Abstract
Anaerobic digestion (AD) involves a consortium of microorganisms that convert substrates into biogas containing methane for renewable energy. The technology has suffered from the perception of being periodically unstable due to limited understanding of the relationship between microbial community structure and function. The emphasis of this review is to describe microbial communities in digesters and quantitative and qualitative relationships between community structure and digester function. Progress has been made in the past few decades to identify key microorganisms influencing AD. Yet, more work is required to realize robust, quantitative relationships between microbial community structure and functions such as methane production rate and resilience after perturbations. Other promising areas of research for improved AD may include methods to increase/control (1) hydrolysis rate, (2) direct interspecies electron transfer to methanogens, (3) community structure–function relationships of methanogens, (4) methanogenesis via acetate oxidation, and (5) bioaugmentation to study community–activity relationships or improve engineered bioprocesses.
Keywords: anaerobic digestion, bioaugmentation, methanogenesis, microbial community
Introduction
Anaerobic digestion (AD) is a microbial process that converts organic matter to biogas containing CH4 and CO2 in an anaerobic environment.1 Although the technology has been employed for decades, it has received renewed attention as it provides a more sustainable alternative to waste treatment over energy-intensive methods of the past.2,3 Compared with traditional aerobic treatment, AD has several potential advantages such as lower operational costs from lack of aeration requirements, energy production from biomethane, significantly less biomass production, which reduces handling and disposal costs, and the ability to degrade certain pollutants, which cannot be aerobically removed.4
Anaerobic conversion of organics to biogas involves a multistep process involving interactions among many different bacterial and archaeal species. With the increasing application of AD, there is a steady effort by both engineers and microbiologists working in this field to increase the existing knowledge of the complex, interacting microbial community that governs the overall AD process. New knowledge is crucial in order to develop better models and design improved AD systems. One key area requiring new knowledge involves the quantitative relationship between microbial community structure and AD process function. The aim of this review is to provide insight into the microbiology of anaerobic digesters and recent studies describing both qualitative and quantitative relationships between microbial community and digester function. The gaps in current knowledge and suggestions for future research are also described.
Phases in AD Process
Conceptually, the microbial processes of AD can be described by the sequential steps of hydrolysis, acidogenesis, acetogenesis, and methanogenesis.5 Each of these steps is accomplished by a guild of microorganisms, and it is critical to maintain a balanced reaction rate among the steps or guilds to ensure rapid and stable digestion. As described above, balanced essentially means that acid- and H2-consuming reactions are fast or potentially faster than acid- and H2-producing steps. Buildup of H2 partial pressure to more than 10−4 atm inhibits the destruction of propionate and butyrate intermediates.6 Accumulation of these volatile fatty acid (VFA) intermediates can drop the pH of the digester and slow or stop methanogenesis. In addition, the rate of one of these steps limits the overall rate of methanogenesis, and the identity of the rate-limiting step can differ among systems based on substrate chemical structure and other parameters. Most importantly, increasing the rate of the rate-limiting step will increase methane production rates, whereas improving other steps will have a little impact.
Hydrolysis
Hydrolysis involves the breakdown of polymeric substrates, such as polysaccharides, lipids, and proteins, to their respective monomers or oligomers using extracellular enzymes. These enzymes generally include amylase, cellulase, lipase, pectinase, and protease.7 Hydrolytic bacteria are phylogenetically diverse; however, two phyla, namely, Bacteroidetes and Firmicutes, include most of the known species. Compared with methanogens, hydrolytic bacteria grow rapidly and have lower sensitivity to changes in environmental factors, such as pH and temperature. For relatively recalcitrant substrates, such as lignin, hydrolysis is often the rate-limiting step for CH4 production. In addition to substrate chemical structure, hydrolysis rate depends on factors such as particle size, pH, enzyme production, diffusion, and adsorption of enzymes on the substrate particles.8–10 Methods to increase hydrolysis rate using mechanical, chemical, and biological processes have been developed,11 but a thorough review is outside the scope of this document.
Acidogenesis and syntrophic acetogenesis
In acidogenesis, products of hydrolysis are converted primarily to VFAs, which include acetate, propionate, isobutyrate, butyrate, valerate, and isovalerate. Besides VFAs, other products of acidogenesis include alcohols, lactate, formate, CO2, and H2. Bacteroidetes, Chloroflexi, Firmicutes, and Proteobacteria are phyla that contain most identified species of acidogenic bacteria.12–15 Acidogenesis is generally rapid, which can lead to accumulation of VFAs and a drop in pH when acid utilization is inhibited or too slow due to organic overload, toxicants, or rapid temperature change. The pH drop can inhibit or stop methanogenesis completely.
Although methanogens can directly use acetate, formate, H2/CO2, and methyl compounds, other intermediates formed via acidogenesis, such as propionate, butyrate, isobutyrate, valerate, isovalerate, and ethanol, have to be further biodegraded by other microorganisms before methanogens can utilize them to produce methane. Syntrophic acetogenesis is the process in which these intermediates are further biotransformed to form acetate, H2, and CO2. Fermentation of propionate via syntrophic acetogenesis is critical because approximately 30% of complex substrates in municipal wastewater solids and other wastes can be shuttled through propionate to CH4 during AD under typical conditions.16 Most of the medium- to long-chain fatty acids resulting from hydrolysis of lipids and lignocellulosic compounds are also biotransformed to acetate, H2, and CO2 through this process.
Under standard conditions, syntrophic acetogenesis is thermodynamically unfavorable and only proceeds if the partial pressure of H2 is lower than 10−4 atm.6,17 Hydrogenotrophic methanogens and/or other H2 utilizers live in syntrophy with acetogens, consuming H2 released from the latter.18 The syntrophic relationship makes acetogenesis thermodynamically feasible. Formic acid (HCOOH) and formate are similar to H2 since they are essentially H2 associated with CO2 (ie, H2 + CO2 = HCOOH). Therefore, interspecies formate transfer has also been observed to play a critical syntrophic role. In addition, acetogenesis from other higher molecular weight organic acids also relies on syntrophy with H2 or formate utilizers. This syntrophy is based on H2/formate transfer from H2-producing to H2-consuming microorganisms, which is commonly referred to as interspecies H2 transfer.19 The H2 also can be thought of as protons (H+) with associated electrons, and interspecies hydrogen/formate transfer is also interspecies electron transfer. Interestingly, a recent study has shown that some microorganisms perform direct interspecies electron transfer using electrically conductive pili and electrons can be shuttled in this way from, for example, Geobacter to Methanosaeta.20–23 This interspecies electron transfer is rapid and may prove to be an important mechanism for stable AD in the future; more research is warranted to more fully understand direct interspecies electron transfer mechanisms and how it can be encouraged in engineered systems.
