Abstract
Dehydrogenases are an important target for the development of cancer therapeutics. Dehydrogenases either produce or consume NAD(P)H, which is fluorescent but at a wavelength where many compounds found in chemical libraries are also fluorescent. By coupling dehydrogenases to diaphorase, which utilizes NAD(P)H to produce the fluorescent molecule resorufin from resazurin, the assay can be red-shifted into a spectral region that reduces interference from compound libraries. Dehydrogenases that produce NAD(P)H, such as isocitrate dehydrogenase 1 (IDH1), can be read in kinetic mode. Dehydrogenases that consume NAD(P)H, such as mutant IDH1 R132H, can be read in endpoint mode. Here, we report protocols for robust and miniaturized 1,536-well assays for WT IDH1 and IDH1 R132H coupled to diaphorase, and the counterassays used to further detect compound interference with the coupling reagents. This coupling technique is applicable to dehydrogenases that either produce or consume NAD(P)H, and the examples provided here can act as guidelines for the development of high-throughput screens against this enzyme class.
Introduction
Interest in targeting dehydrogenases for the treatment of diseases, such as cancer, has been growing.1,2 Dehydrogenases play a role in metabolism and, as such, have an effect on the proliferation of rapidly dividing cells. Thus, there is a need to be able to rapidly develop dehydrogenase assays that efficiently detect hits while minimizing interference from compounds in the chemical libraries. A dehydrogenase is an enzyme that catalyzes the removal of hydrogen from a substrate and the transfer to an acceptor in an oxidation–reduction reaction, usually utilizing NAD+ or NADP+. NADH and NADPH levels can be measured directly in biochemical assays by virtue of their intrinsic fluorescence (excitation = 340 ± 30 nm, emission = 460 ± 50 nm). However, profiling the fluorescence properties of a large chemical library revealed that a significant percentage of library compounds were fluorescent in the spectral region of NAD(P)H, which leads to assay interference, while a much lower percentage of compounds would interfere at longer wavelengths.3 Fluorescence interference in assay design is discussed in more detail in this issue by Hall et al.4 When possible, assays can be designed to minimize interference from a library, and one technique is to red-shift the assay to longer wavelengths. The diaphorase/resazurin assay is such a red-shifted assay that can be used as a primary assay (the authors' preference) or secondary confirmatory assay. Diaphorase utilizes NAD(P)H to convert the dye resazurin to the fluorescent molecule resorufin. As resorufin fluoresces at 585 nm, the coupled diaphorase/resazurin reaction can be used to red-shift dehydrogenase biochemical assays into a spectral region that reduces interference from blue-fluorescent compound library members.
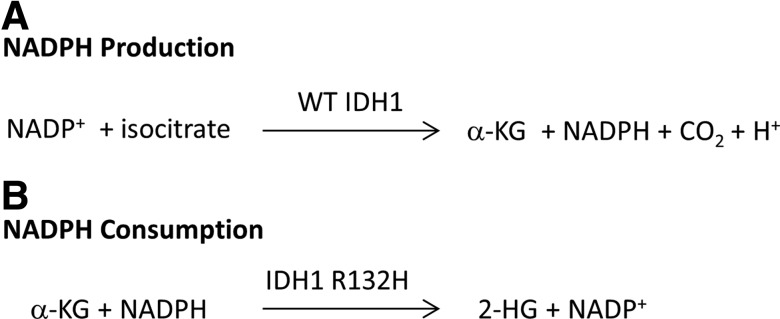
We will describe the development of diaphorase/resazurin-coupled assays with the target isocitrate dehydrogenase 1 (IDH1). Mutations in IDH1 at position R132 have been shown to lead to a neomorphic production of 2-hydroxyglutarate, an oncometabolite (Fig. 1).1 Mutations in IDH1 are found in a variety of cancers, including gliomas, and have therefore been the subject of inhibitor discovery programs.5 We and others have previously reported the development of assays for screening mutant and wild-type (WT) IDH1.6–8 While WT IDH1 produces NADPH, the mutant enzyme consumes NADPH, and in both cases we utilized the diaphorase/resazurin coupling reaction. This work led to the development and characterization of an alpha-ketoglutarate (α-KG)-competitive probe inhibitor of IDH1 R132H and IDH1 R132C that was WT sparing.6,7 This inhibitor was characterized with a 384-well format assay for the WT and mutant IDH1 enzymes.
