Summary
The difficulty in detecting rare infected cells immediately after mucosal HIV transmission has hindered our understanding of the initial cells targeted by the virus. Working with the macaque-simian immunodeficiency virus (SIV) vaginal challenge model, we developed methodology to identify discrete foci of SIV (mac239) infection 48 hours after vaginal inoculation. We find infectious foci throughout the reproductive tract, from labia to ovary. Phenotyping infected cells reveals that SIV has a significant bias for infection of CCR6+ CD4+ T cells. SIV infected cells expressed the transcriptional regulator RORγt confirming that the initial target cells are specifically of the Th17 lineage. Furthermore, we detect host responses to infection, as evidenced by apoptosis, cell lysis, and phagocytosis of infected cells. Thus, our analysis identifies Th17 lineage CCR6+ CD4+ T cells as primary targets of SIV during vaginal transmission. This opens new opportunities for interventions to protect these cells and prevent HIV transmission.
Introduction
Developing effective prevention measures for HIV is hindered by incomplete knowledge of the phenotype and localization of the initial target cells of infection after mucosal viral exposure. Previous studies examining the earliest events after vaginal transmission have been limited by their inability to reliably detect rare infected cells using general surveys of exposed tissues. Thus, the earliest targets of SIV/HIV mucosal transmission remain an area of debate. Studies utilizing a variety of approaches have differentially implicated all CD4+ cells as the earliest targets of infection after vaginal challenge in macaques or human tissue explant models (Blauvelt et al., 2000; Gupta et al., 2002; Hladik et al., 2007; Hu et al., 2000; Miller and Hu, 1999; Peters et al., 2015; Reece et al., 1998; Zhang et al., 1999). A small number of studies have attempted to identify the cells infected by SIV in the first days after vaginal inoculation in rhesus macaque (RM) models. Utilizing the SIVmac251 virus swarm, Langerhans cells were identified as the major viral targets 18–24 hours post infection (Hu et al., 2000; Miller and Hu, 1999). Likewise, studies identifying infected cells with in situ PCR implicated dendritic cells as primary targets in the female reproductive tract (FRT) 2 days post challenge with SIVmac251 (Spira et al., 1996). In contrast, another study with SIVmac251 found infected T cells in the endocervix of RMs after 3 days, although the paucity of infected cells identified by in situ hybridization prevented complete definition of infected cell phenotype (Zhang et al., 1999). Studies quantifying cells infected with SIVmac251 at time points of 4 days or longer, with a focus on the endocervix, found T cells are principle targets of infection (Li et al., 2009; Zhang et al., 1999). To advance our understanding of transmission and the relevant target cells it is clear that more studies of the earliest time points after vaginal challenge with SIV are required.
We have previously shown, through vaginal inoculation of RMs with a high titer SIV-based dual reporter vector (LICh) that expresses luciferase, that initial infection events can be widespread throughout the FRT and highly variable in their localization (Stieh et al., 2014). Using LICh as a guide gives us the ability to systematically identify and study small foci of infection events in situ 48 hours after viral challenge. By mixing wild-type SIV with LICh, we utilize the reporter system to identify discrete sites of susceptibility to infection and determine if SIVmac239 infection is also established. Utilizing this approach to SIV challenge, we routinely identify infected cells and their fates in the FRT 48 hours after vaginal challenge. By phenotyping infected cells, we find that primary targets of infection are Th17 cells. This expands upon the previously reported susceptibility of Th17 cells to infection and their early depletion by SIV/HIV infection following vaginal transmission (Cecchinato and Franchini, 2010; Cecchinato et al., 2008). Knowing the preference for Th17 cells during transmission paves the way to unprecedented characterization of host-virus interactions taking place during the earliest events in transmission, and ultimately designing more effective treatment and prevention strategies.
Results
LICh reporter reveals SIV infection
We hypothesized that our single round non-replicating LICh reporter could be used as a macroscopic guide to identify sites susceptible to SIV infection shortly after inoculation, enabling identification of sites where transmission occurred. To test this, a 3ml mixed vaginal challenge of SIVmac239 and high titer LICh was administered to 5 female mature RMs and the animals were sacrificed 48 hours later as described in the Experimental Procedures. The 48-hour time point provides a “snapshot” into the state of SIV infection very early after challenge, before widespread systemic response to the virus. This time point was chosen as it was the earliest interval at which we could reliably detect reporter expression (Stieh et al., 2014). Reproductive tracts were removed, soaked in luciferin to reveal luciferase expression and sites of infection were detected using an in vivo imaging system (IVIS) as previously described (Stieh et al., 2014).
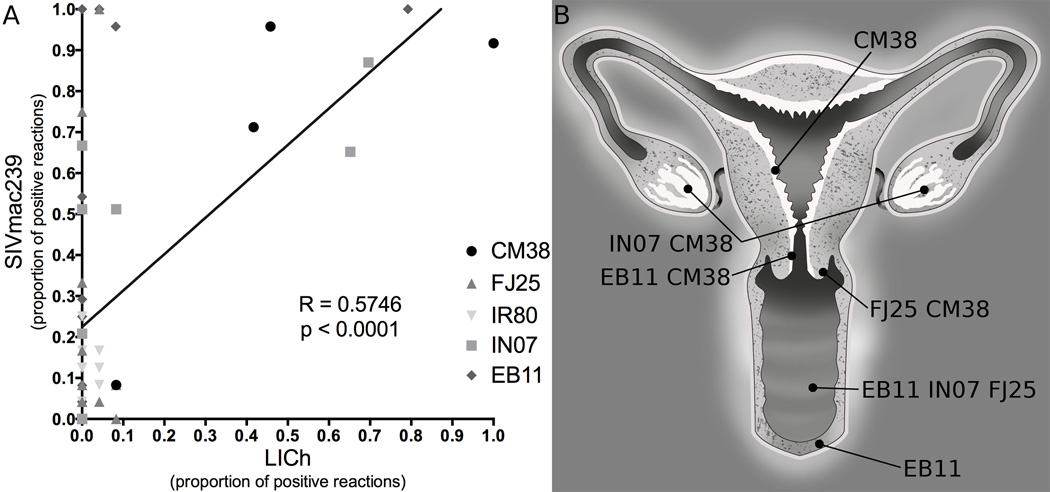
To test for SIVmac239 infection at each site in the FRT in the mixed challenge, we extracted DNA from luciferase positive and random tissue blocks to probe for reverse transcribed DNA specific to the LICh LTR or SIV gag with single copy sensitivity. Nested PCR was performed on 56 tissues, representing diverse sites throughout the FRT. Comparing the proportion of reactions that were positive for LICh and SIVmac239 revealed that their localization is highly correlated (Figure 1A, Pearson R = 0.5746, p < 0.001). SIVmac239 infected cells were present at all sites where there was detectable luciferase signal from LICh. The sites where infection was found by PCR and confirmed by microscopic analysis were widespread throughout the FRT and highly variable between animals (Figure 1B). These findings recapitulate the dispersed distribution of vector susceptible sites found previously with LICh, where we detected transduction from the vaginal vault to ovary. Most foci of infection were found in vaginal tissue, where infection was identified in 3 animals. Detection of SIVmac239 infection in the ovary of two animals also confirms our finding that the ovary is a site of early infection after vaginal challenge. Additionally, we found infection in the uterus of one animal (CM38).
Figure 1.