Most commonly observed syntrophic acetogens in anaerobic digesters involved in propionate degradation belong to the genera such as Pelotomaculum, Smithllela, and Syntrophobacter.24–26 Oxidation of butyrate and other fatty acids are performed by acetogens belonging to the genera Syntrophus and Syntrophomonas.26–28 Syntrophic acetogenesis is a critical and, often, rate-limiting step to maintain rapid, stable AD operation because some of the VFAs, particularly propionate, inhibit methanogenesis at high concentrations even at neutral pH.1,29–31 Syntrophic acetogens play a critical role in the overall AD process, but have not been thoroughly studied, in part, due to the difficulty in maintaining pure cultures and lack of primers to identify them in mixed cultures using molecular techniques.32
Methanogenesis
The final step in biomethane production, methanogenesis, is performed by a specialized group of microorganisms belonging to the domain archaea, called methanogens. There are three known types of methanogens: acetoclastic, hydrogenotrophic, and methylotrophic. Acetoclastic methanogens convert acetate to CH4 and CO2, hydrogenotrophic methanogens use H2 or formate to reduce CO2 to CH4, and methylotrophic methanogens produce CH4 from methyl compounds, such as methanol, methylamines, and methylsulfides.33 In typical municipal anaerobic digesters, about 70% of the CH4 is produced from acetate, and the rest from H2 and CO2. Only a minimal amount of CH4 is produced via methylotrophic methanogenesis.34
Hydrogenotrophic methanogens are critical for AD process owing to their ability to scavenge H2 and maintain the partial pressure low. Methanobacterium, Methanobrevibacter, Methanoculleus, Methanospirillum, and Methanothermobacter are the most commonly observed hydrogenotrophic methanogens in anaerobic digesters.35–38
Acetoclastic methanogens belong to two genera: Methanosaeta and Methanosarcina. Methanosaeta are obligate acetoclastic methanogens and are only known to use acetate or direct electron transport as the substrate or electron donor. Methanosaeta have a relatively slow growth rate but possess a high affinity for acetate and hence dominate at low acetate concentration.33 Methanosarcina are facultative acetoclastic methanogens. Most Methanosarcina species can use H2/CO2, and C-1 compounds in addition to acetate.33,39,40 In addition to its wider range of substrates, Methanosarcina has a higher growth rate and lower affinity for acetate so it can dominate over Methanosaeta in digesters, where the concentration of acetate is high.38–40 The filamentous morphology of Methanosaeta can play an important role in granule formation since the filaments act as binders to help hold the granule together. Many AD configurations, such as the upflow anaerobic sludge blanket, rely on the formation of these microbial agglomerations, called granules that are 1–5 mm particles, containing microbes that settle rapidly. When granulation does not occur in these bioreactors, the process is difficult to maintain and can fail.
Syntrophic acetate-oxidizing bacteria
Under certain conditions, an alternative pathway for CH4 production from acetate has been observed in some anaerobic digesters. This pathway combines the conversion of acetate to H2 and CO2 by acetate-oxidizing bacteria that are subsequently converted to CH4 by hydrogenotrophic methanogens.41 Only few species of microorganisms have been identified that perform syntrophic acetate oxidization in conjunction with H2-consuming methanogens, which include strain AOR (ie, Reversibacter), Clostridium ultunense, Thermacetogenium phaeum, Tepidanaerobacter acetatoxydans, Thermotoga lettingae, and Syntrophaceticus schinkii.42–45 This pathway is not believed to be a typical AD pathway for CH4 production because acetoclastic methanogens outcompete syntrophic acetate-oxidizing bacteria in most digesters; however, more work is required to understand the importance of the process in AD systems.46 Under conditions that might be inhibitory to acetoclastic methanogens, such as high ammonia (>3 g/L NH3-N) or sulfate concentration and/or high temperature, this pathway can be critical for biogas production.42,46–51 Studies have also shown that a long hydraulic retention time along with a low abundance of Methanosaeta can promote a shift toward syntrophic acetateoxidizing pathway from acetoclastic methanogenesis.52,53
Environmental Parameters Affecting Digester Microbial Community
Many studies have reported the influence of environmental parameters on the microbial community structure of a digester, primarily focusing on the methanogenesis pathway since it plays a direct role in reducing the pollutant load and producing CH4 as a renewable energy source.54 Compared with bacteria, methanogens have a lower growth rate and are sensitive to environmental disturbances, such as pH decline, high VFA, and ammonia concentrations.55,56 Environmental parameters such as pH, temperature, substrate concentration, substrate composition, and presence of toxic or inhibitory compounds can cause a shift in the methanogenic community structure and affect the overall digestion process.55
Compared with thermophilic temperature (55°C), the methanogenic community exhibits higher diversity at mesophilic temperature (37°C).33 Lowering the temperature further to psychrophilic values may shift the community from acetoclastic to hydrogenotrophic methanogens, but the relationship is currently unclear and requires additional research.57,58 Substrate disturbances, which include changes in the substrate concentration and composition, can affect the methanogenic community and its activity.59 Different substrates can lead to development of different methanogenic communities, for example, manure versus wastewater sludge56 and glucose versus whey permeate and sewage sludge.60 Higher acetate concentration can lead to Methanosarcina being selected as a dominant acetoclastic methanogen over Methanosaeta.61,62
In most large-scale industrial or municipal anaerobic digesters, changes in substrate concentration or substrate overload due to the variability in wastewater streams are the most common causes of digester instability. Of the four trophic phases, hydrolysis and acidogenesis can proceed at a faster rate than acetogenesis and methanogenesis.63 During substrate overload, the rate of formation of VFA intermediates is higher than that of their conversion to methane. Therefore, the VFAs accumulate to high concentrations in the digester, causing a pH decrease from the typical optimal values of pH 7–8 for efficient methanogenic activity.63 Apart from lowering the pH, VFAs can inhibit methanogenesis at high concentrations, and the inhibition is much stronger at lower pH values.64 The pH influences the ratio of undissociated to dissociated forms of VFAs, and the former is more toxic to microorganisms as the undissociated form can diffuse through cell membrane and cause damage by decreasing the intracellular pH.65
Many studies have investigated a wide range of environmental and nutrient factors that might severely inhibit the methanogenic process. Comprehensive reviews published by Blum and Speece66 and Chen et al55 provide detailed summaries of factors that might cause inhibition of anaerobic processes, which include specific organic chemicals, ammonia, sulfate/sulfite, and toxicity due to light metal ions (Na, K, Mg, Ca, and Al) and heavy metal ions (Fe, Zn, Ni, Co, Mo, Cu, etc.). However, it is important to note that metal ions are also essential trace elements for methanogenesis and are required at adequate concentrations, below inhibitory levels, for sustained methanogenesis.67
Relating Microbial Community Structure to Digester Stability and Function
Despite numerous reports describing the effect of environmental parameters on the microbial community structure, the reverse approach describing the influence of microbial community structure on digester function and its stability has been studied less. Researchers have just begun to utilize the information pertaining to microbial community structure to understand or predict its underlying influence on digester performance.