Fig. 1.
Assays can be designed for dehydrogenases that produce or consume NAD(P)H. (A) NADPH production by wild-type IDH1. (B) NADPH consumption by IDH1 R132H. IDH1, isocitrate dehydrogenase 1.
The technical protocol reported here provides a detailed description of the 1,536-well format quantitative high-throughput screening (qHTS)9 IDH1 R132H assay that was used to screen compounds (PubChem AID 602179) from the Molecular Libraries Small Molecule Repository (MLSMR) library and the WT IDH1 assay that was used as a counterscreen (PubChem AID 623995). These two examples show how kinetic assays can be developed for NADPH producing assays such as WT IDH1 and how endpoint assays can be developed for NADPH consuming assays such as IDH1 R132H. While diaphorase coupling strategies for IDH1 have been utilized previously,6–8 the intention herein is to highlight the general importance of this strategy and describe in detail how assays can be designed for dehydrogenases using WT IDH1 and IDH1 R132H as examples that produce and consume NADPH, respectively. In addition, diaphorase is able to utilize both NADPH and NADH as a substrate.10,11 Therefore, the protocols reported here using diaphorase for coupling can be used for almost any dehydrogenase enzyme, irrespective of the enzyme direction being assayed or the preference for NADH versus NADPH.
Materials and Methods
Reagents and Consumables
NADPH, NADP+, resazurin, isocitrate, α-KG, protease-free BSA, β-mercaptoethanol, and diaphorase (Clostridium kluyveri) are from Sigma Aldrich (St. Louis, MO). Magnesium chloride, Tris pH 7.5, and sodium chloride are from Quality Biological (Gaithersburg, MD). Protein was acquired from beryllium (Bedford, MA) and stored at −80°C until use. WT IDH1 = residues 1–414 and IDH1 R132H is the same but with the single R132H point mutation. Black solid medium-binding 1,536-well plates (Greiner, Monroe, NC) were used for all assays described herein.
Compound Plates
Compounds were dissolved and titrated in dimethyl sulfoxide (DMSO) and plates were prepared as described previously with the library formatted into columns 5–48.12 A DMSO control plate was made using columns 1–4 and a CyBi-Well (CyBio, Jena, Germany) to transfer compounds from a 384 plate into the 1,536-well plate. The final concentration range of the compounds in the IDH1 R132H assay was 76–0.15 μM for the cherrypick plates and 76 μM to 2 nM for the original qHTS plates, and the final concentration range of the compounds in the WT IDH1 assay was 114–0.2 μM for the cherrypick plates.
