti-Sox1 (R&D, AF3369, 1:100), goat anti-Sox2 (Santa Cruz sc-17320, IgG, 1:200), mouse anti-Nestin (Abcam AB6142, IgG1, 1:200), goat anti-Glast1/Eaat1 (Santa Cruz SC-7757, IgG, 1:200) , rabbit anti-Brn4/Pou3f4 (Millipore, IgG, 1:200) chicken anti-GFP/EYFP (AVES, 1:300, GFP-1020) and anti-mouse cardiac troponin (cTnT) (DSHB, CT3, 1:1000). Secondary antibodies used were A11056 Alexa Fluor 546 donkey anti-goat IgG, A21123 Alexa Fluor 546 goat anti-mouse IgG1, A10040 Alexa Fluor donkey anti-rabbit IgG and A11055 Alexa Fluor 488 donkey anti-goat IgG, all at a 1:250 dilution. 488 goat anti-chicken IgG (A-11039, Life technologies,) or Rabbit anti chicken IgY (F4137, Sigma) were used at a dilution of 1:1000. We employed anti-EGFP/EYFP antibodies to stain for EYFP when co-immunostaining colonies for Sox1 or cTnT since the endogenous EYFP protein is quenched by PFA treatment during the immunostaining protocol.
Antibody live staining
For live staining of cells, PE-conjugated anti-mouse PeCAM1 (CD31) antibody (12–031182 eBioscience, 1:200) or PE-conjugated anti-mouse CD326 EpCAM (clone G8.8 12–5791-83, eBioscience 1:200) were added directly to the cells. Cells were then incubated at 37C for an hour, washed twice with 1xPBS and visualized for surface marker expression.
Flow cytometry analysis of surface markers
Cells were harvested using 0.25% trypsin and resuspended in 1xPBS buffer containing 2% FBS (“FACS buffer”). The appropriate conjugated antibodies were added at similar ratios (1:200); anti-mouse PE-conjugated CD326 EpCAM (1:200, eBioscience) and anti-mouse eFluor 450-conjugated CD90 Thy1.2 (1:200, eBioscience). Cells were incubated on ice for 20 minutes, washed in 1xPBS, resuspended in FACS buffer, filtered and then analyzed on a MACSQuant flow cytometer (Miltenyi Biotec). Analysis was performed using the software FlowJo.
RNA extraction and microarrays
RNA (DNase-treated) was extracted using RNAeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and subjected to hybridization to the GeneChip Mouse 2.0 ST arrays (Affymetrix) at the Partners Center for Personalized Genetic Medicine. Expression Console (Affymetrix) was used for RMA. Hierarchical clustering and dendrogram analysis were performed using Expander software (EXPression Analyzer and DisplayER). Scatter plot, analysis of linear regression coefficients and TTEST analysis were performed using Excel. Classification and annotations of upregulated genes were performed using the DAVID online functional annotation tool (http://david.abcc.ncifcrf.gov/). RNA was extracted from expanded OKSM or BKSM-iNSC clusters that have been propagated for several passages. Expression data for MEFs and pluripotent stem cells were previously published20. The microarray data has been deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE65951.
Alkaline phosphatase staining
To detect iPSC colonies, alkaline phosphatase kit (Vector Labs) was used according to the manufacturer’s recommendations.
Teratoma formation
Approximately 2x106 BKSM-iPSCs were injected under the flanks of SCID-beige mice. 3–4 weeks post-injection, mice were sacrificed to harvest tumor masses. Paraffin section and hematoxylin and eosin (H&E) staining were performed at the Brigham and Women’s Hospital pathology core.
Generation of chimeric mice
Chimeras were generated by blastocyst injections as described previously20. Briefly, superovulation was initiated by injecting five female B6D2F1/J mice (Jackson Laboratory) with pregnant mare serum (Sigma) by intraperitoneal injection. Two days later, human chorionic gonadotropin (Sigma) was administered via intraperitoneal injection, and female mice were mated with B6D2F1/J males. After three days, developing blastocysts were collected and injected with the indicated cell types. Injected blastocysts were transplanted into the uterus of pseudo-pregnant Swiss Webster female mice (Jackson Laboratory; approximately 2 months of age) and allowed to develop to midgestation or term. All procedures involving animals were performed according to a mouse protocol approved by the MGH Subcommittee on Research Animal Care.