Detection of SIV infection and LICh transduction in female reproductive tract tissues by nested PCR. A, gDNA extracted from macaque tissue was probed for the presence of gag and LICh LTR. Each symbol represents one piece of tissue, sampled 24 times. Sites where LICh transduction was identified indicated those where SIV infection was found. Pearson correlation between the proportion of positive PCR reactions for each challenge component was highly significant, R = 0.5748, p < 0.0001. B, Distribution of sites of SIV infection throughout the female reproductive tract of each animal which were confirmed by PCR and microscopic analysis.
Validating SIV infected cells by detection of altered surface protein expression
Having identified that sites of SIVmac239 infection span the FRT by PCR, we sought to identify individual infected cells in tissues. Because the wild type virus was not tagged, we identified infected cells utilizing antibodies to SIV antigens. To gain insights into the appearance of SIV infected cells and identify optimal reagents, we optimized detection of SIV Gag and Env in RM PBMCs infected with SIVmac251 infectious molecular clone 766 (Figure S1) (Del Prete et al., 2013). Structured-illumination super-resolution microscopy showed Gag and Env expression had distinct localizations. Gag was typically detected in puncta consistent with assembling virions (Ivanchenko et al., 2009; Jouvenet et al., 2008), while Env primarily associated with internal cellular membranes and the plasma membrane of the cell surface (Hunter and Swanstrom, 1990; Rowell et al., 1995). Denucleated, apoptotic bodies were also identified in these cultures (Figure S1H).
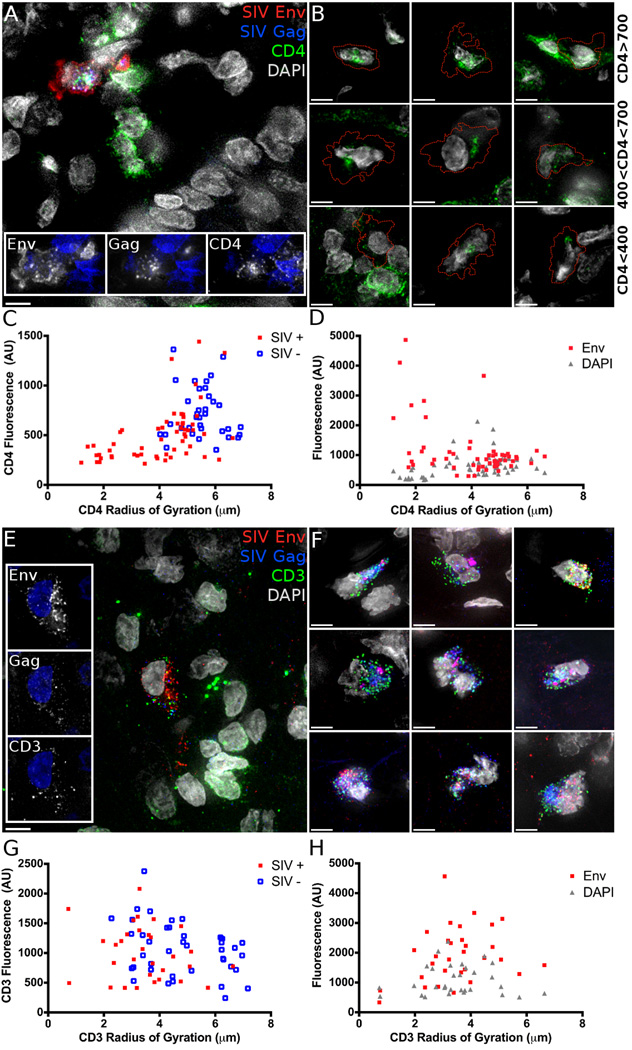
Staining tissue sections from luciferase positive tissue identified SIV Gag and Env positive cells with anticipated expression patterns. To further validate that we were detecting SIV infected cells within tissue, we attempted to detect known changes in cellular protein levels and localization within infected cells. To this end, we costained for CD4, which is internalized and degraded in infected cells (Garcia and Miller, 1991; Vincent et al., 1993). In uninfected cells, CD4 localizes primarily to the plasma membrane, while in SIV infected cells CD4 is internalized and fluorescence intensity decreases (Figure 2A). CD4 signal in infected cells varies from levels seen in uninfected cells (Figure 2B, top row) to just above background levels (Figure 2B, bottom row) as degradation becomes complete. To determine the spatial distribution of CD4, internalization was quantified by measuring the intensity weighted size distribution of the CD4 fluorescence, calculated as the radius of gyration (RoG) of CD4 positive pixels. We examined the relationship between CD4 fluorescence intensity and RoG to quantify the difference between SIV infected and uninfected cells (Figure 2C). We employed a repeated measures model including SIV status, RoG and the interaction between these factors as predictors of CD4 intensity and found both SIV status and the interaction of SIV status and RoG are significant predictors (Parameter estimates ± SE; SIV: −1.30±0.50, p=0.027; Interaction: 0.21±0.09, p=0.011) and the CD4 RoG is lower (−1.47±0.25, p<0.001). There is a negative correlation between Env fluorescence and CD4 RoG (Fig. 2d; −0.14±0.05, p=0.010). The cells with the smallest CD4 RoG were those with the highest levels of Env fluorescence, consistent with greater levels of viral protein expression potentially reflecting longer duration of infection.
Figure 2.
Cell surface CD3 and CD4 are internalized and CD4 is degraded in SIV infected cells. Tissue stained for A, B CD4 (Green) or E, F CD3 (Green), shows the localization of SIV Env (Red) and Gag (Blue) positive cells below the basement membrane of the vaginal epithelium. A, CD4 and E, CD3 are internalized unlike nearby uninfected cells. B, Example images showing CD4 fluorescence at high (>700 AU, top row), intermediate (400 to 700 AU, middle) and dim (<400 AU, bottom row) levels. Dashed line indicates the border of an infected cell. C, CD4 or G, CD3 fluorescence intensity compared to the spatial distribution, measured as the radius of gyration (RoG) of the CD4 or CD3 signal in SIV positive and negative cells. D, CD4 or H, CD3 RoG in SIV infected cells correlated with fluorescence intensity of SIV Env or DAPI as a control. Scale bars measure 5µm. Animal codes: IN07 and FJ25. See also Figure S2.
Similarly, CD3 internalization without subsequent degradation has been demonstrated in vitro (Bell et al., 1998; Swigut et al., 2003). CD3 expression in SIV infected cells shows punctate intracellular localization, while uninfected cells have CD3 on the cell surface (Figure 2E and 2F). In a similar analysis as was used to analyze CD4, only CD3 RoG was a significant predictor of CD3 intensity (Figure 2G; −139.01±48.86, p=0.006). The RoG is decreased (−1.36±0.32, p<0.001) in infected cells while fluorescence intensity is unchanged. Unlike the CD4 results, no correlation is seen between Env intensity and CD3 RoG (Figure 2H, 0.003±0.065, p=0.968). The observation of CD4 internalization and degradation and the internalization but not degradation of CD3 in SIV Gag/Env expressing cells validates that we are observing SIV infected cells within the FRT, 48 hours post challenge.
CCR6+ T cells are preferentially targeted by SIV
Having established that we could identify small foci of SIVmac239 infected cells in discreet sites in the FRT, we sought to phenotype infected cells by immunofluorescent (IF) staining of their surface marker expression. We first examined CD3 expression in 4 animals where foci of infection were identified. Of 229 SIVmac239 infected cells characterized, 75.1% (172) of were CD3+, consistent with our previous study (Stieh et al., 2014). This indicates a significant portion of CD3 negative target cells (24.9%) including macrophages and dendritic cells (DCs) are infected during or shortly after transmission.