qHTS Protocol WT IDH1
WT IDH1 (3 μL) was added to the black solid bottom 1,536-well assay plate using a flying reagent dispenser (FRD; Beckman Coulter, Fullerton, CA). A pintool (Kalypsys, San Diego, CA) was used to transfer 23 nL of compound solution (library and control) to the 1,536-well assay plates. After 30 min of incubation at room temperature, 1 μL of buffer containing isocitrate, NADP+, diaphorase, and resazurin was added to initiate the reaction. The final concentration of DMSO in the 4 μL reaction was 0.57%. The plate was rapidly transferred to a ViewLux (PerkinElmer, Waltham, MA) and the fluorescence product resorufin was measured (excitation = 525 nm, emission = 598 nm) in kinetic mode. For individual experiments, the timing of reads varied depending on the number of plates. In initial optimization experiments, the plates were read continuously from t = 0 to t = 30 min at ∼30-s intervals to determine the linear range. For the high-throughput screening (HTS), the plates were read within the predetermined linear range at t = 0 and t = 10 min. The robotic system used has been described previously.13
qHTS Protocol IDH1 R132H
IDH1 R132H (3 μL) was added to the black solid bottom 1,536-well assay plate using an FRD (Beckman Coulter). A pintool (Kalypsys) was used to transfer 23 nL of compound solution (library and control) to the 1,536-well assay plates. After 30 min of incubation at room temperature, 3 μL of buffer containing NADPH and α-KG was added to initiate the reaction. The final concentration of DMSO in the 6 μL reaction was 0.38%. After 60 min of incubation, 3 μL of diaphorase/resazurin was added leading to a total volume of 9 μL. The plate was transferred to an Envision (PerkinElmer) after 5 min and the fluorescence product resorufin was measured (excitation = 540 nm, emission = 590 nm) in endpoint mode. The robotic system used has been described previously.13
Analysis of qHTS Data
Screening data were corrected and normalized, and using in-house algorithms, concentration–response curves were generated.9 Percent activity was computed from the delta at 10 min (i.e., subtracting the fluorescence at t = 10 min from t = 0 min) after checking that the assay was linear up to 15 min for WT IDH1, and from the endpoint data for IDH1 R132H. The no-enzyme column with DMSO was normalized as 100% inhibited, and the column with all components and DMSO was normalized as 0% inhibited.
Results and Discussion
In this technical brief, we describe the application of coupling diaphorase to two dehydrogenases (WT IDH1 and IDH1 R132H). In one case, we were able to use kinetic mode (WT IDH1) and in the other, we utilized endpoint mode (IDH1 R132H).
WT IDH1 can be run in kinetic mode because it produces NADPH, the substrate for diaphorase. Assays can be designed to bias hits toward particular mechanisms of action.14 Here compounds that bound outside of the NADP+ pocket were desired, so the concentration of IDH1 substrate NADP+ was in excess of the Km and the concentration of isocitrate was set to just above the Km. The Kms for WT IDH1 had been previously determined using a stopped flow spectrophotometer and were 49 μM for NADP+ and 65 μM for isocitrate.1 The Km for diaphorase for NADPH was reported as 1.1 μM for rat,15 but the exact Km for the diaphorase utilized herein was not measured. The amount of WT IDH1 enzyme was selected during initial assay design to have a good assay window (t = 10 min to t = 0 min) and a linear response at 10 min. A no-enzyme negative control (0×) was used to normalize the data along with full reaction (1×) positive control. 0× enzyme values corresponded to full inhibition (100% inhibitory activity) and 1× WT IDH1 enzyme values were used to normalize to no inhibition (0% inhibitory activity). The detailed protocol for WT IDH1 is displayed in Table 1.
Table 1.
Protocol for Wild-Type IDH1 qHTS Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Dispense enzyme | 3 μL | WT IDH1 |
| 2 | Pin-transfer controls | 23 nL | 1:3 dilution |
| 3 | Pin-transfer library compounds | 23 nL | 115–0.2 μM dilution series |
| 4 | Read fluorescence | Endpoint | ViewLux, fluorescence |
| 5 | Incubation time | 30 min | Room temperature |
| 6 | Dispense substrate/detection | 1 μL | NADP+, isocitrate, resazurin, diaphorase |
| 7 | Read fluorescence | 10 min, kinetic | ViewLux, fluorescence |
Step Notes
1. 0.06 μg/mL WT IDH1, 2.67 mM β-mercaptoethanol, 150 mM NaCl, 20 mM Tris pH 7.5, 10 mM MgCl2, 0.05% protease-free BSA.
2. Pintool (V&P Scientific) transfer (tip wash sequence, DMSO, IPA, 3-s vacuum dry).
3. Pintool (V&P Scientific) transfer (tip wash sequence, DMSO, IPA, 3-s vacuum dry).
4. 525 nm excitation, 598 nm emission, bodipy mirror, 0.5-s exposure, single read.