Qualitative structure–function relationships
Microbial diversity, specifically quantified as species richness (number of species) and evenness (relative abundance of species), has been shown to play an important role in both natural and engineered ecosystem function.68–73 Ecosystems containing more than one organism capable of performing a specific function (high richness) with a relatively equal abundance (high evenness) have a higher probability of functional redundancy or functional stability. It is a form of functional insurance for an ecosystem to have high richness and evenness based on compensatory growth. If the population of one species within a functional group is reduced or lost due to system perturbation, then another species from the same functional group, but higher resistance to the perturbation, may rapidly take its place if originally present in enough numbers.68–70,74
Engineered biological systems, such as anaerobic digesters, are often prone to and criticized for functional instability; therefore, studies involving functional resistance and resilience of biological treatment systems after perturbation have focused on relating species richness and evenness to overall functional stability. Although not a methanogenic system, Wittebolle et al72 working on denitrifying bacteria reported that communities with higher evenness exhibited higher rates of denitrification when exposed to salt toxicity compared with communities with low evenness. In parallel papers, Fernandez et al68 and Hashsham et al69 studying perturbation of methanogenic digesters using glucose overload concluded that greater functional stability was observed in communities exhibiting multiple microorganisms within the same functional group.
Apart from qualitatively linking species richness and evenness to digester stability during perturbation, studies have shown the relationship between microbial community structure and digester activity under nonperturbed conditions. Clustering analysis performed by Carballa et al75 using two molecular techniques, denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP), showed similar results, with a clear separation between the mesophilic and thermophilic communities. The bacterial and mesophilic communities were more diverse and even than the archaeal and thermophilic communities. The study also concluded that a digester with a higher evenness and diversity in its bacterial community resulted in a higher biogas/methane production.75 Tale et al76 measured specific methanogenic activity (SMA) against propionate for 14 different biomass samples from full-scale anaerobic digesters. Microbial community analysis was performed to elucidate only the methanogenic community structure defined by DGGE banding pattern of a gene ubiquitous to methanogens, the methyl coenzyme M reductase (mcrA) gene. Using the band intensities as a quantitative measure and employing principal component analysis, Tale et al76 showed that the digesters with high SMA values clustered together on the principal component analysis plot and correlated linearly with the DGGE banding patterns. Upon excising and sequencing the gel bands, the presence of hydrogenotrophic methanogens, Methanospirillum hungatei and Methanobacterium beijingense, was prominent in digesters with high propionate SMA. In another multiyear study, Werner et al73 looked at the bacterial communities of nine full-scale digesters treating brewery wastewater by employing 454 pyrosequencing to sequence the 16S rRNA gene. Using principal coordination analysis, they observed that digesters with higher SMA against acetate correlated with high community evenness.
Quantitative structure–function relation
Though the general understanding of the relationship between digester function and microbial community structure is increasing rapidly, the relationship is still difficult to quantitatively model. Current AD mathematical models consider biomass to be one independent population that is viewed as a black box77 or, as in the case of models such as AD Model 1 (ADM1) and others,78 as trophic groups containing one member. The lack of microbial community descriptors that may quantify, for example, functional redundancy in models is an obvious hurdle to improving design and operation of anaerobic digesters. The very important, but underappreciated, work of Ramirez et al79 began to tackle this issue by including microbial diversity concepts into an extended ADM1 model. However, more work is required to improve AD models using microbial community descriptors.
A few studies have reported a direct correlation between methanogenic activity and microbial community descriptors. Although not an anaerobic digester, Freitag and Prosser80 observed a linear correlation between the methanogenic activities of peat soil samples and mcrA gene copy numbers quantified using qPCR. The mcrA gene copy number has also been shown to linearly correlate with SMA values against H2/CO2 in four biomass samples.81 Regueiro et al82 reported that higher hydrolytic and methanogenic activities were correlated with higher Bacteroidetes and Archaea abundances. The linear correlation observed in these studies is encouraging; however, multiple linear regression (MLR) models, when performed using a small sample size and a high number of independent variables (10 DGGE bands in the case for Tale et al),76 are overfitted and not predictive.83 Therefore, a large number (ie, >30) of different microbial communities must be analyzed to develop statistically relevant empirical correlations, which is one issue that has limited the development of structure–function relationships.
