Abstract
In this issue of Blood, Kanakry et al analyzed Epstein-Barr virus (EBV) DNA expression in peripheral blood cellular compartments from 2146 patients with various malignancies and found that cell-free plasma EBV DNA was a better marker of EBV+ disease than EBV DNA from peripheral blood mononuclear cells (PBMCs).1
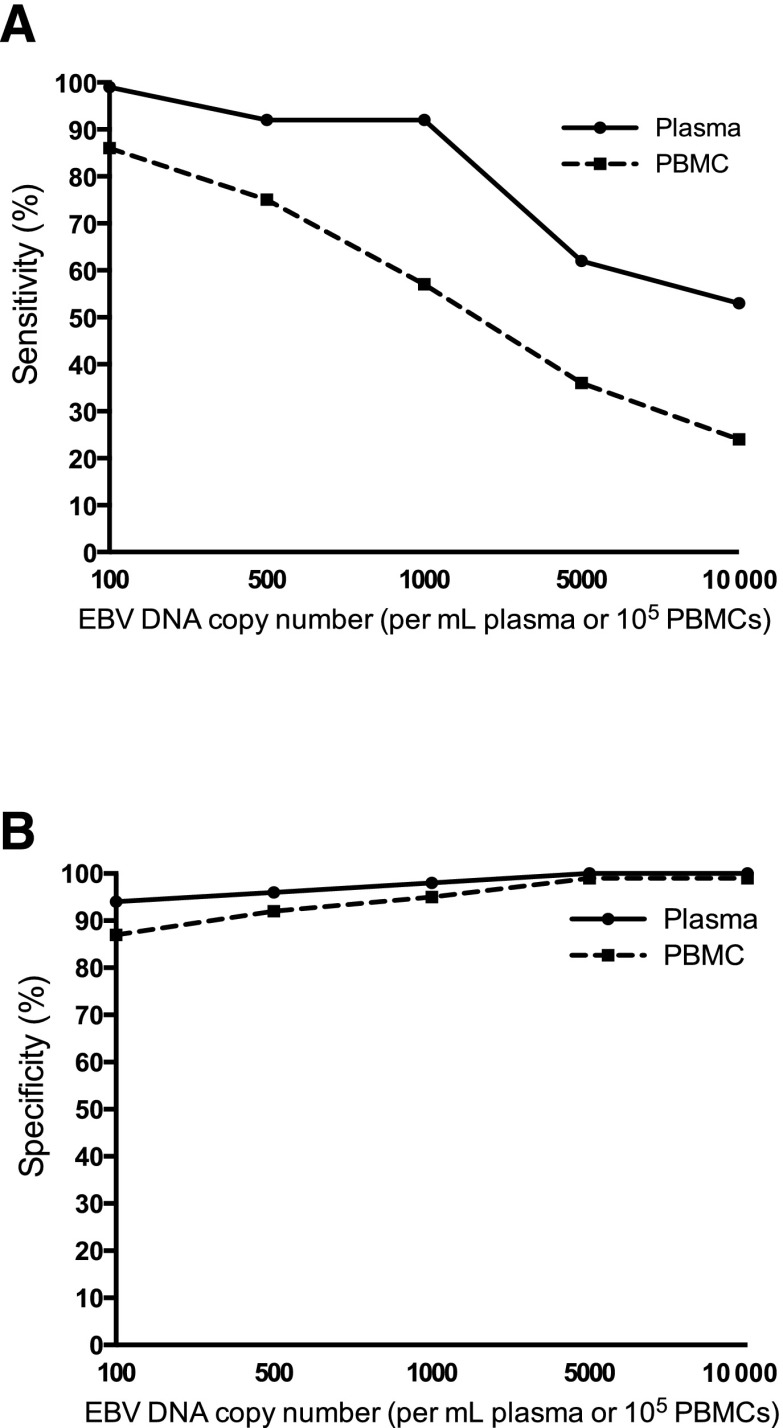
EBV DNA in plasma had better sensitivity (A) and specificity (B) than PBMCs in distinguishing between patients with active, systemic EBV+ disease and patients that did not have prior, current, or subsequent history of EBV+ disease. See Figure 2B-C in the article by Kanakry et al that begins on page 2007.
EBV is a human herpesvirus which primarily infects B cells and epithelial cells. It is one of the most common viruses, with >90% of adults having evidence of previous infection. In children, the symptoms (if any) are usually mild and flulike, whereas in adolescents and young adults, EBV can cause infectious mononucleosis (IM), also known as glandular fever or “kissing disease.” Once infected with EBV, the virus remains latently present in B cells in the blood for the rest of the individual’s life. In addition, EBV infection has been associated with the development of a variety of human malignancies, including several types of lymphoma, lymphoproliferative disorder, and nasopharyngeal carcinoma (NPC).2
In the present study, plasma and PBMC samples of 2146 patients with various diseases, including IM, hemophagocytic lymphohistiocytosis, posttransplantation lymphoproliferative disorder, non-Hodgkin lymphoma, Hodgkin lymphoma, lymphoproliferative disorder, NPC, and oral hairy leukoplakia, are analyzed for EBV DNA expression and its association with EBV+ disease. The authors show that EBV DNA in plasma has a higher sensitivity and specificity for distinguishing patients with an active, systemic EBV+ disease from patients with no prior, current, or subsequent history of EBV+ disease than PBMCs (see figure). They describe a positive correlation between EBV DNA copy number and active, systemic EBV+ disease, compared with patients with EBV+ disease in remission and those with no EBV+ disease, both in PBMCs and plasma. Finally, in patients with no EBV+ disease, EBV DNA is more often present in PBMCs than in plasma. These data lead to the conclusion that cell-free (plasma) EBV DNA performs better than cellular (PBMC) EBV DNA as a marker for EBV+ disease.
