Abstract
Co-evolution of ticks and the vertebrate immune system has led to the development of immunosuppressive molecules that prevent immediate response of skin-resident immune cells to quickly fend off the parasite. Herein, we demonstrate that the tick-derived immunosuppressor sialostatin L restrains IL-9 production by mast cells while degranulation and IL-6 expression are both unaffected. In addition, the expression of IL-1β and IRF4 is strongly reduced in the presence of sialostatin L. Correspondingly, IRF4- or IL-1 receptor-deficient mast cells exhibit strong impairment in IL-9 production demonstrating the importance of IRF4 and IL-1 in the regulation of the Il9 locus in mast cells. Furthermore, IRF4 binds to the promoters of Il1b and Il9 suggesting that sialostatin L suppresses mast cell-derived IL-9 preferentially by inhibiting IRF4. In an experimental asthma model, mast cell-specific deficiency in IRF4 or administration of sialostatin L results in a strong reduction of asthma symptoms demonstrating the immunosuppressive potency of tick-derived molecules.
Introduction
Development and reproduction of hard ticks (Ixodidae) depend on blood meals that require a close attachment to their host for several days. As a consequence, the penetration of the skin will initially provoke innate immune reactions in addition with an adaptive immune response when a vertebrate host is repeatedly infested. Suppression of such an immune reaction is therefore of vital importance for these parasites and therefore tick saliva contains a multitude of immune-modulating ingredients that ensure an adequate supply with blood (1). The cystatin sialostatin L (sialoL) was isolated from Ixodes scapularis saliva and was shown to inhibit the maturation of dendritic cells and the antigen-specific stimulation and proliferation of CD4+ T cells as well (2, 3). Recently, we could demonstrate that primary IL-9 production of Th9 cells is strongly suppressed by sialoL in vitro, and its administration in vivo inhibited airway hyperresponsiveness and eosinophilia in mice suffering from experimental asthma (4). These results suggested that suppression of Th9-derived IL-9 by sialoL prevents the development of asthmatic symptoms.
Initially, IL-9 was found to have mast cell growth-enhancing activity and IL-9-induced mastocytosis was shown to be essentially involved in protective immune responses during parasitic worm infections (5). Interestingly, mast cells probably represent also the most important innate source of IL-9 thus amplifying such reactions in an autocrine fashion. Mast cells preferentially reside beneath epithelial surfaces which are considered as interfaces between host and environment. This localization and the expression of a great variety of Toll-like receptors (TLRs) (6, 7) predestine mast cells to perceive invading parasites like ticks and to boost an immediate early immune response. In particular, their ability to rapidly release mediators such as histamine and leukotrienes and to serve as a primary source of cytokines including TNF-α, IL-1β, and IL-9 enables mast cells to finely shape the fate and magnitude of an anti-parasitic immune response (8). Concerning IL-9 our analyses revealed that IL-1, IL-10, ckit-ligand or the TLR4-ligand LPS synergistically enhance the production of this cytokine by mast cells (9–11). At the transcriptional level GATA-1 was found to regulate the expression of the Il9 gene upon IgE-mediated activation of mast cells (12). However, the detailed molecular mechanisms underlying Il9 gene regulation in mast cells are far from being definitive.
In this study, we demonstrate that the tick saliva protein sialoL represses mast cell-derived IL-9 by inhibiting the expression of IL-1β and the transcription factor IRF4, as well.
Materials and Methods
Mice
Mice of strain C57BL/6 were obtained from Charles River Laboratories (Sulzfeld, Germany) and bred in our own animal facility. Irf4-deficient mice (Irf4−/−) (13) on a C57BL/6 background were a gift from Prof. Magdalena Huber, Marburg, Germany. Il1r1−/− (14) and Il1a/b double-deficient mice (15) on C57BL/6 background were a gift from Prof. Esther von Stebut-Borschitz, Mainz, Germany. Mice were bred in our own animal facility.
Genetically mast cell-deficient KitW-sh/W-sh mice on a C57BL/6 background (16) were initially obtained by Prof. Marcus Maurer (Department of Dermatology, Charite, Berlin, Germany). Males and females were used at the age of 6–12 weeks. Animal procedures were conducted in accordance with the institutional guidelines.
Generation of mast cells and reconstitution of mast cell-deficient mice
For the generation of mast cells, mice were sacrificed by cervical dislocation. Intact femurs and tibias were removed and bone marrow was harvested by repeated flushing with MEM supplemented with 2% FCS. Mast cell cultures were established at a density of 2×106 cells/ml in IMDM supplemented with 10% FCS (previously inactivated at 56°C), 2mM L-glutamine, 1mM pyruvate, 100U/ml penicillin and 100μg/ml streptomycin, 20U/ml murine (m)IL-3, 50U/ml mIL-4, and 200ng/ml SCF. Non-adherent cells were transferred to fresh culture plates every 2–3 days for a total of at least 28 days to remove adherent macrophages and fibroblasts.