Based on the studies by Tale et al76 and Morris et al,81 a study by Bocher et al84 utilized MLR modeling and addressed the issue of overfitting by increasing the sample size (49 samples) and reducing the number of independent variables (5 DGGE bands) to develop the MLR equations relating community and functional descriptors. Methanogenic microbial communities were assayed for methanogenic activity against glucose and propionate and the methanogenic community structure was quantified using mcrA gene DGGE band intensities. Of the 49 microbial samples, 30 were randomly selected and used as a training set to develop MLR equations relating propionate and glucose SMA values to band intensities [equations (1) and (2)]. The maximum correlation coefficient (R2) value was observed using a minimum of five bands. The MLR equations derived were then used to predict the activity of remaining 19 samples (the test set). In conclusion, the MLR equation described a regression with good quantitative predictability with the validation parameter (q2) value higher than the threshold value of 0.5 for glucose (q2 = 0.53) and propionate (q2 = 0.52) relationships.
| (1) |
| (2) |
SMAp and SMAg are the SMA values against propionate and glucose, respectively (mL CH4/mg iATP-h), and Xn is the demeaned, normalized band intensity value for band n on a DGGE gel of amplified mcrA products.84
To the best of authors’ knowledge, this is the only study that has reported a quantitative, predictive model between methanogenic community structure and anaerobic biomass activity. The model as described by equations (1) and (2) shows, for example, that the presence of methanogens represented by DGGE bands X10 and X15 for SMAp and X15 for SMAg positively correlates with higher SMA. This kind of information could be used in the future to select or design microbial communities to seed or bioaugment anaerobic digesters for more rapid methane production. Similarly, methanogens represented by bands X4, X8, and X14 for SMAp and bands X4, X7, and X11 for SMAg negatively correlate with higher SMA.
This is a first step and does not describe the many ways microbial community structure relates to digester function. In the future, however, these and other, more robust quantitative structure–activity relationships (QSARs) could be used to develop specific cultures that could increase process performance via digester seeding or bioaugmentation. The recently developed next-generation sequencing technologies may provide a breakthrough, as they allow sequencing of a large number of 16S rRNA gene PCR amplicon samples and have a rapid turnover time. At the same time, instead of analyzing for a specific functional or taxonomical group, next-generation sequencing can be used to thoroughly describe the digester microbial community, either by using a metagenomic approach, employing universal 16S rRNA gene primers, or by other approaches.85
Bioaugmentation as a tool to study structure–function relationships
Bioaugmentation is the practice of adding specialized or a mixed community of microorganisms to a system to obtain a desired process function.86 A recent review published by Herrero and Stuckey87 broadly covers the application of bioaugmentation in wastewater treatment. Bioaugmentation of anaerobic digesters has now been demonstrated in studies for reactor start-up,88 odor reduction,89 and degradation of organic compounds, including 3-chlorobenzoate,90 pentachlorophenol,91 tetrachloroethene,92 benzene,93 selenate and nitrate,94 phenol and cresol,95 fat, oil, and grease,96 and oleate,97 and to aid in the recovery of upset digesters.76,98,99
The relevance of bioaugmentation to study structure–function relationships comes from the underlying hypothesis that addition of an exogenous culture ostensibly alters the original microbial community that may, in turn, change digester function. Hence, if the microbial community structure of the augment culture, the original digester biomass, and their mixture are well characterized, then their functional activities could be used to relate function and community structure. This concept was tested in a study performed by Bocher,100 who compared the rates of propionate conversion to CH4 before and after bioaugmentation with a propionate degrading, methanogenic augment. Nine different biomass samples, each with a different microbial community, were collected from different full-scale anaerobic reactors. Bioaugmentation was done by mixing the augment with each digester biomass sample at an iATP ratio of 1:5 (augment:biomass). Six of the nine biomass samples assayed showed a statistically significant increase in the SMA after bioaugmentation. The bioaugmentation results were correlated with the dissimilarity (calculated as 1—Pearson’s correlation coefficient) between the methanogenic community structure of the augment and original digester biomass cultures (Fig. 1). The results of bioaugmentation were measured as the percentage increase in SMA against propionate, before and after bioaugmentation. The dissimilarity between the methanogenic community structure of the digester biomass and the augment culture was quantified as the distance, calculated using 1—Pearson’s correlation coefficient, of the mcrA DGGE banding patterns of each digester biomass sample and the augment culture.
Figure 1.
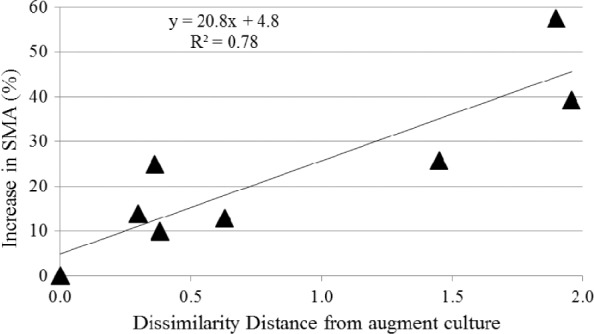
Difference between methanogen community structures in the augmented and biomass samples correlated with percent increase in SMA values.100
A linear correlation was observed and supported the argument that functional improvement (ie, increased rate of propionate degradation) after bioaugmentation is not only a function of the augment culture community structure but also the methanogenic community structure of the original biomass within a digester (ie, how much different it is from the augment culture). This correlation provides a rationale to further study bioaugmentation as a tool to analyze structure–function relationship in AD process. Bioaugmentation will not improve every existing anaerobic biomass, but may improve some, and a method to quantify potential improvement based on microbial community descriptors should exist.
Although linear models have been successful, other structure–activity models may prove to be more appropriate. This is similar to the historical development of quantitative QSARs for drug activity and other physiochemical parameter estimation over the past 80 years; these QSARs initially relied on linear relationships, but were later refined using nonlinear relationships such as artificial neural networks.101,102 In any event, the initial success using empirical, linear relationships encouraged the development of more robust and accurate empirical and mechanistic models. It is probable that more robust models can be developed in the near future to predict the function of engineered microbial processes using microbial community descriptors as well as environmental parameter values. These new, robust models will be very helpful to improve engineered bioprocesses.