5. Lidded plate.
6. 0.24 mM NADP+, 0.08 mg/mL diaphorase, 0.05 mM resazurin, 0.32 mM isocitrate, 150 mM NaCl, 20 mM Tris pH 7.5, 10 mM MgCl2, 0.05% protease-free BSA.
7. 525 nm excitation, 598 nm emission, bodipy mirror, 0.5-s exposure, read at time = 0 and time = 10 min.
DMSO, dimethyl sulfoxide; IDH1, isocitrate dehydrogenase 1; qHTS, quantitative high-throughput screening; WT, wild type.
Briefly, enzyme is added to the plate followed by compounds, which are allowed to equilibrate with enzyme for 30 min before addition of the substrates and detection reagents. During this time, a preread can be performed to assess the inherent fluorescence of the compounds being tested. Addition of the substrates initiates the enzymatic reaction (t = 0), and the plate is immediately transferred to a plate reader to begin kinetic measurements. When multiple plates are being run, the kinetics can be measured with just two reads—an initial read (t = 0) and a final read (t = 10 min; in the linear region). This allows subsequent plates to be read during the 10-min incubation, optimizing the total assay run time. If only one plate is being run (or during assay optimization), collecting multiple data points (t = 0, 0.5, 1, …, 10 min) allows the signal to be plotted to ensure the delta (change in signal) is obtained in the linear region. Active inhibitors result in a decrease in fluorescence production over time relative to inactive compounds, concordant with a decrease in WT IDH1 enzyme activity and less formation of NADPH product, which is the substrate needed by diaphorase for the production of the fluorescent resorufin product. To apply this type of protocol to another dehydrogenase that produces NADH or NADPH, it is important to determine the (enzyme) that produces a linear response in a time frame that is manageable to one's assay handling system, be it robotic or manual. Additional considerations are to ensure that both diaphorase and resazurin are in excess and not rate limiting, so that the assay is indeed reporting on the kinetics of the enzyme of interest.
In contrast to WT IDH1, IDH1 R132H was run in endpoint mode, because both IDH1 R132H and diaphorase use NADPH as their substrate. As such, if added simultaneously, they would be competing for substrate. Instead, the mutant IDH1 enzyme assay is run until ∼80% completion, at which time the diaphorase/resazurin detection reagents are added. Three controls are used, a no-enzyme control (represents 100% inhibition), a control with no enzyme and no NADPH (represents reaction run to 100% completion), and the full reaction components along with DMSO vehicle control (represents the full uninhibited reaction). These three controls allow the % conversion to be readily calculated. For each new lot of IDH1 R132H enzyme, the concentration of enzyme and/or the time of reaction should be reassessed to ensure that the reaction conditions achieve ∼80% completion. This represents an optimal conversion for signal to background, but avoids inflating the measured IC50 by overcooking the reaction.14 As for WT IDH1, compounds that bound outside of the NADPH pocket were desired, so the concentration of NADPH used was well in excess of the Km, and the concentration of α-KG was set to just above the Km.14 The Kms have been previously reported for IDH1 R132H using a stopped flow spectrophotometer in two studies and were 820 or 965 μM for α-KG and 0.044 or 0.44 μM for NADPH.1,6 The amount of IDH1 R132H enzyme was selected such that 80% conversion was reached at 60 min as determined by an NADPH standard curve. The detailed protocol is displayed in Table 2, and the experiment was run at room temperature. Briefly, IDH1 R132H is added to the plate and incubated for 30 min with compounds. During this time, a preread can be done to assess the inherent fluorescence of the compounds being tested. Then, NADPH and α-KG are added to initiate the reaction. After 60 min of reaction, diaphorase and resazurin are added and the plate is read after 5 min.
Table 2.