Although T helper cells represent the major target of SIV, not all subsets are equally susceptible to infection and are not depleted at the same rate from the mucosa (Alvarez et al., 2013; Brenchley et al., 2008; Cartwright et al., 2014; Cecchinato and Franchini, 2010; Guillot-Delost et al., 2012; Paiardini et al., 2011; Prendergast et al., 2010; Rodriguez-Garcia et al., 2014; Veazey et al., 2000). Th17 cells have been identified as being highly susceptible in vitro and selectively depleted from the gut and FRT (Canary et al., 2013; Klatt et al., 2012; Xu et al., 2012; Xu et al., 2014). We hypothesized that the virus would also preferentially target these cells during transmission. Infected cells were stained for Gag and Env simultaneously, combined with staining for CD3 and CCR6 or CCR10 and CCR6 (Figure 3A–C). This established approach can identify a distinct population containing Th17 cells (Acosta-Rodriguez et al., 2007; Annunziato et al., 2007; Duhen et al., 2009; Gosselin et al., 2010; Hirota et al., 2007; Wan et al., 2011). Diverse cells were infected by SIV, including CD3-CCR6+ (Figure 3B) and CD3-CCR6- (Figure 3C) cells, which could be immature DCs and macrophages, respectively.
Figure 3.
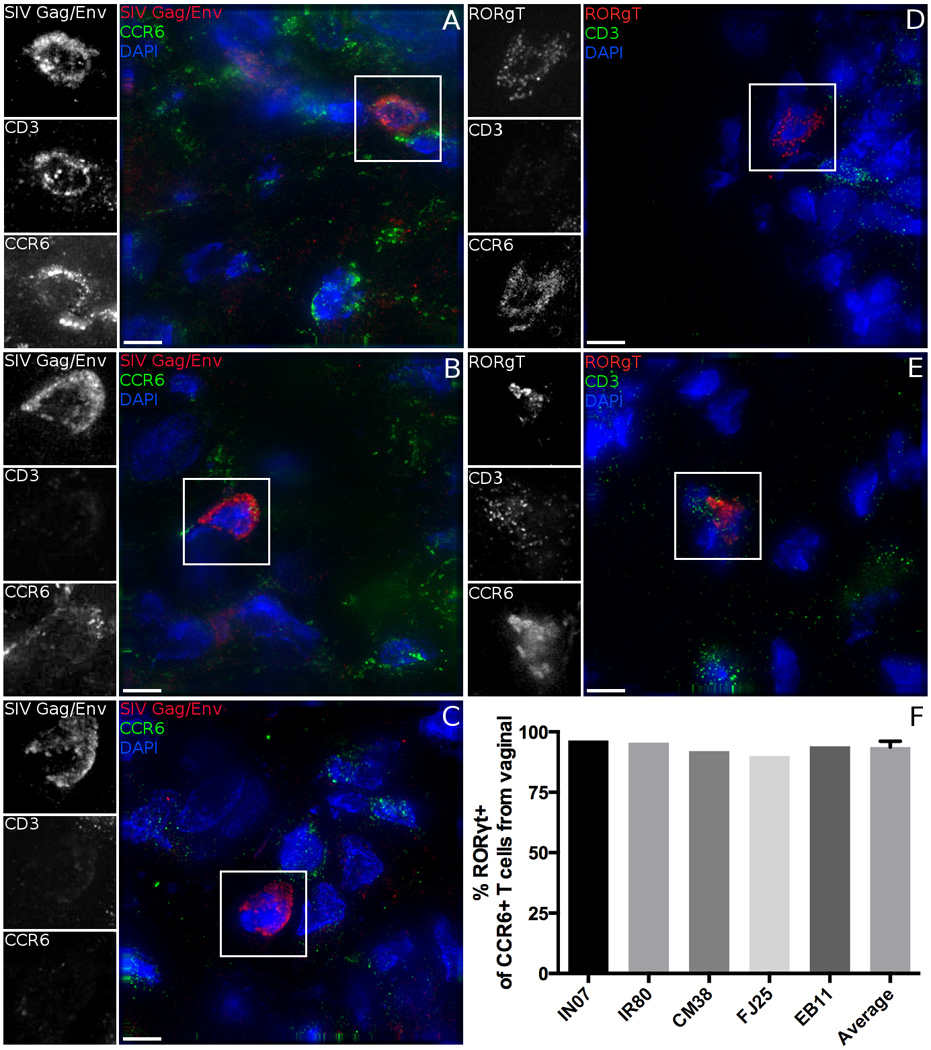
Diverse cell phenotypes are susceptible to SIV infection. A-C, Tissues are stained for SIV antigens Gag and Env (Red and inset), CCR6 (Green and inset), CD3 (inset only), and DAPI (Blue). A, CD3+CCR6+ B, CD3-CCR6+ and C, double negative cells are infected by SIV in the female reproductive tract. D, E Tissues are stained for RORγt (Red and inset), CD3 (Green and inset), CCR6 (inset only), and DAPI (Blue). Both CD3+ and CD3- cells express RORγt, but there is near universal expression of RORγt in CCR6+ cells. F, Quantification of the % of CCR6+CD3+ cells that also express transcription factor RORγt. Error bar indicates SD. Scale bars measure 5µm. Animal codes: FJ25 and EB11. See also Figure S2, S3 and S4.
Remarkably, 84.3% (145) of the 172 SIV infected T cells expressed CCR6, indicating they were most likely Th17 cells (Table 1 and 2). Regardless of the animal (Table 1), the anatomic site (Table 2) or the staining approach employed (CD3 and CCR6, Figure 3A–C or CCR10 and CCR6, data not shown), the CCR6+ T cell population represented the majority of cells with productive SIV infection. Infected cells in all sites identified including vagina, ectocervix, endocervix, uterus, and ovary showed a major preference for infection of CD3+CCR6+ Th17-like cells. Other T cells totaled only 11.8% (27 out of 229) of infected cells.
Table 1.
Proportion of SIV infected cells with a CD3+CCR6+ Th17-like phenotype in each animal with the infected tissues examined indicated.
| Animal | CD3 and CCR6 Phenotyping Analysis1 | Cells phenoty ped |
CD3+ CCR6+ Cells |
% Th17 Cells |
||||
|---|---|---|---|---|---|---|---|---|
| Vagina | Ectoce rvix |
Endoce rvix |
Uterus | Ovary | ||||
| FJ25 | X | X | 90 | 51 | 56.7 | |||
| IN07 | X | X | 69 | 50 | 72.5 | |||
| CM38 | X | X | X | X | 23 | 16 | 69.6 | |
| EB11 | X | 47 | 28 | 59.6 | ||||
| Total: | 229 | 145 | 63.3 | |||||
Infected cells in the labia and lymph node are not included in the analysis of CCR6 expression because the foci of infection in these tissues were depleted in our immune cell phenotyping studies.
Table 2.