Conclusion
As a biological treatment process, both efficiency and stability of AD technology depend fundamentally on the complex microbial communities and their activities in digesters. Owing to this, over the years, scientists and engineers working in this field have focused their attention to answering the central questions: (1) which microorganisms are present, (2) how many different types of microorganisms are present, (3) which microorganisms are active and growing, (4) how do microorganisms behave under certain environmental conditions, and (5) how does the microbial community structure relate to digester function.
Considerable progress has been made in the last decade to identify the key groups of microorganisms that influence the trophic phases of AD as well as how various environmental conditions affect the microbial community structure and digester function. Yet, more work is required to realize quantitative, predictive relationships between the complex microbial community structure and the digester functional output. A robust quantitative microbial structure–function relationship would be a holy grail for engineers and scientists who are looking to develop new predictive models that can be used to improve the design and operation of anaerobic digesters for waste treatment and renewable energy generation. However, for a valid quantitative relationship, it is essential to analyze the microbial community structure and monitor the functional and environmental parameters for a large sample of different anaerobic digester communities, and this has limited model development. Future experimental work can be envisioned, in which a large number of different microbial communities from various, controlled anaerobic digesters are analyzed using next-generation sequencing technology. The community and functional data could then be used to determine predictive, empirical, or mechanistic relationships between community structure and digester function descriptors, including CH4 production rate. It would be a worthwhile endeavor and an important step forward in this field.
Other promising areas of research for improved AD processes may include (1) methods to increase hydrolysis rate, (2) direct interspecies electron transfer to methanogens via conductive pili or other mechanisms, (3) community structure and function relationships of methanogenic communities, (4) methanogenesis via acetate-oxidizing bacteria, and (5) bioaugmentation to study microbial community–activity relationships or improve engineered bioprocesses.
Footnotes
ACADEMIC EDITOR: Professor Raúl Rivas, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2074 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: KV, BB, JM, and DZ. Analyzed the data: KV, BB, JM, and DZ. Wrote the first draft of the manuscript: KV and DZ. Contributed to the writing of the manuscript: KV, BB, JM, and DZ. Agreed with manuscript results and conclusions: KV, BB, JM, and DZ. Jointly developed the structure and arguments for the paper: KV, BB, JM, and DZ. Made critical revisions and approved the final version: KV, BB, JM, and DZ. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Pullammanappallil PC, Svoronos SA, Chynoweth DP, Lyberatos G. Expert system for control of anaerobic digesters. Biotechnol Bioeng. 1998;58(1):13–22. doi: 10.1002/(sici)1097-0290(19980405)58:1<13::aid-bit2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 2.Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol. 2004;22(9):477–485. doi: 10.1016/j.tibtech.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Lettinga G. Challenges of a feasible route towards sustainability in environmental protection. Front Environ Sci Eng China. 2010;4(2):123–134. [Google Scholar]
- 4.Suryawanshi PC, Chaudhari AB, Kothari RM. Mesophilic anaerobic digestion: first option for waste treatment in tropical regions. Crit Rev Biotechnol. 2010;30(4):259–282. doi: 10.3109/07388551.2010.487047. [DOI] [PubMed] [Google Scholar]
- 5.Bitton G. Wastewater Microbiology. 3rd ed. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- 6.McCarty PL, Smith DP. Anaerobic wastewater treatment. Environ Sci Technol. 1986;20(12):1200–1206. [Google Scholar]
- 7.Singh OV, Harvey SP. Sustainable Biotechnology. Dordrecht: Springer; 2010. [Google Scholar]
- 8.Noike T, Endo G, Chang JE, Yaguchi J, Matsumoto J. Characteristics of carbohydrate degradation and the rate-limiting step in anaerobic digestion. Biotechnol Bioeng. 1985;27(10):1482–1489. doi: 10.1002/bit.260271013. [DOI] [PubMed] [Google Scholar]
- 9.Mata-Alvarez J, Macé S, Llabrés P. Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. Bioresour Technol. 2000;74(1):3–16. [Google Scholar]
- 10.Vidal G. Influence of the content in fats and proteins on the anaerobic biodegradability of dairy wastewaters. Bioresour Technol. 2000;74(3):231–239. [Google Scholar]
- 11.Ariunbaatar J, Panico A, Esposito G, Pirozzi F, Lens PNL. Pretreatment methods to enhance anaerobic digestion of organic solid waste. Appl Energy. 2014;123:143–156. [Google Scholar]
- 12.Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36(1):1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 13.Balk M, Weijma J, Stams AJM. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol. 2002;52(pt 4):1361–1368. doi: 10.1099/00207713-52-4-1361. [DOI] [PubMed] [Google Scholar]
- 14.Dong X, Xin Y, Jian W, Liu X, Ling D. Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester. Int J Syst Evol Microbiol. 2000;50(pt 1):119–125. doi: 10.1099/00207713-50-1-119. [DOI] [PubMed] [Google Scholar]
- 15.Yamada T, Sekiguchi Y, Hanada S, et al. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol. 2006;56(pt 6):1331–1340. doi: 10.1099/ijs.0.64169-0. [DOI] [PubMed] [Google Scholar]
- 16.Speece RE, Boonyakitsombut S, Kim M, Azbar N, Ursillo P. Overview of anaerobic treatment: thermophilic and propionate implications. Water Environ Res. 2006;78(5):460–473. [PubMed] [Google Scholar]