Protocol for IDH1 R132H qHTS Assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Dispense enzyme | 3 μL | IDH1 R132H |
| 2 | Pin-transfer controls | 23 nL | 1:3 dilution series |
| 3 | Pin-transfer library compounds | 23 nL | 115–0.2 μM dilution series |
| 4 | Read fluorescence | Endpoint | ViewLux, fluorescence |
| 5 | Incubation time | 30 min | Room temperature |
| 6 | Dispense substrate | 3 μL | NAPDH/α-KG |
| 7 | Incubation time | 60 min | Room temperature |
| 8 | Dispense detection reagents | 3 μL | Diaphorase/resazurin reagents |
| 9 | Incubation time | 5 min | Room temperature |
| 10 | Read fluorescence | Endpoint | Envision, fluorescence |
Step Notes
1. 0.5 μg/mL IDH1 R132H, 4 mM β-mercaptoethanol, 150 mM NaCl, 20 mM Tris pH 7.5, 10 mM MgCl2, 0.05% BSA into columns 2–3, 5–48; same buffer but with no enzyme in column 1 and 4.
2. Pintool (V&P Scientific) transfer (tip wash sequence, DMSO, IPA, 3-s vacuum dry).
3. Pintool (V&P Scientific) transfer (tip wash sequence, DMSO, IPA, 3-s vacuum dry).
4. 525 nm excitation, 598 nm emission, bodipy mirror, 0.5-s exposure, single read.
5. Lidded plate.
6. 0.016 mM NADPH, 2 mM α-KG, 150 mM NaCl, 20 mM Tris pH 7.5, 10 mM MgCl2, 0.05% BSA into columns 1–3, 5–48; same buffer but no substrates into column 4.
7. Lidded plate.
8. 0.06 mg/mL diaphorase, 0.036 mM resazurin, 150 mM NaCl, 20 mM Tris pH 7.5, 10 mM MgCl2, 0.05% BSA into all columns.
9. Lidded plate.
10. 540 nm excitation, 590 nm emission, bodipy mirror, 10 flashes.
When very large quantities of plates are being run, the reagents should be kept on ice until dispensed, but the plates are still incubated at room temperature. It is important to allow NADPH powder to warm to room temperature before opening to prevent wetting. In addition, since NADPH can oxidize, it is preferable to prepare it immediately before an assay, or to store aliquots frozen until use. The solution of NADPH should be nearly colorless and becomes visibly yellow upon oxidation. This color change is the basis for the original colorimetric NAD(P)H assay. Bottles containing NADPH were protected from light during the assay.
While red-shifting the assay away from the NAD(P)H excitation and emission spectral regions will limit some of the artifacts caused by fluorescent compounds, it does not limit the possibility of interference entirely, so counterassays are crucial. As for all assays, it is important to consider what other phenomena, other than the desired biology, could contribute to an apparent hit in these assays. Compounds can interfere with an assay by fluorescing at the detection wavelength or quenching the fluorescence, as detailed in Hall et al. in this issue.4 To detect library compound fluorescence, the assay plate can be preread before adding the substrate to initiate the reaction. To detect quenching, resorufin (available commercially) alone can be plated in assay buffer, and compounds of interest can be added to determine whether they are able to quench the fluorescence of the product. Last, it is possible that compounds may interfere by inhibiting or activating the coupling enzyme, diaphorase, a phenomenon studied extensively for another coupling enzyme, luciferase.16 To assess this, one can run a counterassay in which the protein of interest (WT IDH1 or IDH1 R132H) is omitted and instead a mixture of NADPH/NADP+ at the approximate % conversion anticipated from the reaction is added to a plate in assay buffer (e.g., 80% conversion of IDH1 R132H, the counterassay would be run with 80% NADP+ and 20% NADPH), compounds are added, and then the diaphorase/resazurin detection is added and the plate is read after 5 min. Compounds that show an effect in this counterassay would indicate that they interfered with one or more of the detection assay components and can be triaged.