Distribution and proportion of SIV infected cell types arranged by site throughout the female reproductive tract. Count and proportion of each cell type within each tissue is shown.
| Tissue | Cell Count (% per tissue) | Total | |||
|---|---|---|---|---|---|
| CCR6+ CD4 T cells |
Other CD4 T cells |
CCR6+ Non- T cells |
Other CCR6- | ||
| Vagina | 108 (63) | 21 (12) | 25 (15) | 17 (10) | 171 |
| Ectocervix | 12 (63) | 2 (11) | 4 (21) | 1 (5) | 19 |
| Endocervix | 7 (64) | 1 (9) | 3 (27) | 0 (0) | 11 |
| Uterus | 3 (75) | 0 (0) | 1 (25) | 0 (0) | 4 |
| Ovary | 15 (62) | 3 (13) | 5 (21) | 1 (4) | 24 |
| Total | 145 (63) | 27 (12) | 38 (17) | 19 (8) | 229 |
Although all Th17s are included within the CCR6+ CD4 T cell population, not all CCR6+ fit the definition of IL-17 producing cells (Wan et al., 2011). While chemokine receptor expression is commonly used to define T helper subsets, the most conclusive mechanism to determine a cell’s functional phenotype is measurements of cytokine production. However, stimulation techniques are unavailable to studies of frozen tissue. An alternative approach is to examine the expression of lineage specific transcription factors: one such Th17-specific factor is the retinoic acid receptor-related orphan receptor gamma T (RORγt) (Yang et al., 2008). We performed fluorescent in situ hybridization for the RORc transcription variant encoding RORγt, combined with IF labeling of CD3 and CCR6. Both CD3+ and CD3- cells can express RORγt, as expected (Figure 3D, E). Further, we find that the vast majority (93.7%) of CCR6+ T cells are also RORγt+ in all animals that received the SIV and LICh mixed challenge (Figure 3F). CD3-CCR6+ cell populations expressing RORγt are presumably type 3 innate lymphoid cells (Sawa et al., 2010).
In order to confirm that what we classified as Th17 cells have the potential to be functional, we looked for evidence of IL-17A expression by SIV infected cells. Intracellular bodies of IL-17A were found in a subset of SIV infected cells (Figure S2A). Mitogenic stimulation of PBMC or vaginally derived CD4 T cells from uninfected RMs demonstrated that all IL-17 producing cells are from the CCR6+ population and that their frequency in vaginal tissue is substantially enriched relative to the periphery (Figure S2B, C), consistent with previous human and murine studies.
If infection were occurring stochastically in all susceptible cell types, the frequency of infected cells would mirror the frequency of SIV susceptible cells in the reproductive tract, or at least at the specific sites where focal infection occurs. To test this, vaginal and ectocervical tissue adjacent to sites where SIV infection was identified was stained for chemokine receptor expression to measure the makeup of the T helper cell populations. Th1-like cells (defined as CD3+CD4+CXCR3+) were 51±2.1% of CD3+CD4+ cells (Figure S3). In contrast, the Th17 cells (CD3+CD4+ CCR6+) were far less frequent in tissues, making up 19.1±2.8% of CD4 T cells. The density of the Th17 cells (CD3+CD4+ CCR6+) counted in Figure S3 were further evaluated for enriched localization at sites of either high T cell density, representative of the sites where SIV infection was most commonly found, and at low T cell density, more typically seen throughout the FRT (Figure S4). In both vaginal (18.1±2.3%) and ectocervical tissue (20.2±2.1%), no large change in the relative proportion of Th17-like cells was observed. This generally agrees with the proportion of Th17 cells throughout the human FRT as described by Rodriguez-Garcia et al (Rodriguez-Garcia et al., 2014). Finally, Th2-like cells (CD3+CD4+CCR4+) were less abundant than either Th1 or Th17 cells (9.6±3.8%, Figure S3). Other T cell populations were not quantified based on local lymphocyte density.
With random targeting, out of the 172 SIV infected T cells (Table 2), we would expect 88 infected cells to be Th1 cells, and only 33 cells to be Th17s because they represent 19% of the FRT CD4 T cell population (Figure S3). Instead, Th17 infected cells totaled 84.3% of all T cell infections. There was a 4.4-fold enhancement in their frequency of being infected relative to their proportion of T helper cells. This represents a highly significant deviation from stochastic targeting (p < 0.001), demonstrating a strong selective preference for infection of Th17 cells by SIVmac239 in the first 48 hours after vaginal exposure. Accounting for all CD4+CCR5+ cells and not only T cells would demonstrate an even greater preferential targeting of Th17 cells during transmission, but this analysis is not achievable with 4-color IF in tissue sections.
Further, SIV infected cells were often found in areas of increased immune cell density below the basal layer of the epithelium, similar to lymphoid aggregates previously reported in the FRT (Kobayashi et al., 2002). We find SIV infection occurs in small foci and is not evenly distributed. Immune cells more densely populate sites of SIV infection relative to the tissue average. However, not all lymphocyte clusters are sites of infection. When we stain for SIV antigens combined with CD3 and DC marker CD11c, we typically see SIV infection associated with a subset of lymphocyte aggregates, while nearby ones have limited evidence of infected cells (Figure S5). This is similar to previous studies of transmission, where infection was found in focal regions at later time points of 4 days and beyond (Zhang et al., 1999). The presence of these regions of dense immune cell localization so early after infection may indicate that they are pre-existing sites of inflammation.
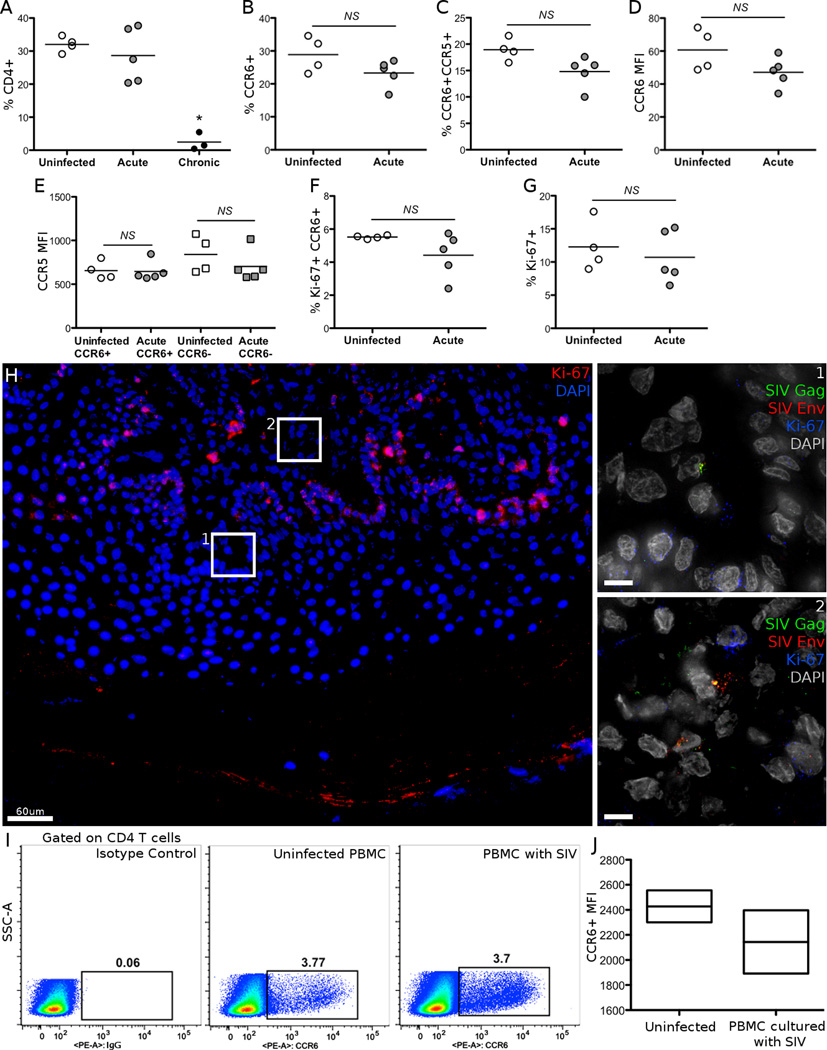
Other cellular factors have been shown to be key determinants of susceptibility to infection, including co-receptor expression and the replicative state of T cells. To relate these previously defined parameters to our observations of preferential infection of CCR6+ T cells, we measured CCR5 and Ki-67 expression on vaginally derived CD4+ T cells from a different cohort of 4 naïve, 5 acutely infected (7–14 days) and 3 chronically infected (>3 months) RMs by flow cytometry (Figure 4). CD4 T cell frequency was maintained early in vaginal tissue, but almost completely eliminated during chronic infection (Figure 4A). This depletion prevented us from further characterizing vaginal T cells from chronically SIV infected RMs. The frequency of CCR6+ CD4 T cells was similar in naïve and acutely infected RMs and over 60% of their CCR6+ CD4 T cells also expressed CCR5, regardless of infection status (Figure 4B, C). Expression levels of CCR6 in vaginal lymphocytes were also unchanged during acute infection. The levels of CCR5 expression on CCR6+ and CCR6- vaginal CD4 T cells were similar between uninfected and acutely infected animals, indicating that receptor expression levels are not the reason for preferential infection (Figure 4E).