KitW-sh/KitW-sh mice were systemically reconstituted with 5×106 mast cells derived from either C57BL/6, Irf4−/− mice or Il1r1−/− mice by intravenous injection 8 weeks before the experiments were conducted. To assess reconstitution efficiencies, lungs were fixed by inflation (1ml), immersed in 4% formalin or Carnoy’s solution and embedded in paraffin. Tissue sections were used for avidin-based mast cell stainings (17). Slides were examined in a blinded fashion with a microscope (BX40; Olympus, Hamburg, Germany). For the assessment of mast cell numbers in each slide, mast cells were counted by a blinded investigator in five different fields and in each field the lung area was measured using an image analysis system (Soft Imaging System; Olympus). Numbers of mast cells are expressed per square centimeter.
Stimulation of mast cells
Stimulation of mast cells with either ionomycin or IgE/ anti-IgE was performed in IMDM supplemented with 10% FCS (previously inactivated at 56°C), 2mM L-glutamine, 1mM pyruvate, 100U/ml penicillin and 100μg/ml streptomycin.
Lymphokine assays
Cytokine secretion was assessed by specific ELISA. To detect IL-9 we used mAb 229.4 (1μg/ml) and biotinylated mAb C12 (1μg/ml) which was a gift from Prof. J. van Snick, Brussels, Belgium. This ELISA detects biologically active murine IL-9, as confirmed by using an IL-9-specific bioassay (18). For the detection of murine IL-6 we used Mouse IL-6 ELISA Set (BD Pharmingen) according to the manufacturers’ manual and for the analysis of IL-1β content in lysates of mast cells the mouse IL-1β Elisa Ready Set Go kit (eBioscience) was used. Lysates from 2×105 mast cells were prepared by using 60μl 0.5% Triton X-100 in water and subsequently 50μl were used for the detection of IL-1β by ELISA. ELISAs were evaluated according to reference standard curves by using known amounts of the specific cytokine.
Degranulation/β-Hexosaminidase release
Degranulation of mast cells was quantified by assaying the release of β-hexosaminidase. Mast cells were stimulated using 2.5μg/ml IgE-anti-DNP (clone SPE-7) for 72h at 37°C. Crosslinking of receptor bound IgE was performed by 1μg/ml anti-IgE for 30min (clone EM95.3) (Baniyash and Eshhar, 1984) in Tyrode (106 cells per ml). The cells were lysed in 20μl 0.5% Triton X-100. Three aliquots of 20μl from each supernatant and the corresponding mast cell lysate were transferred to separate plates. Fifty microliters of substrate solution (1.3mg/ml p-nitrophenyl-N-acetyl-β-D-glucosamine in 0.1M sodium citrate, pH 4.5) were added, and the plates were incubated for 90min at 37°C. The reaction was stopped by adding 150μl of 0.2 M glycine (pH 10.7). Hydrolysis of the substrate was measured at 410nm. Activity of β-hexosaminidase in supernatant and lysate were summarized and defined as total enzyme content. The results are expressed as percentage of β-hexosaminidase activity released into the medium.
Surface staining
IL-1R1
To determine IL-1R1 expression on mast cells we used anti-mouse CD121a (clone JAMA-147, Biolegend).
CD117 and FcεR1
FACS analyses using anti-mouse CD117 (c-Kit) (clone ACK2, eBioscience) and anti-mouse FcεRI (clone MAR1, eBioscience) revealed that the resulting cell populations used consisted of >95% mast cells (data not shown).
Intracellular staining
For intracellular staining of IL-9, IL-1β and IL-6 murine mast cells were stimulated with ionomycin (1μM) for 6h (IL-6), 24h (IL-1β) or 48h (IL-9). For the last 3–5h of stimulation Monensin (eBioscience) was added to the cells. Cells were harvested and washed with PBS. Fixation and permeabilization were performed with buffers from FoxP3 staining kit (eBiosciences). Cells were stained for IL-9 (RM9A4 and rat IgG isotype controle, Biolegend), IL-1β Proform (NJTEN3 and rat IgG isotype control, eBioscience) and IL-6 (MP5-20F3 and rat IgG isotype control, eBioscience).