- 17.Lowe SE, Jain MK, Zeikus JG. Biology, ecology, and biotechnological applications of anaerobic bacteria adapted to environmental stresses in temperature, pH, salinity, or substrates. Microbiol Rev. 1993;57(2):451–509. doi: 10.1128/mr.57.2.451-509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61(2):262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stams AJM, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7(8):568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 20.Rotaru AE, Shrestha PM, Liu F, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci. 2014;7(1):408. [Google Scholar]
- 21.Shrestha PM, Rotaru AE, Summers ZM, Shrestha M, Liu F, Lovley DR. Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl Environ Microbiol. 2013;79(7):2397–2404. doi: 10.1128/AEM.03837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita M, Malvankar NS, Franks AE, et al. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. MBio. 2011;2(4):e159–e111. doi: 10.1128/mBio.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Z, Zhang Y, Wang L, Quan X. Potential for direct interspecies electron transfer in an electric-anaerobic system to increase methane production from sludge digestion. Sci Rep. 2015;5:11094. doi: 10.1038/srep11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Balkwill DL, Aldrich HC, Drake GR, Boone DR. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int J Syst Bacteriol. 1999;49(pt 2):545–556. doi: 10.1099/00207713-49-2-545. [DOI] [PubMed] [Google Scholar]
- 25.de Bok FA, Stams AJ, Dijkema C, Boone DR. Pathway of propionate oxidation by a syntrophic culture of Smithella propionica and Methanospirillum hungatei. Appl Environ Microbiol. 2001;67(4):1800–1804. doi: 10.1128/AEM.67.4.1800-1804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imachi H, Sakai S, Ohashi A, et al. Pelotomaculum propionicicum sp. nov., an anaerobic, mesophilic, obligately syntrophic, propionate-oxidizing bacterium. Int J Syst Evol Microbiol. 2007;57(pt 7):1487–1492. doi: 10.1099/ijs.0.64925-0. [DOI] [PubMed] [Google Scholar]
- 27.Jackson BE, Bhupathiraju VK, Tanner RS, Woese CR, McInerney MJ. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol. 1999;171(2):107–114. doi: 10.1007/s002030050685. [DOI] [PubMed] [Google Scholar]
- 28.Sousa DZ, Smidt H, Alves MM, Stams AJM. Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with Methanobacterium formicicum. Int J Syst Evol Microbiol. 2007;57(pt 3):609–615. doi: 10.1099/ijs.0.64734-0. [DOI] [PubMed] [Google Scholar]
- 29.Barredo MS, Evison LM. Effect of propionate toxicity on methanogen-enriched sludge, Methanobrevibacter smithii, and Methanospirillum hungatii at different pH values. Appl Environ Microbiol. 1991;57(6):1764–1769. doi: 10.1128/aem.57.6.1764-1769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demirel B, Yenigun O. The effects of change in volatile fatty acid (VFA) composition on methanogenic upflow filter reactor (UFAF) performance. Environ Technol. 2002;23(10):1179–1187. doi: 10.1080/09593332308618336. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen H, Uellendahl H, Ahring B. Regulation and optimization of the bio-gas process: propionate as a key parameter. Biomass Bioenergy. 2007;31(11–12):820–830. [Google Scholar]
- 32.Mathai PP, Zitomer DH, Maki JS. Quantitative detection of syntrophic fatty acid degrading bacterial communities in methanogenic environments. Microbiology. 2015;161(6):1189–1197. doi: 10.1099/mic.0.000085. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 34.Ferry JG, editor. Methanogenesis Ecology, physiology, biochemistry and genetics. Chapman & Hall; New York: 1993. [Google Scholar]
- 35.Cuzin N, Ouattara AS, Labat M, Garcia JL. Methanobacterium congolense sp. nov., from a methanogenic fermentation of cassava peel. Int J Syst Evol Microbiol. 2001;51(pt 2):489–493. doi: 10.1099/00207713-51-2-489. [DOI] [PubMed] [Google Scholar]
- 36.Savant DV, Shouche YS, Prakash S, Ranade DR. Methanobrevibacter acididurans sp. nov., a novel methanogen from a sour anaerobic digester. Int J Syst Evol Microbiol. 2002;52(pt 4):1081–1087. doi: 10.1099/00207713-52-4-1081. [DOI] [PubMed] [Google Scholar]
- 37.Leclerc M, Delgènes J-P, Godon JJ. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol. 2004;6(8):809–819. doi: 10.1111/j.1462-2920.2004.00616.x. [DOI] [PubMed] [Google Scholar]
- 38.Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl Environ Microbiol. 2006;72(2):1623–1630. doi: 10.1128/AEM.72.2.1623-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westermann P, Ahring BK, Mah RA. Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria. Appl Environ Microbiol. 1989;55(2):514–515. doi: 10.1128/aem.55.2.514-515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conklin A, Stensel HD, Ferguson J. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res. 2006;78(5):486–496. doi: 10.2175/106143006x95393. [DOI] [PubMed] [Google Scholar]
- 41.Zinder SH, Koch M. Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Arch Microbiol. 1984;138(3):263–272. [Google Scholar]
- 42.Hattori S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 2008;23(2):118–127. doi: 10.1264/jsme2.23.118. [DOI] [PubMed] [Google Scholar]
- 43.Fotidis IA, Karakashev D, Angelidaki I. Bioaugmentation with an acetateoxidising consortium as a tool to tackle ammonia inhibition of anaerobic digestion. Bioresour Technol. 2013;146:57–62. doi: 10.1016/j.biortech.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 44.Westerholm M, Roos S, Schnürer A. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol Lett. 2010;309(1):100–104. doi: 10.1111/j.1574-6968.2010.02023.x. [DOI] [PubMed] [Google Scholar]
- 45.Westerholm M, Roos S, Schnürer A. Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst Appl Microbiol. 2011;34(4):260–266. doi: 10.1016/j.syapm.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Rui J, Qiu Q, Lu Y. Syntrophic acetate oxidation under thermophilic methanogenic condition in Chinese paddy field soil. FEMS Microbiol Ecol. 2011;77(2):264–273. doi: 10.1111/j.1574-6941.2011.01104.x. [DOI] [PubMed] [Google Scholar]
- 47.Schnurer A, Zellner G, Svensson BH. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol Ecol. 1999;29(3):249–261. [Google Scholar]
- 48.Schnurer A, Nordberg A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci Technol. 2008;57(5):735–740. doi: 10.2166/wst.2008.097. [DOI] [PubMed] [Google Scholar]