Figure 4.
Characterization of markers of SIV susceptibility. A-G, Vaginal lymphocytes from uninfected, acutely and chronically SIVmac251-infected RMs were stained by CD3, CD4, CD8, CCR5, CCR6 and Ki-67. B-G, All cells analyzed were gated on the live CD4+CD8-CD3+ cell populations. Bars indicate the mean. H, Vaginal tissue from animal EB11 was stained for SIV antigens Gag (Green) and Env (Red), and Ki-67 (Red or Blue in inset) to measure whether SIV infected cells were actively dividing. Scale bars measure 10µm unless indicated. I-J, The frequency and MFI of CCR6+ cells was measured on the CD4 T cell population after 5 days of infection. PBMC from uninfected RMs were cultured with or without SIVmac251. Box plots indicate the median and interquartile range. * p < 0.05.
The Ki-67+CCR6+ fraction of CD4 T cells was not changed during acute infection, and averaged 5.2% in uninfected animals (Figure 4F). Similarly, the frequency of Ki-67 positive cells within CCR6+ vaginal CD4 T cells averaged 11.9% and did not change during acute infection relative to uninfected animals. In a limited survey of SIV infected cells from animal EB11, no infected cells found expressed Ki-67, unlike basal karatinocytes which were proliferating (Figure 4H). While this does not confirm that these cells were not dividing at the time infection was established, we do see that SIV proteins are actively expressed in non-proliferating cells. Thus, the exact metabolic state of cells that permits establishment of mucosal SIV infection remains an open question. Separately, in PBMC from uninfected RMs we looked for any increase in expression of CCR6 after in vitro SIV infection, which may offer an alternate explanation of infection in CCR6+ T cells. However, neither the frequency nor MFI of CCR6 on the CD4 T cell populations increased over 3 (data not shown) or 5 days in the presence of a spreading SIV infection (Figure 4I, J), and instead had a slight but insignificant decrease.
SIV infected cell fates in the FRT
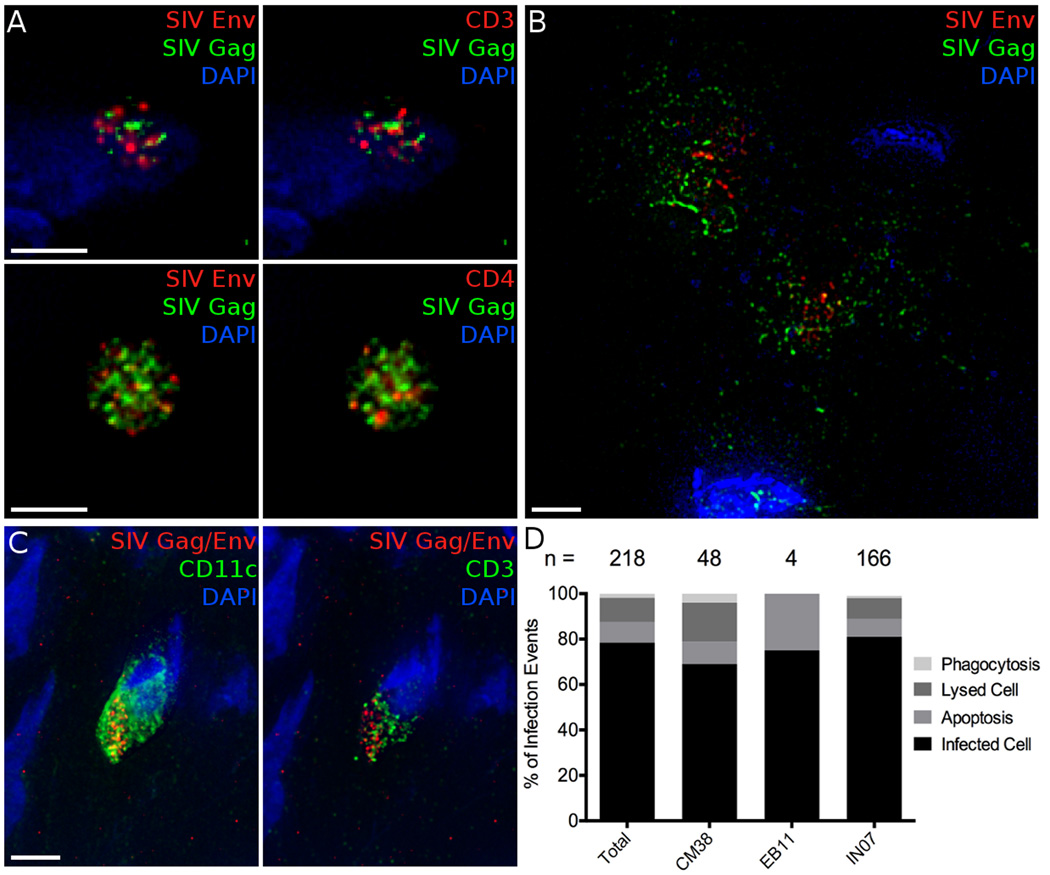
In addition to finding infected cells that still appear to be viable and nucleated, we also identified other apparent fates of SIV infected cells. First, similar to our experience in PBMCs infection (Figure S1H), we find denucleated apoptotic bodies in vivo where Gag, Env and cellular proteins, such as CD3 and CD4, are still detectable (Figure 5A). These apparent apoptotic bodies are typically small, measuring 2–5µm in diameter. Secondly, we observe cell lysis where SIV Gag and Env are spread over larger areas and not associated with any particular cell (Figure 5B, Supplemental Movie 1). Finally, we see phagocytosis of infected T cells by myeloid cells including macrophages and DCs (Figure 5C). This was initially suggested by discrete localization of SIV Gag and Env in cells lacking CD3. Phagocytosis of infected cells was demonstrated in two ways: first, where SIV Gag and Env localized in one dense cluster within a CCR10+ or CD11c+ cell, or secondly, where SIV antigens and CD3 colocalize within a CD11c+ cell, indicating a T cell contained within a DC (Figure 5C). This has been reported to represent the main mechanism by which macrophages and DCs acquire viral antigens during SIV infection (Calantone et al., 2014).
Figure 5.
Alternate fates of SIV infected cells. A, Denucleated, apoptotic bodies stained for SIV antigens Gag (Green) and Env (Red, left) and CD3 (Red, top right) or CD4 (Red, bottom right) and nuclei (Blue). B, 2 lysed cells stained by SIV antigens Gag (Green) and Env (Red) are not associated with any live cells. C, Phagocytosis of an SIV infected T cell visualized by staining for SIV antigens Gag and Env (Red), CD11c (Green, left), and CD3 (Green, right). CD3 and SIV antigens are localized within one domain of a CD11c+ phagocytic cell. D, Quantification of the relative proportion of infected cell fates. The number of total infection events observed in each animal and the total is indicated. Scale bars measure 5µm. Animal codes: IN07, CM38. See also Figure S5.