Westernblot analyses
After stimulation with ionomycin (1μM) for 24h or 48h mast cells (106) were lysed in 50μl RIPA buffer and sonicated for 10min (30sec on/ 30sec off; high power) using the Bioruptor Plus Sonication device (Diagenode). Lysates were fractionated by SDS-PAGE. After transfer on PVDF membrane protein was analyzed by immunoblot with goat polyclonal anti-IRF4 Ab (M17; Santa Cruz Biotechnology) and donkey anti-goat IgG-HRP (SC2020; Santa Cruz Biotechnology). Detection of β-Actin (clone AC-16 coupled to HRP, Sigma-Aldrich) served as control. Densitometric analysis was performed using Image Lab 4 software (BioRad).
Chromatin Immunoprecipitation (ChIP)
ChIP analyses were performed using the High Cell ChIP Kit with Protein G-coated paramagnetic beads (Diagenode). After stimulation of 2×107 mast cells with 1μM ionomycin for 24h cells were washed twice with PBS and then protein-DNA complexes were crosslinked using formaldehyde (Thermo Scientific, 1% final concentration) for 7 minutes. After incubation, fixation was quenched by adding glycine to a final concentration of 125mM. Isolation of nuclei (3×106 cells in 300μl per 1.5ml TPX tube) was performed as described in the manual using Lysis buffer 1 and Lysis buffer 2. Sonication of chromatin was conducted at 4°C in Shearing buffer S1 in the presence of a protease inhibitor cocktail (Roche) using the Bioruptor Plus sonication device (Diagenode) set to high power. Four cycles of 10min with 30sec pulse followed by 30sec with no pulse were performed leading to a distinct 100bp–500bp chromatin fraction. Chromatin immunoprecipitation was conducted using 6.3μg anti-IRF4 (M-17, Santa Cruz) or normal-goat IgG (Santa Cruz) as isotype control bound to protein G coated magnetic beads according to the manufacturers’ instructions. After elution of DNA with DIB (DNA isolation buffer; Diagenode) conventional PCR (35 cycles) was performed with oligonucleotides flanking the desired IRF4 binding site within the murine Il9 promoter: Il9 prom (−285 to −265).for 5′-TTTTAAAGGGGGTTGGGGCT-3′ and Il9 prom (−174 to −194).rev 5′-AGGCTGTCTTATGCCAGGAA-3′ or the murine Il1b promoter: Il1b prom (−1222 to −1202).for 5′-GCTCCCTCAGCTTAAGCACA-3′ and Il1b prom (−1034 to −1054).rev 5′-ATCGTGGTGGAAATGGGCAT-3′ and afterwards PCR products were loaded onto a 2% agarose gel for visualization.
Luciferase reporter assay, plasmids and transfection
The 5′-region of the murine Il9 gene encompassing nucleotides −610 to +32 (11) or Il1b gene encompassing nucleotides −758 to −1308 were cloned into the promoterless pGL3 basic luciferase reporter gene vector (Promega). Mutageneses of two potential IRF4 binding sites (Il1b prom Δ−1060bp to −1043bp, and Il9 prom Δ−243bp to −255bp upstream from translational start site) were performed using the QuickChange site-directed mutagenesis kit (Agilent Laboratories) and were verified by DNA sequencing. 3×106 mast cells in 200μl serum free IMDM were transfected with 8μg of the Il1b or Il9 promoter reporter vectors or 8μg of the mutated Il1b or Il9 promoter reporter vectors in combination with 300ng of pRL-TK plasmid (Dual luciferase reporter assay system, Promega) as internal control to exclude differences in transfection efficiency. Additionally, cells were co-transfected with 4μg of a plasmid coding for IRF4 or empty vector pcDNA3.1; Invitrogen). Transfections were performed by electroporation in 0.2-cm cuvettes at room temperature using a Gene Pulser II (Bio-Rad) set to 290 V and 600μF. Cells were allowed to recover for 3h in mast cell culture medium and were then stimulated with 0.5μM ionomycin. After 16h of stimulation, cells were harvested, washed with PBS and lysed according to the manufacturers’ instructions. Relative luciferase activity was measured using a luminometer (TD20/20, Turner Designs).
Isolation of total RNA
5×106 mast cells were stimulated with ionomycin (1μM) in the presence or absence of sialoL (3μM) for 24h. Total RNA was prepared by using Trizol (Invitrogen) and RNA quality (RNA integrity number ≥8) was determined on a Bioanalyzer 2100 using a RNA 6000 Nano Chip (Agilent).