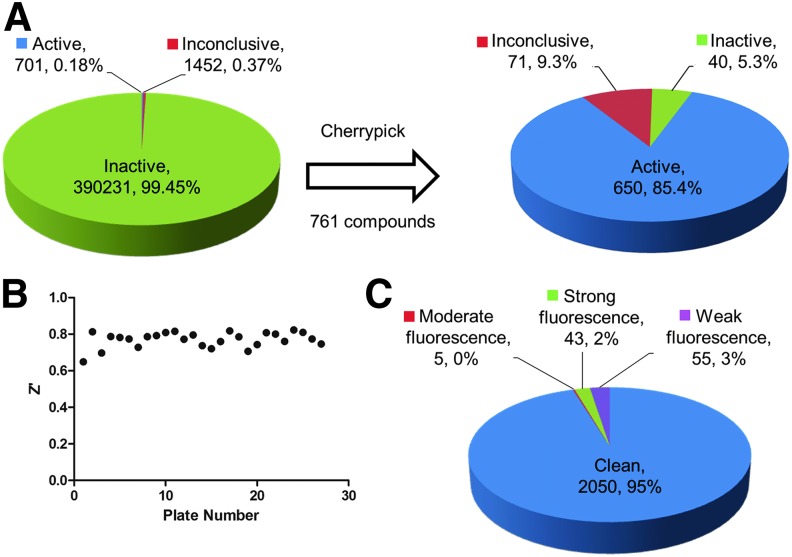
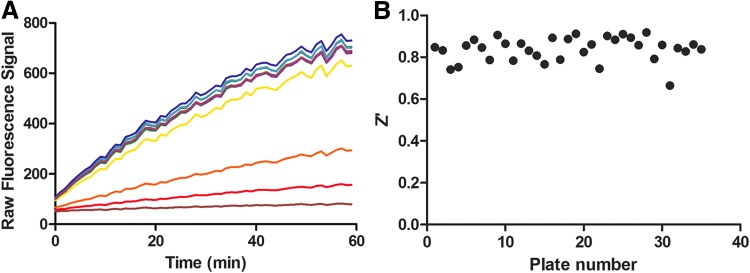
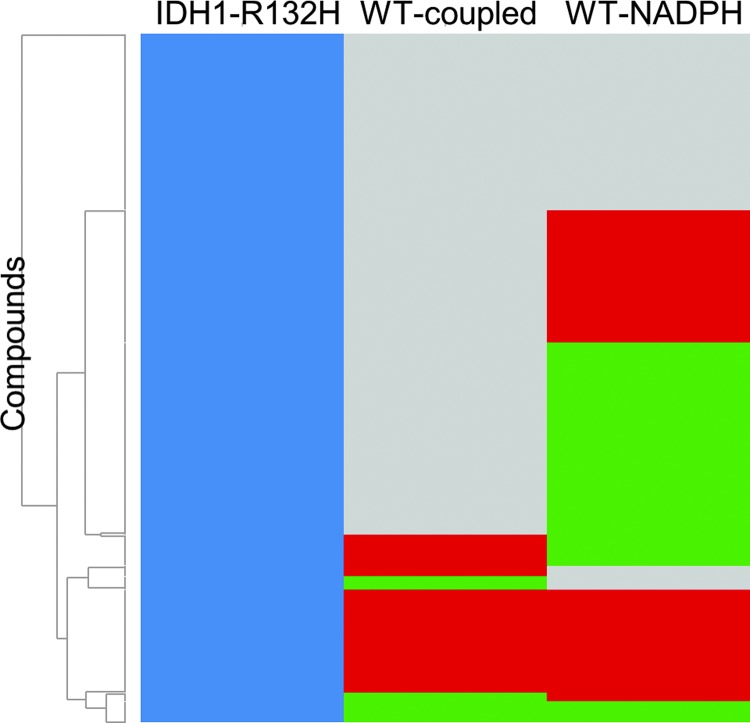
As mentioned earlier, the protocol for IDH1 R132H was used to screen the MLSMR library (PubChem AID 602179). Figure 2A shows the overall hit rate for the original diaphorase-coupled IDH1 R132H assay screened against over 300,000 compounds and the hit rate of the 761 compounds that were cherrypicked and retested, and Figure 2B shows the Z′ obtained from the automated assay validation across multiple plates. The assay has a respectable Z′ (0.77 ± 0.04) and the hit rate was manageable for follow-up studies. During the assay, after addition of compounds to the enzyme, the fluorescence is measured (i.e., a preread) to determine the inherent fluorescence of the compounds themselves. The 2,153 initial compounds that were designated as active or inconclusive (either had single-point activity or a low R2 combined with low efficacy) were assessed for inherent fluorescence during the preread (Fig. 2C) and were designated as strongly fluorescent, moderately fluorescent, or weakly fluorescent based on their fold increase in fluorescence over the basal level. Assessing WT IDH1 activity was important for the project, so hits from the IDH1 R132H screen were cherrypicked and tested against WT IDH1. A representative timecourse for a compound active against WT IDH1 is shown in Figure 3A. As the concentration of the active inhibitor increases, the rate of increase in signal is decreased. The signal is linear throughout the time measured. The WT IDH1 assay also has a respectable Z′ (0.83 ± 0.06) as shown in Figure 3B, which depicts Z′ data from 14 small independent experiments. For WT IDH1, both coupled and uncoupled HTS assay strategies were considered initially. Figure 4 shows a heatmap of the inhibitors of IDH1 R132H confirmed to be active in the cherrypick experiment alongside the results from the WT IDH1 counterscreen run in coupled and uncoupled formats. Compounds that were themselves fluorescent in the WT IDH1 assay or that decreased the signal are highlighted. True hits of WT IDH1 would be found within the compounds that decrease the signal in both the WT IDH1 coupled and