The relative frequency of these infectious outcomes was evaluated, only including events where SIV Gag and Env were stained for in separate channels. This ensured discrimination between apoptotic bodies or lysed cells and non-specific fluorescence, while also seeing infected cells and phagocytosis. These events were not easily distinguished in the infected cells where staining was focused on Th17 phenotyping and SIV Gag and Env were stained with the same fluorophore. In total, 218 infection events met these criteria (Figure 5D). Infected, nucleated cells were the most common, but lysed and apoptotic cells made up 10.6 and 9.2% of infection events, respectively. Phagocytosis of infected cells comprised only 1.9% of infection events, but this potentially under-represents the actual frequency due to difficulty in identifying SIV Gag and Env positive regions within antigen presenting cells. The subsequent degradation of viral proteins in phagolysosomes will remove the ability to detect phagocytosed cells by IF imaging. An additional confounder of this analysis is that the half-life of SIV infected cell fates in vivo is not defined.
Discussion
Utilizing a mixed vaginal challenge model with LICh and SIVmac239 allows routine identification of SIV infected cell foci 48 hours after exposure. The initiation of mucosal infection in such foci has been reported by others (Miller et al., 2005). Characterization of these foci provides insights into the earliest steps of vaginal transmission of wild type SIV. By identifying multiple small foci of infection, we can phenotype and quantify the diverse cell types susceptible to infection and determine the potential outcomes of SIV infection at sites of transmission. Over the course of 48 hours, viral replication can occur and the cells identified here likely include others than those infected by the initial inoculum. In total, data describing over 500 infection events are included in this study. We show SIV infection is found throughout the FRT, from the labia to the ovaries, demonstrating that prevention measures must protect the entire organ system to afford maximal protection.
Phenotyping infected cells reveals a significant bias for infection of Th17-like, CCR6+ cells during vaginal infection. Confirmation that the CCR6+ CD4 T cells that we find infected by SIV express the transcriptional regulator RORγt provides further evidence that the cells targeted are specifically of the Th17 lineage. A variety of reports indicate that Th17 cells are acutely susceptible to SIV and HIV infection and are systematically depleted during early infection. The reasons for increased susceptibility of these cells are not yet clear. Th17 cells have been reported to express high levels of CCR5, facilitating entry. However, enhanced susceptibility to SIV is independent of route of entry, with VSVg pseudotyped virions also preferentially infecting Th17 cells (El Hed et al., 2010; Monteiro et al., 2011). We find similar CCR5 levels when comparing CCR6+ and CCR6- vaginally derived CD4 T cells in naïve or acutely SIV infected RMs, in agreement with others (Alvarez et al., 2013). A potential caveat in comparing the SIVmac239 based studies here to previous work with SIVmac251 is potential difference in target cell tropism. The clonal SIVmac239 virus has been reported to have a preference to infect T cells while the SIVmac251 swarm can better infect macrophages (Hu et al., 2000; Miller and Hu, 1999). It is notable that we do detect 25% of our infections in dendritic cells and macrophages with SIVmac239. Future studies are required to determine how SIV and HIV envelope tropism influences target cell preference after vaginal challenge.
Preferential infection of Th17 cells extending to the earliest events of transmission is unexpected, and has important implications. These cells are critical for maintaining mucosal epithelial integrity, and their loss may play a role in the ensuing inflammation that drives further viral replication, facilitating dissemination. Sites which are susceptible to SIV infection in this challenge model comprise only a small proportion of the entire FRT. Most sites have intact epithelial or mucus barriers that prevent viral ingress (Carias et al., 2013), but where barrier integrity is weakened, immune cells are recruited. In particular, the role of Th17 cells in maintaining epithelial integrity through the CCL20/CCR6 axis is an important mechanism for stimulating keratinocyte production (Martin et al., 2013). This establishes a model of transmission where epithelial sites with weakened barrier function that are susceptible to viral entry sense increased antigen levels and increase production of CCL20. In response, these same sites are locations to which highly susceptible Th17 cells are recruited and stimulated. Rather than relying on the broadcasting of a small founder population of infected cells, SIV instead can most easily cross the epithelium where a pre-existing cluster of target cells are activated. Further, rapid elimination of these cells prevents epithelial repair, permitting additional antigenic exposure and further recruitment of target cells, rapidly facilitating spread of infected foci.
The studies presented here reveal a rapid innate and intrinsic antiviral response to SIV infection in the FRT. Which alternatives are beneficial to the spread of infection and which, if any, result in improved control remains an open question. Rapid host responses are consistent with developing models of transmission suggesting transmitted viruses are more resistant to interferon responses and that PAMP receptors detect SIV very early. Apparent lysed cells have been previously described in lymphoid tissues during acute SIV infection (Wang et al., 2009), and may reflect cell death by pyroptosis, defined by cell lysis in response to signals from pathogens (Fink and Cookson, 2005; Lamkanfi and Dixit, 2010). There have been other recent reports of immunodeficiency viruses inducing bystander cell death by pyroptosis, unlike the lysed infected cells observed here (Doitsh et al., 2014). Lytic cells at the portal of infection suggest pro-inflammatory cell death begins very early after infection. In contrast we also find apoptosis, which is a highly controlled non-inflammatory cell death mechanism and avoids elicitation of inflammatory responses.
Defining the location and phenotype of SIV infected cell foci and early host responses informs the development of interventions designed to decrease HIV acquisition. Preferential infection of cells from the Th17 lineage could explain the known conditions that increase HIV acquisition, including sexually transmitted infections and bacterial vaginosis. How these conditions precisely influence mucosal barrier function or the density of target cells remains to be determined. However, the system presented here provides essential sampling of these foci, facilitating characterization of the earliest host responses to SIV/HIV infection.
Experimental Procedures
Ethics Statement
All animal studies were conducted in accordance with protocols approved by Northwestern University and Tulane National Primate Research Center local Institutional Animal Care and Use Committees (IACUC), protocol P0153. This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH) and with the recommendations of the Weatherall report; “The use of non-human primates in research”. All procedures were performed under anesthesia using ketamine hydrochloride and all efforts were made to minimize stress, improve housing conditions, and to provide enrichment opportunities. Animals were euthanized by sedation with ketamine hydrochloride injection followed by intravenous barbiturate overdose in accordance with the recommendations of the panel on Euthanasia of the American Veterinary Medical Association.
Reporter vector, virus production and characterization
SIVmac251 infectious molecular clone 766 (SIVmac766) was generated as previously described by transfection of 239T cells with pSIVmac766.4 complexed with Polyethyleneimine (PEI)(Del Prete et al., 2013).
LICh reporter was produced as previously reported(Stieh et al., 2014). Briefly, reporter virus is produced by transfection of 293T cells with 4 plasmids complexed with PEI: LICh reporter genome, SIV3+ packaging vector, REV expression plasmid DM121 and JRFL envelope. Virus containing supernatants were collected 48 hours post-transfection, purified through 0.22µm filters, concentrated over sucrose cushions and resuspended in DMEM. Concentrated virus was stored at −80°C.