Next-generation sequencing, RNA sequencing (RNA-Seq)
Barcoded mRNA-seq cDNA libraries were prepared from 5ug of total RNA using a modified version of the Illumina mRNA-seq protocol. With the aid of Seramag Oligo(dT) magnetic beads (Thermo Scientific) mRNA was isolated and subsequently fragmented using divalent cations and heat resulting in fragments ranging from 160–220bp. Fragmented mRNA was converted to cDNA using random primers and SuperScriptII (Invitrogen) followed by second strand synthesis using DNA polymerase I and RNaseH. cDNA was end repaired using T4 DNA polymerase, Klenow DNA polymerase and 5′ phosphorylated using T4 polynucleotide kinase. Blunt ended cDNA fragments were 3′ adenylated using Klenow fragment (3′ to 5′ exo minus). 3′ single T-overhang Illumina multiplex specific adapters were ligated using a 10:1 molar ratio of adapter to cDNA insert using T4 DNA ligase. cDNA libraries were purified and size selected at 200–220bp using the E-Gel 2% SizeSelect gel (Invitrogen). Enrichment, adding of Illumina six base index and flow cell specific sequences were done by PCR using Phusion DNA polymerase (Finnzymes). Clean-up was done using 1,8x volume of AgencourtAMPure XP magnetic beads. All quality controls were done using Invitrogen’s Qubit HS assay and fragment size was determined using Agilent’s 2100 Bioanalyzer HS DNA assay. Barcoded RNA-Seq libraries were clustered on the cBot using Truseq SR cluster kit v2.5 using 7pM and 50bps were sequenced on the Illumina HiSeq2000 using Truseq SBS kit-HS 50bp. The raw output data of the HiSeq was preprocessed according to the Illumina standard protocol. This includes filtering for low quality reads and demultiplexing. Sequence reads were aligned to the reference genomic sequence (Mouse Genome Sequencing Consortium et al., 2002) using bowtie (19) (Parameters for the alignment to the reference genome: -v2–best). The alignment coordinates were compared to the exon coordinates of the RefSeq transcripts (20) and for each transcript the counts of overlapping alignments were recorded. Sequence reads not alignable to the genomic sequence were aligned to a database of all possible exon-exon junction sequences of the RefSeq transcripts (Parameters for the alignment to the splice junction database: default). The counts of reads aligning to the splice junctions were aggregated with the respective transcript counts obtained after alignment and normalized to RPKM (number of reads which map per kilobase of exon model per million mapped reads (21) for each transcript. Alternatively, CLC Genomics Workbench (Qiagen) was used to analyze results of the RNA sequencing.
RNA-Seq results were validated employing qRT-PCR upon stimulation of 1×106 mast cells with 1μM ionomycin for 24h in the presence or absence of 3μM sialostatinL using following primers. mIL-9.for: 5′-CTGATGATTGTACCACACCGTGC-3′, mIL-9.rev: 5′-GCCTTTGCATCTCTGTCTTCTGG-3′; mIRF4.for: 5′-GCCCAACAAGCTAGAAAG-3′, mIRF4.rev: 5′-TCTCTGAGGGTCTGGAAACT-3′; mIL-1b.for: 5′-CAACCAACAAGTGATATTCTCCATG-3′, mIL-1b.rev: 5′-GATCCACACTCTCCAGCTGCA-3′; mIL-6.for: 5′-CTGCAAGAGACTTCCATCCAG-3′, mIL-6.rev: AGTGGTATAGACAGGTCTGTTGG-3′ ; mHPRT.for: 5′-GTTGGATACAGGCCAGACTTTGTTG -3′, mHPRT.rev: 5′-GAGGGTAGGCTGGCCTATAGGCT-3′.
SialoL preparation and LPS decontamination
SialoL protein was expressed in Escherichia coli, and the corresponding active protein was purified, as previously described (2). LPS contamination was removed by Arvys Protein using detergent extraction. The presence of endotoxin was estimated as <4×10−5 endotoxin U/μg protein (approximately <3 × 10−14 g endotoxin per microgram protein) with a sensitive fluorescent-based endotoxin assay (PyroGene recombinant factor C endotoxin detection system; LonzaBiologics).
Asthma experimental protocol
Ten to 12-wk-old C57BL/6 mice or KitW-sh/W-sh mice were sensitized by injection of 20μg OVA i.p. (grade V; Sigma-Aldrich) in a total volume of 100μl on day 1 and 14. Mice were challenged (20min) for three consecutive days (day 27, 28, 29) with OVA (1% in saline) using ultrasonic nebulization (NE-U17; Omron). SialoL (i.v. 10μg/ challenge) and IL-9 (i.n. 500ng/ challenge) were applied thirty minutes before each challenge. Bronchoalveolar lavage (BAL) fluid was collected and airway reactivity was measured 24h after the last challenge. Schematic representation is given in Supp. Fig. 1.