Allele-specific real-time PCR for X-linked marker gene
DNase-treated RNA was extracted using the RNAeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Conversion of RNA to cDNA was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche) as per manufacturer’s instructions. mRNA levels were measured using Brilliant iii SybrGreen kit (Agilent) on a Stratagene Mx3005P qRT-PCR system (Agilent). mRNA levels were normalized to GAPDH. We used the following primer sequences for qRT-PCR: Uba1 129/mus musculus-specific primer – TGAGCTCCTCAAGACGGCAG. Uba1 mus castaneous-specific primer – TGAGCTCCTCAAGACGGCCA. Common Uba1 reverse primer - CCTTTTTGCCCAGACCTATF.
Supplementary Material
Acknowledgments
We thank members of the Hochedlinger lab for critical evaluation and discussion of this manuscript. We thank Effie Apostolou and Sihem Cheloufi for helpful comments and Nimet Maherali for suggesting the idea of X chromosome reactivation. We are also grateful to Bernhard Payer and Jeannie Lee for providing tail-tip fibroblasts carrying an X-linked GFP reporter and to Andrew Brack for sharing Rosa26-lsl-DTA mice. We are grateful to Laura Prickett, Meredith Weglarz and Kat Folz-Donahue at the MGH/HSCI flow cytometry core for their constant assistance and support. O.B-N. is supported by a Gruss-Lipper postdoctoral fellowship from the EGL foundation. B.A.S has been supported by a T32 (5-T32-CA-9216-33) grant and through a Postdoctoral Fellowship Award by the MGH Executive Committee on Research Fund for Medical Discovery. J.B. and I.L. are supported by Ruth L. Kirschstein F32 Post-doctoral Fellowships from NIH (1F32HD078029-01A1 to J.B and 1F32HD079225-01A1 to I.L.). Support to K.H. is from the NIH (R01HD058013) and HHMI.
Footnotes
Accession codes: NCBI Gene Expression Omnibus (GEO): microarray data have been deposited under accession number GSE65951.
Author contributions
O.B-N. and K.H. conceived the experiments, interpreted results and wrote the manuscript. O.B.-N. conducted all reprogramming experiments, performed statistical analysis, bioinformatics analysis of expression data and generated figures; C.V. assisted in most reprogramming experiments and interpretation of results; A.G.S and G.M generated the BKSM lentiviral cassette,; J.B. generated chimeric animals by blastocyst injections; B.A.S conducted reprogramming experiments from FACS-sorted intermediates; I.L. performed allele-specific real-time PCR for an X-linked marker gene; A.J.H. confirmed specificity of the Oct4-CreER lineage-tracing system in vivo.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Molecular cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature cell biology. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 3.Han DW, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell stem cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li K, et al. Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell stem cell. 2014;14:228–236. doi: 10.1016/j.stem.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margariti A, et al. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thier M, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell stem cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhu S, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014 doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nature methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell stem cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeVeale B, et al. Oct4 is required ∼E7.5 for proliferation in the primitive streak. PLoS genetics. 2013;9:e1003957. doi: 10.1371/journal.pgen.1003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 15.Greder LV, et al. Analysis of endogenous Oct4 activation during induced pluripotent stem cell reprogramming using an inducible Oct4 lineage label. Stem cells. 2012;30:2596–2601. doi: 10.1002/stem.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell stem cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz BA, Bar-Nur O, Silva JC, Hochedlinger K. Nanog is dispensable for the generation of induced pluripotent stem cells. Current biology : CB. 2014;24:347–350. doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-Nur O, et al. Small molecules facilitate rapid and synchronous iPSC generation. Nature methods. 2014;11:1170–1176. doi: 10.1038/nmeth.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissbein U, Ben-David U, Benvenisty N. Virtual karyotyping reveals greater chromosomal stability in neural cells derived by transdifferentiation than those from stem cells. Cell stem cell. 2014;15:687–691. doi: 10.1016/j.stem.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, et al. Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell research. 2011;21:884–894. doi: 10.1038/cr.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 26.van Oosten AL, Costa Y, Smith A, Silva JC. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nature communications. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 29.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Non-invasive sexing of preimplantation stage mammalian embryos. Nature genetics. 1998;19:220–222. doi: 10.1038/893. [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.