The clinical importance of this study lies in the fact that it was done on a very large set of patients, and that it helps to bring clarity to the conflicting data that have been published so far on which blood component is best to be used for EBV DNA expression as a marker for EBV+ disease.3,4 Several additional studies on smaller sets of patients were recently published, reporting that EBV DNA positivity was associated with worse survival in chronic lymphocytic leukemia (CLL).5,6 All these together support the concept that development of vaccines against EBV in patients at risk for aggressive disease could be a future prophylactic approach for the prevention of the deadly metastatic disease or of Richter transformation. Furthermore, EBV-derived biomarkers can be used to predict the natural history of an EBV+ cancer.
Although Kanakry et al found that detection of EBV DNA in plasma was uncommon in the absence of EBV+ disease, there were still 6% of patients in which EBV DNA was detected in plasma although they did not present with EBV+ disease. Therefore, it might be worth considering markers other than EBV DNA to screen for EBV-associated diseases or for evaluating response to therapy. In this regard, Epstein-Barr viral microRNAs (miRNAs) may be of use. EBV miRNAs are located within 2 transcripts of the viral genome, the BamHI fragment H rightward open reading frame 1 (BHRF1) and the BamHI A rightward transcripts (BARTs).7 Recently, the involvement of EBV viral miRNAs in some EBV-associated malignancies was described. For example, viral miRNA BHRF1-1 is expressed significantly higher in plasma of patients with CLL when compared with normal individuals, and BHRF1-1 expression in B cells of patients with CLL is associated with shorter overall survival.8 Similarly, Zhang et al9 found expression of BART7 and BART13 viral miRNAs in plasma of patients with NPC, whereas these miRNAs are only rarely expressed in healthy individuals and non-NPC tumor patient controls. In addition, higher EBV miRNA expression is associated with advanced disease stages, and the expression is reduced upon radiotherapy.9 Also in NPC, BART17 was found to be highly expressed in plasma samples from NPC patients, when compared with non-NPC controls.10
Whether expression of EBV miRNAs can be used as a biomarker for other EBV- associated diseases still remains to be determined, but the first reports seem promising and the research worth continuing.
Footnotes
Conflict-of interest disclosure: The authors declare no competing financial interests.
REFERENCES
- 1.Kanakry JA, Hegde AM, Durand CM, et al. The clinical significance of EBV DNA in the plasma and peripheral blood mononuclear cells of patients with or without EBV diseases. Blood. 2016;127(16):2007–2017. doi: 10.1182/blood-2015-09-672030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Hakim H, Gibson C, Pan J, et al. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J Clin Microbiol. 2007;45(7):2151–2155. doi: 10.1128/JCM.02308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai DE, Luskin MR, Kremer BE, et al. A pilot trial of quantitative Epstein-Barr virus polymerase chain reaction in patients undergoing treatment for their malignancy: potential use of Epstein-Barr virus polymerase chain reaction in multiple cancer types. Leuk Lymphoma. 2015;56(5):1530–1532. doi: 10.3109/10428194.2014.963577. [DOI] [PubMed] [Google Scholar]
- 5.Grywalska E, Roliński J, Pasiarski M, et al. High viral loads of Epstein-Barr virus DNA in peripheral blood of patients with chronic lymphocytic leukemia associated with unfavorable prognosis. PLoS One. 2015;10(10):e0140178. doi: 10.1371/journal.pone.0140178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang JH, Gao R, Xia Y, et al. Prognostic impact of Epstein-Barr Virus (EBV)-DNA copy number at diagnosis in chronic lymphocytic leukemia. Oncotarget. 2015;7(2):2135–2142. doi: 10.18632/oncotarget.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klinke O, Feederle R, Delecluse HJ. Genetics of Epstein-Barr virus microRNAs. Semin Cancer Biol. 2014;26:52–59. doi: 10.1016/j.semcancer.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Ferrajoli A, Ivan C, Ciccone M, et al. Epstein-Barr virus microRNAs are expressed in patients with chronic lymphocytic leukemia and correlate with overall survival. EBioMedicine. 2015;2(6):572–582. doi: 10.1016/j.ebiom.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136(5):E301–E312. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 10.Gourzones C, Ferrand FR, Amiel C, et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients--evidence of non-exosomal transport. Virol J. 2013;10:119. doi: 10.1186/1743-422X-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]