Bronchoalveolar lavage
After assessment of airway function, cells were isolated by lavage of the lungs via a tracheal tube with PBS (1ml). Numbers of living cells were counted by using trypan blue dye exclusion. Differential cell counts were made from cytocentrifuged preparations fixed and stained with the Microscopy Hemacolor Set (Merck).
Measurement of airway reactivity
Measurement of the airway resistance was performed on anesthetized, intubated, and mechanically ventilated mice (FlexiVent; Scireq, Montreal, QC, Canada) in response to increasing doses of inhaled methacholine (ranging from 3.125 to 50mg/ml). Measurement of airway resistance was performed every 15s following each nebulization step until a plateau phase was reached (Reuter and Taube, 2008).
Statistical analysis
Statistical evaluations were performed by GraphPad Prism software (Version 6.0f) using unpaired student t test. The p values ≤0.05 were considered statistically significant.
Results
The tick salivary protein sialoL decisively modulates mast cell effector function
Due to their location beneath interfaces between host and environment, mast cells are probably the first cells sensing ticks upon bite. We have previously demonstrated that sialoL possesses immunomodulatory properties (3, 4). To analyze the impact of sialoL on mast cell effector functions, we stimulated mast cells in the presence and absence of this cystatin and measured their ability to produce IL-9 and IL-6 as well as to degranulate (Fig. 1). IL-9 production by mast cells was found to be severely impaired (Fig. 1A–C) while secretion of IL-6 (Fig. 1D) and degranulation (Fig. 1E) in response to IgE sensitization and subsequent anti-IgE treatment were minimally affected. Notably, the viability of mast cells was not reduced in the presence of sialoL thus excluding nonspecific toxic effects (Fig. 1F). These findings led us to conclude that sialoL is an immune modulator that selectively blocks mast cell-derived IL-9 production while leaving other mast cell effector functions untouched (IL-6, degranulation).
Figure 1. SialoL impairs mast cell-derived production of IL-9 without affecting degranulation, IL-6 secretion and cell viability.
Mast cells were stimulated with ionomycin (Iono, 1μM) in the presence or absence of different concentrations of sialoL (1.5, 3 and 6.0μM). Production of IL-9 was determined by intracellular flow cytometry (A, B), and ELISA (C) after 48h of stimulation. IL-6 production was measured by ELISA after 24h of stimulation (D). Degranulation of mast cells was assessed after sensitization with IgE and crosslinking with the aid of anti-IgE as described in the materials and methods section (E). Cell viability was measured via FACS using “Fixable Viability Dye” (eBioscience) (F). Except for panel (A) (one representative from three independent experiments) the mean of three independent experiments (±SD) is shown. * P≤0.05; **≤P 0.01; ***≤P 0.001 (two- tailed unpaired t-test). ns= not significant.
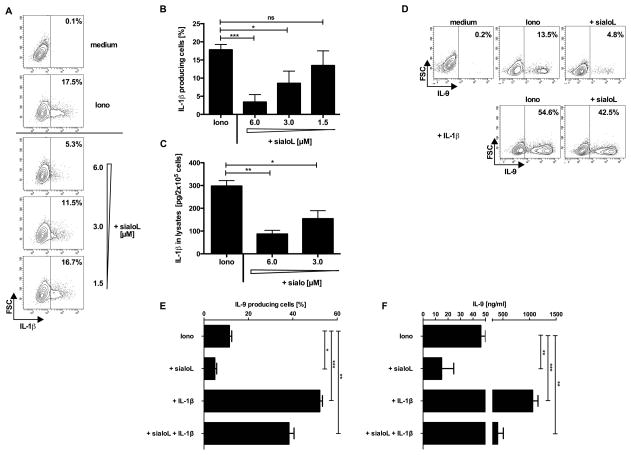
SialoL inhibits autocrine production of the IL-9-promoting cytokine IL-1
Previously, we demonstrated that production of IL-9 by mast cells is strongly enhanced in the presence of IL-1β (9). Therefore, we analyzed the influence of sialoL on the expression of IL-1β by mast cells. Intracellular FACS analyses demonstrated that IL-1β expression was strongly reduced upon stimulation of mast cells in the presence of sialoL (Fig. 2A and 2B). Since IL-1β could not be detected in the supernatants of these mast cells we presumed that its concentration was below the detection level of the applied ELISA. Therefore, IL-1β was titrated on activated IL-1α and IL-1β double-deficient and WT mast cells. Supp. Fig. 2 shows that 37.5 pg/ml of IL-1β induced production of IL-9 in IL-1α and IL-1β double-deficient mast cells (white bars) resulting in a concentration in the supernatant that is comparable to the concentration measured in the supernatant of WT mast cells in the absence of exogenous IL-1β (Iono: black bar). Regardless of cytokine consumption, one can conclude that activated WT mast cells produce approximately 37.5 pg/ml IL-1β. Alternatively, activated mast cells were lysed and intracellular IL-1β was assessed revealing that the IL-1β content of mast cells was considerably reduced in the presence of sialoL (Fig. 2C). In order to test whether inhibition of IL-1β production at least partially accounts for the observed sialoL-mediated reduction of IL-9 expression we stimulated mast cells in the presence and absence of sialoL in combination with exogenous IL-1β. Notably, exogenous IL-1β was able to almost totally compensate for the sialoL-mediated suppression of IL-9 production by mast cells on a single cell level (Fig. 2D and 2E) and in the resulting supernatants as well (Fig. 2F).