uncoupled assays. As had been seen with other libraries,3 there is a much higher proportion of compounds with the potential to have fluorescence interference that will impact assay quality using the NADPH readout versus the red-shifted resorufin readout. As it is still possible that, but not very likely, a compound could interfere with the assay in other ways (e.g., aggregation), confirming direct binding of a particular compound of interest to the target is important and example methods that have been used for IDH1 R132H include surface plasmon resonance and microscale thermophoresis.6
Fig. 2.
Assay characteristics for IDH1 R132H. (A) Pie charts representing the results from screening diaphorase-coupled IDH1 R132H against the MLSMR collection (PubChem AID 602179). The hit rate of the initial IDH1 R132H primary screen is depicted on the left where compounds are classified as active, inactive, or inconclusive (i.e., single-point activity or low efficacy and low R2). A total of 761 active and inconclusive compounds were cherrypicked and retested, and their activity breakdown is shown on the right. (B) Plot of Z′ versus plate number for the online IDH1 R132H validation assay. (C) The fluorescence properties of the cherrypick compounds themselves. The primary IDH1 R132H assay contained a preread step after compound addition and before starting the enzyme reaction to identify compounds that were themselves fluorescent and would interfere with the detection of resorufin fluorescence. Weak fluorescence = less than twofold of basal fluorescence, moderate fluorescence = twofold to fivefold of basal fluorescence, strong fluorescence = greater than fivefold of basal fluorescence, and clean = no interference.
Fig. 3.
Assay characteristics for WT IDH1. (A) Single-well reaction timecourses for WT IDH1 and varying concentrations of an inhibitor. The top concentration of inhibitor is the lowest line on the graph (100% inhibition; 57 μM), and as the inhibitor is diluted, the slope increases as the reaction moves from complete inhibition (red line) to no inhibition (purple line; 0.00097 μM). (B) Plot of Z′ versus plate number compiled from 14 small independent experiments. WT, wild type.
Fig. 4.