SIVmac239 was generated by transfection and expansion in CEMx174 cells. Virus production was monitored by measuring p27 antigen. Cells were maintained with RPMI and addition of new cells every 3 to 4 days. 24 hours before harvest, cells are washed and resuspended in fresh RPMI. Virus containing supernatant is then centrifuged, filtered, aliquoted and stored at −80°C. The p27 concentration of the challenge stock was 1130 pgmL−1. Infection of Ghost-Bonzo cells revealed infection of ∼4% of the cells at 1:2 dilution 48 hours after viral exposure. Infectivity of viral stock was determined by measuring infection of CEMx174 cells by at the limiting viral dilution that was positive for p27 after 21 days of culture; this gave a TCID50 determination of 105.91 per ml.
Non-human Primate Studies
In total 7 female and 2 male rhesus macaques (Macaca mulatta; RM) were challenged in this study, all of which were fully mature. Estimation of SIVmac239 50% animal infectious dose (AID50) was performed by increasing dose mucosal challenge. 2 female macaques were inoculated vaginally and 2 male macaques were inoculated rectally every 2 weeks beginning with 1 in 100,000 dilution and increasing 10-fold. Challenge with 0.5ml of a 1 in 10 dilution resulted in one vaginally inoculated animal and one rectally inoculated animal becoming infected. This allows us to estimate the challenge dose used in subsequent vaginal inoculations at an AID50 of 101.30 per ml.
5 female animals received a 2mm vaginal pinch biopsy in the upper vagina/fornix, and observed for 3–4 min using a vaginal speculum to insure there was no excessive bleeding. The vaginal viral inoculations were performed immediately after using a pliable, blunt no. 8 French pediatric feeding tube attached to the inoculum syringe, inserted 2–3 cm into the vaginal vault and the inoculum is slowly expelled into the vaginal vault. After delivery the tube is flushed with air to ensure that the entire volume of material has been delivered. The feeding tube is slowly removed and the animal is maintained in ventral recumbency with the pelvis elevated for 15 min. The animal is then returned to her cage and monitored until fully recovered from anesthesia. Animals were sacrificed 48 hours after inoculation and their genital tracts were removed and shipped on ice overnight for further processing as previously described (Stieh et al., 2014).
Tissue Staining and Microscopy
Antibody clones and suppliers used are as follows: SIV Gag (Ag3.0, hybridoma supernatant), SIV Env (KK41 and KK42, AIDS Reagent Repository), CD3 (SP7, Abcam), CD4 (OKT4, hybridoma supernatant), CCR4 (551121, BD Pharmigen), CXCR3 (ab125255, Abcam), CCR6 (53103, R & D Systems), CCR10 (6588-5, BioLegend), IL-17 (H-132, Santa Cruz Biotechnology), CD11c (3.9, Life Technologies). All secondary antibodies were raised in donkey and supplied by Jackson Immuno Research. PBMC were adhered to coverslips with CellTak then fixed in 1% formaldehyde in PIPES buffer and stained with anti-SIV Gag and Env antibodies in 0.1% Triton X-100. Cryosections of tissue (15µm thick) were fixed in 1% formaldehyde in PIPES buffer. When multiple primary antibodies of the same species were used, one was labeled with a Xenon IgG labeling kit using AlexaFluor fluorochromes. After antibody staining, cells and tissue were incubated with 4’,6-diamidino-2-phenylindole (DAPI) for visualization of nuclei. Fluorescent in situ hybridization (FISH) to detect RORc transcript variants encoding RORγt were performed with CAL Fluor Red 590 conjugated RNA probes generated using Stellaris RNA FISH probe designer. FISH staining was performed according to the manufacturer’s recommendations for sequential IF and FISH (Biosearch Technologies, CA). Secondary only antibody control images were used to set specificity thresholds for each channel, as previously documented (Stieh et al., 2014). Specificity of the fluorescent signal is established by comparing images to that of control slides to identify patterns of autofluorescent background that are present. 4-color image stacks containing 30 sections in the Z plane in 0.5µm steps were acquired and deconvolved using softWoRx software (Applied Precision) on a DeltaVision inverted microscope. Quantitative image analysis of viral antigens, CD3 and CD4 fluorescence signal and radius of gyration was performed in IDL 7.1.
Statistical Analysis
Analyses of CD3 and CD4 behavior on infected cells were performed using repeated measures models accounting for repeats within animal assuming a compound symmetric covariance structure. Assumptions were assessed by examining the distribution of the outcome and model diagnostics to decide on the outcomes functional form and fit of the model. For tissues stained for SIV antigens and CD4, we evaluated if the data suggest relationships between the following variables: 1) log-transformed fluorescence intensity outcome with SIV status, radius of gyration (RoG), and their interaction; 2) outcome of RoG with SIV status; 3) fluorescence intensity log-transformed outcome with RoG stratified by signal type (Env and DAPI). These analyses were repeated for CD3. The models were performed in SAS 9.4 using a significance level of 0.05. Chi-Square test in Microsoft Excel was used to test for non-random infection of Th17 cells.
Supplementary Material
Acknowledgments
We would like to acknowledge Robin Shattock for a reading the manuscript, Ann Carias for assistance with graphical design and Brandon Keele for providing the SIVmac766.4 construct. We also thank the NIH AIDS Reagent Program for antibodies KK41 and KK42 and GHOST Bonzo+ cells. This study was supported by the NIH grants R01AI094584, P50GM0825545, and 1S10OD010777 to TJH and F32HD080540 to DJS. This work was also supported by the Viral Pathogenesis core of Third Coast Center for AIDS Research (P30 AI 117943) supported by NIAID, NHLBI, NIA, NIMHD, and NIMH. Equipment support was provided by the James B. Pendleton Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DJS performed and designed experiments and wrote the manuscript. EM and HX performed experiments. AJF performed statistical analysis. PAM generated and characterized SIVmac239. RSV and JLB performed animal challenges and dissection. TJH designed the study, interpreted results and critically revised the manuscript.