Figure 2. SialoL-mediated suppression of mast cell-derived IL-9 can be partially reversed by exogenous IL-1β.
Mast cells were stimulated with ionomycin (Iono, 1μM) in the presence or absence of different concentrations of sialoL (1.5, 3 and 6μM). Production of IL-1β was determined by intracellular flow cytometry (A, B) or in cell lysates (C) after 48h of stimulation. IL-9 production of mast cells was measured by intracellular FACS analysis (D, E) and ELISA (F) 48h after stimulation with ionomycin in the presence solely of sialoL (3μM) or IL-1β (300pg/ml) or by a combination of both. Except in panel (A) and (D) (one representative from three single experiments) the mean of three independent experiments (±SD) is shown. *≤P 0.05; **≤P 0.01; ***≤P 0.001 (two- tailed unpaired t-test). ns= not significant.
SialoL inhibits IL-1-mediated signal transduction and IRF4 expression to suppress IL-9 production
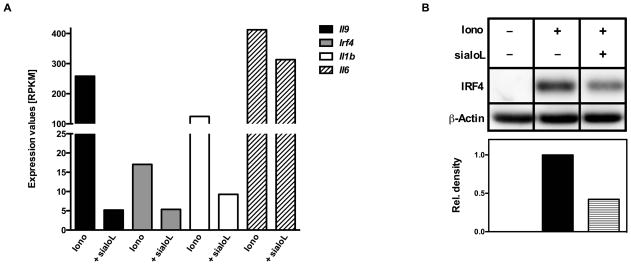
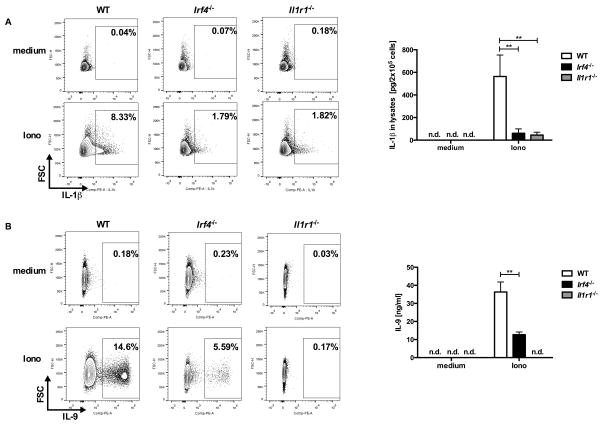
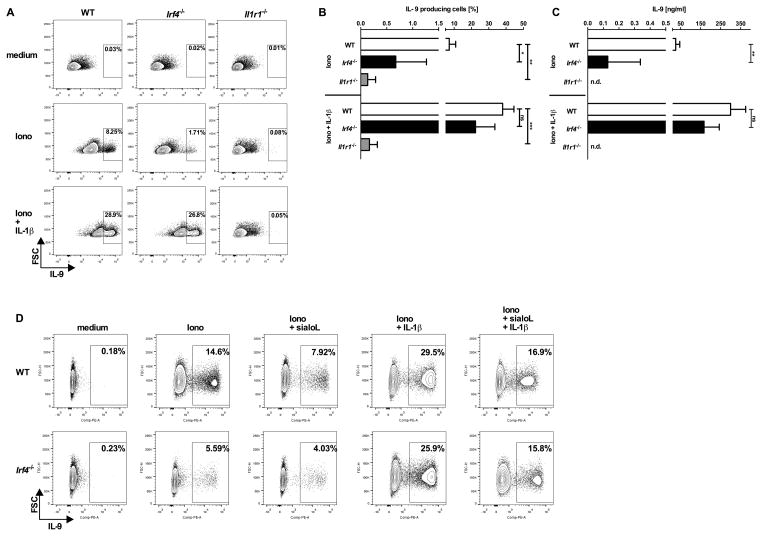
In an attempt to identify the molecular mechanism responsible for the sialoL-mediated inhibition of IL-9 production we performed Next-Generation-Sequencing (NGS) of mRNA. To this end, we stimulated mast cells in the presence and absence of sialoL using ionomycin for 24h. In silico analyses of the raw data obtained by this approach revealed very strong inhibition of Il9 and Il1b gene expression and a significant reduction in Irf4 gene expression while Il6 gene expression was only marginally inhibited (Fig. 3A). These findings were confirmed by applying qRT-PCR (see Supp. Fig. 3). In analogy to IL-1 and IL-9 the inhibition of Irf4 gene expression could also be validated on the protein level (Fig. 3B). We have previously shown that IRF4 is crucial for IL-9 production by Th9 cells (22). In addition, IRF4 was shown to be involved in the regulation of Il1b gene expression in macrophages (23, 24). To test a potential causal correlation between the sialoL-mediated inhibition of IL-9 and IL-1β expression and the suppression of IRF4 expression, we conducted experiments using mast cells from Irf4-deficient (Irf4−/−) as well as Il1r1-deficient (Il1r1−/−) mice. To this end, we comparatively analyzed IL-1β as well as IL-9 production of Irf4−/− mast cells, Il1r1−/− mast cells and the respective littermate controls (WT). Upon 24h of stimulation IL-1β production (Fig. 4A) and after 48h