Heatmap of the cherrypick library. The left column shows the compounds that inhibited IDH1 R132H (blue). The center column shows the WT IDH1 coupled assay (resorufin detection), and the right column shows the WT IDH1 uncoupled assay (NADPH detection). Compounds that were fluorescent in the WT assays are green, compounds that decreased the assay signal (quenchers or possible true WT IDH1 true hits) are red, and inactives are gray. Hierarchical clustering analysis was done using Spotfire DecisionSite 8.2.
The protocols detailed here describe assays that measure biochemical reactions generating NADPH or utilizing NADPH. These protocols provide a good starting point for the development of assays for other dehydrogenases that make or utilize NAD(P)H. These coupled assays should find utility as either a primary assay or a confirmatory assay in dehydrogenase drug development campaigns.
Abbreviations Used
- α-KG
alpha-ketoglutarate
- FRD
flying reagent dispenser
- IDH1
isocitrate dehydrogenase 1
- MLSMR
molecular libraries small molecule repository
- qHTS
quantitative high-throughput screening
- WT
wild type
Acknowledgments
This research was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research and the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure Statement
No competing financial interests exist.
References
- 1.Dang L, White DW, Gross S, et al. : Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Chai S, Wang P, et al. : Aldehyde dehydrogenases and cancer stem cells. Cancer Lett 2015;369:50–57 [DOI] [PubMed] [Google Scholar]
- 3.Simeonov A, Jadhav A, Thomas CJ, et al. : Fluorescence spectroscopic profiling of compound libraries. J Med Chem 2008;51:2363–2371 [DOI] [PubMed] [Google Scholar]
- 4.Hall MD, Simeonov A, Davis MI: Avoiding fluorescence assay interference—The case for diaphorase. Assay Drug Dev Technol 2016;14:175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, Ye D, Guan K-L, Xiong Y: IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin Cancer Res 2012;18:5562–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MI, Gross S, Shen M, et al. : Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. J Biol Chem 2014;289:13717–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M, Pragani R, Popovici-Muller J, et al. : ML309: A potent inhibitor of R132H mutant IDH1 capable of reducing 2-hydroxyglutarate production in U87 MG glioblastoma cells. Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda, MD: National Center for Biotechnology Information (US); 2010. Available at www.ncbi.nlm.nih.gov/books/NBK153220/?report=classic, 2012 (last accessed February1, 2016) [PubMed] [Google Scholar]
- 8.Popovici-Muller J, Saunders JO, Salituro FG, et al. : Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett 2012;3:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inglese J, Auld DS, Jadhav A, et al. : Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A 2006;103:11473–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boethling RS, Weaver TL: A new assay for diaphorase activity in reagent formulations, based on the reduction of thiazolyl blue. Clin Chem 1979;25:2040–2042 [PubMed] [Google Scholar]
- 11.Li W, Sauve A: NAD+ Content and its role in mitochondria. In: Mitochondrial Regulation, Vol. 1241 Palmeira CM, Rolo AP. (eds.), pp. 39–48. Springer, New York, 2015 [DOI] [PubMed] [Google Scholar]
- 12.Yasgar A, Shinn P, Jadhav A, et al. : Compound management for quantitative high-throughput screening. JALA 2008;13:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michael S, Auld D, Klumpp C, et al. : A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol 2008;6:637–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strelow J, Dewe W, Iversen PW, et al. : Mechanism of action assays for enzymes. In: Assay Guidance Manual. Sittampalam GS, Coussens NP, Nelson H, et al. (eds.), pp. 1–15. Bethesda, MD, 2012. Available at www.ncbi.nlm.nih.gov/books/NBK92001/?report=reader (last accessed February1, 2016) [Google Scholar]
- 15.Kemp MC, Kuonen DR, Roberts PJ: Kinetics of rat brain NADPH-diaphorase. Biochem Soc Trans 1987;15:501 [Google Scholar]
- 16.Auld DS, Southall NT, Jadhav A, et al. : Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem 2008;51:2372–2386 [DOI] [PubMed] [Google Scholar]