- 49.Westerholm M, Levén L, Schnurer A. Bioaugmentation of syntrophic acetateoxidizing culture in biogas reactors exposed to increasing levels of ammonia. Appl Environ Microbiol. 2012;78(21):7619–7625. doi: 10.1128/AEM.01637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu F, Hao L, Guan D, Qi Y, Shao L, He P. Synergetic stress of acids and ammonium on the shift in the methanogenic pathways during thermophilic anaerobic digestion of organics. Water Res. 2013;47(7):2297–2306. doi: 10.1016/j.watres.2013.01.049. [DOI] [PubMed] [Google Scholar]
- 51.Hao LP, Lu F, He PJ, Li L, Shao LM. Predominant contribution of syntrophic acetate oxidation to thermophilic methane formation at high acetate concentrations. Environ Sci Technol. 2011;45(2):508–513. doi: 10.1021/es102228v. [DOI] [PubMed] [Google Scholar]
- 52.Shigematsu T, Tang Y, Kobayashi T, Kawaguchi H, Morimura S, Kida K. Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl Environ Microbiol. 2004;70(7):4048–4052. doi: 10.1128/AEM.70.7.4048-4052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karakashev D, Batstone DJ, Trably E, Angelidaki I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol. 2006;72(7):5138–5141. doi: 10.1128/AEM.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89(6):670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Cheng JJ, Creamer KS. Inhibition of anaerobic digestion process: a review. Bioresour Technol. 2008;99(10):4044–4064. doi: 10.1016/j.biortech.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 56.Karakashev D, Batstone DJ, Angelidaki I. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl Environ Microbiol. 2005;71(1):331–338. doi: 10.1128/AEM.71.1.331-338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enright AM, McGrath V, Gill D, Collins G, O’Flaherty V. Effect of seed sludge and operation conditions on performance and archaeal community structure of low-temperature anaerobic solvent-degrading bioreactors. Syst Appl Microbiol. 2009;32(1):65–79. doi: 10.1016/j.syapm.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 58.Zhang D, Zhu W, Tang C, et al. Bioreactor performance and methanogenic population dynamics in a low-temperature 5–18°C) anaerobic fixed-bed reactor. Bioresour Technol. 2012;104:136–143. doi: 10.1016/j.biortech.2011.10.086. [DOI] [PubMed] [Google Scholar]
- 59.Boe K. Online Monitoring and Control of the Biogas Process [Ph.D. dissertation] Kongens Lyngby: Technical University of Denmark; 2006. Available at: http://www2.er.dtu.dk/publications/fulltext/2006/MR2006-055.pdf. [Google Scholar]
- 60.Lee C, Kim J, Hwang K, O’Flaherty V, Hwang S. Quantitative analysis of methanogenic community dynamics in three anaerobic batch digesters treating different wastewaters. Water Res. 2009;43(1):157–165. doi: 10.1016/j.watres.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 61.Griffin ME, McMahon KD, Mackie RI, Raskin L. Methanogenic population dynamics during start-up of anaerobic digesters. Biotechnol Bioeng. 1998;57(3):342–355. doi: 10.1002/(sici)1097-0290(19980205)57:3<342::aid-bit11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 62.McMahon KD, Stroot PG, Mackie RI, Raskin L. Anaerobic codigestion of municipal solid waste and biosolids under various mixing conditions—II: microbial population dynamics. Water Res. 2001;35(7):1817–1827. doi: 10.1016/s0043-1354(00)00438-3. [DOI] [PubMed] [Google Scholar]
- 63.Čater M, Fanedl L, Logar RM. Microbial community analyses in biogas reactors by molecular methods. Acta Chim Slov. 2013;60(2):243–255. [PubMed] [Google Scholar]
- 64.Deublein D, Steinhauser A. Biogas from Waste and Renewable Resources: An Introduction. Weinheim: Wiley-VCH; 2008. [Google Scholar]
- 65.Kadam PC, Boone DR. Influence of pH on ammonia accumulation and toxicity in halophilic, methylotrophic methanogens. Appl Environ Microbiol. 1996;62(12):4486–4492. doi: 10.1128/aem.62.12.4486-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blum DJW, Speece RE. A database of chemical toxicity to environmental bacteria and its use in interspecies comparisons and correlations. Res J Water Pollut Control Fed. 1991;63(3):198–207. [Google Scholar]
- 67.Speece RE. Anaerobic Biotechnology and Odor/corrosion Control for Municipalities and Industries. Nashiville, TN: Archaea Press; 2008. [Google Scholar]
- 68.Fernandez AS, Hashsham SA, Dollhopf SL, et al. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol. 2000;66(9):4058–4067. doi: 10.1128/aem.66.9.4058-4067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hashsham SA, Fernandez AS, Dollhopf SL, et al. Parallel processing of substrate correlates with greater functional stability in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol. 2000;66(9):4050–4057. doi: 10.1128/aem.66.9.4050-4057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Briones A, Raskin L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol. 2003;14(3):270–276. doi: 10.1016/s0958-1669(03)00065-x. [DOI] [PubMed] [Google Scholar]
- 71.Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105(suppl 1):11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wittebolle L, Marzorati M, Clement L, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458(7238):623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 73.Werner JJ, Knights D, Garcia ML, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci U S A. 2011;108(10):4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naeem S, Li S. Biodiversity enhances ecosystem reliability. Nature. 1997;390(6659):507–509. [Google Scholar]
- 75.Carballa M, Smits M, Etchebehere C, Boon N, Verstraete W. Correlations between molecular and operational parameters in continuous lab-scale anaerobic reactors. Appl Microbiol Biotechnol. 2011;89(2):303–314. doi: 10.1007/s00253-010-2858-y. [DOI] [PubMed] [Google Scholar]
- 76.Tale VP, Maki JS, Struble CA, Zitomer DH. Methanogen community structure-activity relationship and bioaugmentation of overloaded anaerobic digesters. Water Res. 2011;45(16):5249–5256. doi: 10.1016/j.watres.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 77.Lawrence AW, McCarty PL. Unified basis for biological treatment design and operation. J Sanit Eng Div. 1970;96(3):757–778. [Google Scholar]