IL-9 production (Fig. 4B) were measured by intracellular FACS analysis as well as ELISA of cell lysates (IL-1β) and supernatants (IL-9). While WT mast cells produced substantial amounts of IL-1β and IL-9 upon stimulation the production of these cytokines by Irf4−/− as well as Il1r1−/− mast cells was strongly reduced. This indicates that IRF4 and endogenous IL-1β are required for Il9 gene expression. In addition, these findings suggest that an IL-1β-dependent positive feedback loop largely contributes to activation-induced IL-1β production of mast cells. In analogy to sialoL-treated mast cells (Supp. Fig. 4A and B), degranulation and IL-6 expression were not affected by Irf4- as well as Il1r1-deficiency (Supp. Fig. 4C and D). Likewise, IRF4-deficient mast cells did not show reduced expression of IL-1R1 thereby excluding an impaired IL-1 responsiveness of such mast cells (Supp. Fig. 4E). Furthermore, extended comparative analyses of WT and Irf4−/− mast cells revealed no differences with regard to the content of beta-hexosaminidase and the expression of RANTES (CCL5), Eotaxin2 (CCL24), Eotaxin3 (CCL26) and MCP1 (CCL2) thus excluding a general inability to produce mast cell mediators (data not shown). To analyze whether the strongly reduced ability of Irf4−/− mast cells to produce IL-9 is due to the limited capability to produce IL-1β, we stimulated mast cells of the genotypes depicted in Fig. 5 in the presence and absence of exogenous IL-1β and measured IL-9 production on a single cell level by intracellular FACS analyses (Fig. 5A and B) and the overall production of this cytokine by ELISA (Fig. 5C). Remarkably, IL-9 production of Irf4−/− mast cells but not of Il1r1−/− mast cells was partially restored in the presence of exogenous IL-1β indicating an important role for IRF4 in the regulation of the Il1b locus and the Il9 locus as well. In addition, application of sialoL only marginally further reduced the strongly attenuated IL-9 production of Irf4−/− mast cells that could be compensated for in the presence of exogenous IL-1β approximately to the level observed in WT mast cells (Fig. 5D, Irf4−/−: 15.8%, WT: 16.9%). Hence, this comparatively weak influence of sialoL on Irf4−/− mast cell-derived IL-9 in combination with the strong compensatory potency of exogenous IL-1β votes for a causal involvement of IRF4 and IL-1β in sialoL-mediated inhibition of mast cell-derived IL-9 production.