References
- Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Alvarez Y, Tuen M, Shen G, Nawaz F, Arthos J, Wolff MJ, Poles MA, Hioe CE. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J Virol. 2013;87:10843–10854. doi: 10.1128/JVI.01838-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell I, Ashman C, Maughan J, Hooker E, Cook F, Reinhart TA. Association of simian immunodeficiency virus Nef with the T-cell receptor (TCR) zeta chain leads to TCR down-modulation. J Gen Virol. 1998;79(Pt 11):2717–2727. doi: 10.1099/0022-1317-79-11-2717. [DOI] [PubMed] [Google Scholar]
- Blauvelt A, Glushakova S, Margolis LB. HIV-infected human Langerhans cells transmit infection to human lymphoid tissue ex vivo. AIDS. 2000;14:647–651. doi: 10.1097/00002030-200004140-00003. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calantone N, Wu F, Klase Z, Deleage C, Perkins M, Matsuda K, Thompson EA, Ortiz AM, Vinton CL, Ourmanov I, et al. Tissue Myeloid Cells in SIV-Infected Primates Acquire Viral DNA through Phagocytosis of Infected T Cells. Immunity. 2014;41:493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canary LA, Vinton CL, Morcock DR, Pierce JB, Estes JD, Brenchley JM, Klatt NR. Rate of AIDS Progression Is Associated with Gastrointestinal Dysfunction in Simian Immunodeficiency Virus-Infected Pigtail Macaques. Journal of Immunology. 2013;190:2959–2965. doi: 10.4049/jimmunol.1202319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carias AM, McCoombe S, McRaven M, Anderson M, Galloway N, Vandergrift N, Fought AJ, Lurain J, Duplantis M, Veazey RS, et al. Defining the interaction of HIV-1 with the mucosal barriers of the female reproductive tract. J Virol. 2013;87:11388–11400. doi: 10.1128/JVI.01377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright EK, McGary CS, Cervasi B, Micci L, Lawson B, Elliott ST, Collman RG, Bosinger SE, Paiardini M, Vanderford TH, et al. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J Immunol. 2014;192:4666–4673. doi: 10.4049/jimmunol.1303193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinato V, Franchini G. Th17 cells in pathogenic simian immunodeficiency virus infection of macaques. Curr Opin HIV AIDS. 2010;5:141–145. doi: 10.1097/COH.0b013e32833653ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunology. 2008;1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete GQ, Scarlotta M, Newman L, Reid C, Parodi LM, Roser JD, Oswald K, Marx PA, Miller CJ, Desrosiers RC, et al. Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J Virol. 2013;87:4584–4595. doi: 10.1128/JVI.03507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, Valentine F, Littman DR, Unutmaz D. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis. 2010;201:843–854. doi: 10.1086/651021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel MR, Routy JP, et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot-Delost M, Le Gouvello S, Mesel-Lemoine M, Cherai M, Baillou C, Simon A, Levy Y, Weiss L, Louafi S, Chaput N, et al. Human CD90 identifies Th17/Tc17 T cell subsets that are depleted in HIV-infected patients. J Immunol. 2012;188:981–991. doi: 10.4049/jimmunol.1101592. [DOI] [PubMed] [Google Scholar]
- Gupta P, Collins KB, Ratner D, Watkins S, Naus GJ, Landers DV, Patterson BK. Memory CD4(+) T cells are the earliest detectable human immunodeficiency virus type 1 (HIV-1)-infected cells in the female genital mucosal tissue during HIV-1 transmission in an organ culture system. J Virol. 2002;76:9868–9876. doi: 10.1128/JVI.76.19.9868-9876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Yoshitomi H, Hashimoto M, Maeda S, Teradaira S, Sugimoto N, Yamaguchi T, Nomura T, Ito H, Nakamura T, et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J Exp Med. 2007;204:2803–2812. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hladik F, Sakchalathorn P, Ballweber L, Lentz G, Fialkow M, Eschenbach D, McElrath MJ. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity. 2007;26:257–270. doi: 10.1016/j.immuni.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Gardner MB, Miller CJ. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol. 2000;74:6087–6095. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol. 2012;5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Darragh T, Herndier B, Anastos K, Minkoff H, Cohen M, Young M, Levine A, Grant LA, Hyun W, et al. Lymphoid follicles are generated in high-grade cervical dysplasia and have differing characteristics depending on HIV status. American Journal of Pathology. 2002;160:151–164. doi: 10.1016/s0002-9440(10)64359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 2010;8:44–54. doi: 10.1016/j.chom.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DA, Towne JE, Kricorian G, Klekotka P, Gudjonsson JE, Krueger JG, Russell CB. The emerging role of IL-17 in the pathogenesis of psoriasis: preclinical and clinical findings. The Journal of investigative dermatology. 2013;133:17–26. doi: 10.1038/jid.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Hu J. T cell-tropic simian immunodeficiency virus (SIV) and simian-human immunodeficiency viruses are readily transmitted by vaginal inoculation of rhesus macaques, and Langerhans’ cells of the female genital tract are infected with SIV. J Infect Dis. 1999;179(Suppl 3):S413–S417. doi: 10.1086/314795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CJ, Li Q, Abel K, Kim EY, Ma ZM, Wietgrefe S, La Franco-Scheuch L, Compton L, Duan L, Shore MD, et al. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005;79:9217–9227. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, Ancuta P. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol. 2011;186:4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, et al. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nat Med. 2011;17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Gonzalez-Perez MP, Musich T, Moore Simas TA, Lin R, Morse AN, Shattock RJ, Derdeyn CA, Clapham PR. Infection of ectocervical tissue and universal targeting of T-cells mediated by primary non-macrophage-tropic and highly macrophage-tropic HIV-1 R5 envelopes. Retrovirology. 2015;12:48. doi: 10.1186/s12977-015-0176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast A, Prado JG, Kang YH, Chen F, Riddell LA, Luzzi G, Goulder P, Klenerman P. HIV-1 infection is characterized by profound depletion of CD161+ Th17 cells and gradual decline in regulatory T cells. AIDS. 2010;24:491–502. doi: 10.1097/QAD.0b013e3283344895. [DOI] [PubMed] [Google Scholar]
- Reece JC, Handley AJ, Anstee EJ, Morrison WA, Crowe SM, Cameron PU. HIV-1 selection by epidermal dendritic cells during transmission across human skin. J Exp Med. 1998;187:1623–1631. doi: 10.1084/jem.187.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Garcia M, Barr FD, Crist SG, Fahey JV, Wira CR. Phenotype and susceptibility to HIV infection of CD4+ Th17 cells in the human female reproductive tract. Mucosal Immunol. 2014;7:1375–1385. doi: 10.1038/mi.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell JF, Ruff AL, Guarnieri FG, Staveley-O’Carroll K, Lin X, Tang J, August JT, Siliciano RF. Lysosome-associated membrane protein-1-mediated targeting of the HIV-1 envelope protein to an endosomal/lysosomal compartment enhances its presentation to MHC class II-restricted T cells. J Immunol. 1995;155:1818–1828. [PubMed] [Google Scholar]
- Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- Spira AI, Marx PA, Patterson BK, Mahoney J, Koup RA, Wolinsky SM, Ho DD. Cellular targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J Exp Med. 1996;183:215–225. doi: 10.1084/jem.183.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieh DJ, Maric D, Kelley ZL, Anderson MR, Hattaway HZ, Beilfuss BA, Rothwangl KB, Veazey RS, Hope TJ. Vaginal challenge with an SIV-based dual reporter system reveals that infection can occur throughout the upper and lower female reproductive tract. PLoS Pathog. 2014;10:e1004440. doi: 10.1371/journal.ppat.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Greenberg M, Skowronski J. Cooperative interactions of simian immunodeficiency virus Nef, AP-2, and CD3-zeta mediate the selective induction of T-cell receptor-CD3 endocytosis. J Virol. 2003;77:8116–8126. doi: 10.1128/JVI.77.14.8116-8126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Mansfield KG, Tham IC, Carville AC, Shvetz DE, Forand AE, Lackner AA. Dynamics of CCR5 expression by CD4(+) T cells in lymphoid tissues during simian immunodeficiency virus infection. J Virol. 2000;74:11001–11007. doi: 10.1128/jvi.74.23.11001-11007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent MJ, Raja NU, Jabbar MA. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J Virol. 1993;67:5538–5549. doi: 10.1128/jvi.67.9.5538-5549.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, Sundrud MS, Unutmaz D. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6(+) human memory T cells. Journal of Experimental Medicine. 2011;208:1875–1887. doi: 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu H, Gill AF, Pahar B, Kempf D, Rasmussen T, Lackner AA, Veazey RS. Monitoring alpha4beta7 integrin expression on circulating CD4+ T cells as a surrogate marker for tracking intestinal CD4+ T-cell loss in SIV infection. Mucosal Immunol. 2009;2:518–526. doi: 10.1038/mi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Liu DX, Moroney-Rasmussen T, Lackner AA, Veazey RS. IL-17-producing innate lymphoid cells are restricted to mucosal tissues and are depleted in SIV-infected macaques. Mucosal Immunol. 2012;5:658–669. doi: 10.1038/mi.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Veazey RS. Th17 Cells Coordinate with Th22 Cells in Maintaining Homeostasis of Intestinal Tissues and both are Depleted in SIV-Infected Macaques. Journal of AIDS & clinical research. 2014:5. doi: 10.4172/2155-6113.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.