- 78.Chen X, Guo J, Xie G-J, Liu Y, Yuan Z, Ni BJ. A new approach to simultaneous ammonium and dissolved methane removal from anaerobic digestion liquor: a model-based investigation of feasibility. Water Res. 2015;85:295–303. doi: 10.1016/j.watres.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 79.Ramirez I, Volcke EIP, Rajinikanth R, Steyer JP. Modeling microbial diversity in anaerobic digestion through an extended ADM1 model. Water Res. 2009;43(11):2787–2800. doi: 10.1016/j.watres.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 80.Freitag TE, Prosser JI. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl Environ Microbiol. 2009;75(21):6679–6687. doi: 10.1128/AEM.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris R, Schauer-Gimenez A, Bhattad U, et al. Methyl coenzyme M reductase (mcrA) gene abundance correlates with activity measurements of methanogenic H2/CO2 enriched anaerobic biomass. Microb Biotechnol. 2014;7(1):77–84. doi: 10.1111/1751-7915.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Regueiro L, Veiga P, Figueroa M, et al. Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol Res. 2012;167(10):581–589. doi: 10.1016/j.micres.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 83.Tale VP. Bioaugmentation for Recovery of Anaerobic Digesters Subjected to Organic Overload [Ph.D. dissertation] Milwaukee, WI: Marquette University; 2010. Available at: http://epublications.marquette.edu/dissertations_mu/88/ [Google Scholar]
- 84.Bocher BTW, Cherukuri K, Maki JS, Johnson M, Zitomer DH. Relating methanogen community structure and anaerobic digester function. Water Res. 2015;70:425–435. doi: 10.1016/j.watres.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 85.Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol. 2014;27:55–64. doi: 10.1016/j.copbio.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 86.Deflaun M, Steffan R.Bioaugmentation Bitton G.Encyclopedia of Environmental Microbiology Hoboken, NJ: John Wiley & Sons, Inc; New York, NY; 2002434–442. [Google Scholar]
- 87.Herrero M, Stuckey DC. Bioaugmentation and its application in wastewater treatment: a review. Chemosphere. 2015;140:119–128. doi: 10.1016/j.chemosphere.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 88.Saravanane R, Murthy DVS, Krishnaiah K. Bioaugmentation and anaerobic treatment of pharmaceutical effluent in fluidized bed reactor. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2001;36(5):779–791. doi: 10.1081/ese-100103760. [DOI] [PubMed] [Google Scholar]
- 89.Duran M, Tepe N, Yurtsever D, Punzi V, Bruno C, Mehta R. Bioaugmenting anaerobic digestion of biosolids with selected strains of Bacillus, Pseudomonas and Actinomycetes species for increased methanogenesis and odor control. Appl Microbiol Biotechnol. 2006;73(4):960–966. doi: 10.1007/s00253-006-0548-6. [DOI] [PubMed] [Google Scholar]
- 90.Ahring BK, Christiansen N, Mathrani I, Hendriksen HV, Macario AJ, Conway de Macario E. Introduction of a de novo bioremediation ability, aryl reductive dechlorination, into anaerobic granular sludge by inoculation of sludge with Desulfomonile tiedjei. Appl Environ Microbiol. 1992;58(11):3677–3682. doi: 10.1128/aem.58.11.3677-3682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guiot SR, Tartakovsky B, Lanthier M, et al. Strategies for augmenting the pentachlorophenol degradation potential of UASB anaerobic granules. Water Sci Technol. 2002;45(10):35–41. [PubMed] [Google Scholar]
- 92.Horber C, Christiansen N, Arvin E, Ahring BK. Improved dechlorinating performance of upflow anaerobic sludge blanket reactors by incorporation of Dehalospirillum multivorans into granular sludge. Appl Environ Microbiol. 1998;64(5):1860–1863. doi: 10.1128/aem.64.5.1860-1863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kasai Y, Kodama Y, Takahata Y, Hoaki T, Watanabe K. Degradative capacities and bioaugmentation potential of an anaerobic benzene-degrading bacterium strain DN11. Environ Sci Technol. 2007;41(17):6222–6227. doi: 10.1021/es062842p. [DOI] [PubMed] [Google Scholar]
- 94.Lenz M, Enright A, O’Flaherty V, van Aelst A, Lens P. Bioaugmentation of UASB reactors with immobilized Sulfurospirillum barnesii for simultaneous selenate and nitrate removal. Appl Microbiol Biotechnol. 2009;83(2):377–388. doi: 10.1007/s00253-009-1915-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hajji KT, Lépine F, Bisaillon J-G, Beaudet R, Hawari J, Guiot SR. Effects of bioaugmentation strategies in UASB reactors with a methanogenic consortium for removal of phenolic compounds. Biotechnol Bioeng. 2000;67(4):417–423. doi: 10.1002/(sici)1097-0290(20000220)67:4<417::aid-bit5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 96.Cirne DG, Björnsson L, Alves M, Mattiasson B. Effects of bioaugmentation by an anaerobic lipolytic bacterium on anaerobic digestion of lipid-rich waste. J Chem Technol Biotechnol. 2006;81(11):1745–1752. [Google Scholar]
- 97.Cavaleiro AJ, Sousa DZ, Alves MM. Methane production from oleate: assessing the bioaugmentation potential of Syntrophomonas zehnderi. Water Res. 2010;44(17):4940–4947. doi: 10.1016/j.watres.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 98.Schauer-Gimenez AE, Zitomer DH, Maki JS, Struble CA. Bioaugmentation for improved recovery of anaerobic digesters after toxicant exposure. Water Res. 2010;44(12):3555–3564. doi: 10.1016/j.watres.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 99.Tale VP, Maki JS, Zitomer DH. Bioaugmentation of overloaded anaerobic digesters restores function and archaeal community. Water Res. 2015;70:138–147. doi: 10.1016/j.watres.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 100.Bocher BTW. Relating Methanogen Community Structure and Function in Anaerobic Digesters [Ph.D. dissertation] Milwaukee, WI: Marquette University; 2012. Available at: http://epublications.marquette.edu/dissertations_mu/208/ [Google Scholar]
- 101.Hammett LP. Linear free energy relationships in rate and equilibrium phenomena. Trans Faraday Soc. 1938;34:156. [Google Scholar]
- 102.Dearden JC, Rowe PH. Use of artificial neural networks in the QSAR prediction of physicochemical properties and toxicities for REACH legislation. Methods Mol Biol. 2015;1260:65–88. doi: 10.1007/978-1-4939-2239-0_5. [DOI] [PubMed] [Google Scholar]