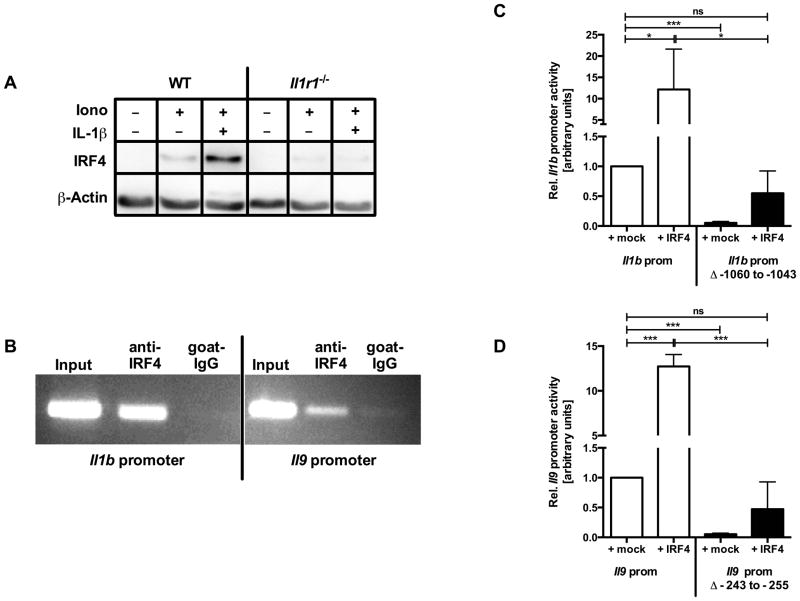
Figure 3. Treatment of mast cells with sialoL strongly reduces the expression of Il9, Irf4 and Il1b.
Mast cells were stimulated with ionomycin (Iono, 1μM) for 24h in the presence or absence of sialoL (+sialoL, 3.0μM). Isolation of total RNA, library preparation and RNA-Seq was conducted as outlined in the materials and methods section. Transcript counts obtained after alignment were normalized to RPKM (number of reads which map per kilobase of exon model per million mapped reads for each transcript) (A; single experiment). The expression of IRF4 was assessed by western blot analysis in parallel to β–actin as loading control after 24h of stimulation; densitometric quantification was conducted as outlined in the materials and methods section (B). In (A) results of a single experiment is shown. Panel (B) shows one representative from three independent experiments.
Figure 4. Mast cells generated from Il1r1−/− or Irf4−/− mice produce strongly reduced amounts of IL-1β and IL-9.
Mast cells generated from C57BL/6 (WT), Il1r1−/− or Irf4−/− mice were cultured in medium (IMDM + 10% FCS) for 24h in the presence or absence of 1μM ionomycin. Subsequently, the production of IL-1β (A) or IL-9 (B) was determined by intracellular FACS analysis using anti-IL-9 or anti-IL-1β antibodies and isotype control. Except for FACS blots in panel A and B (one representative experiment of three (A) or five single experiments (B)) the mean of three (A) or five (B) independent experiments obtained by ELISA (±SD) is shown. Amounts that were not detectable are indicated by n.d.. **≤P 0.01 (two- tailed unpaired t-test).
Figure 5. Impaired production of IL-9 by mast cells from Irf4−/− mice can be partially reversed by exogenous IL-1β.
Mast cells generated from C57BL/6 (WT), Il1r1−/− or Irf4−/− mice were stimulated for 48h with ionomycin (Iono, 1μM) in the presence or absence of IL-1β (300pg/ml) and sialoL [3μM]. Production of IL-9 was assessed by intracellular FACS analysis (A, B, D) and ELISA in the resulting supernatants (C). Amounts that were not detectable are indicated by n.d.. Except for panel (A) (one representative from three independent experiments) and (D) (one representative from two independent experiments) the mean of at least four independent experiments (±SD) is shown. *≤P 0.05; **≤P 0.01; ***≤P 0.001 (two- tailed unpaired t-test). ns= not significant.
IL-1 receptor 1-mediated induction of IRF4 crucially contributes to Il9 and Il1 gene regulation
To investigate the interrelation between IL-1β-mediated signaling and the expression of IRF4 in mast cells we analyzed the expression of IRF4 upon stimulation in the presence and absence of IL-1β. While stimulation of mast cells with the calcium ionophore ionomycin already induced the expression of IRF4, exogenous IL-1β further enhanced IRF4 expression in WT mice while no effect could be observed in mast cells from Il1r1−/− mice (Fig. 6A) Along with the finding that the IL-1β expression of IRF4-deficient mast cells was strongly impaired (see Fig. 4A) these data suggested a mutual positive regulation between IL-1β and IRF4.
Figure 6. IRF4 binds to and transactivates the Il9 and Il1b promoter in mast cells.
Mast cells generated from C57BL/6 (WT) or Il1r1−/− mice were left untreated or stimulated for 48h with ionomycin (Iono, 1μM) in the presence or absence of IL-1β (300pg/ml). IRF4 expression was assessed by western blot analyses (A). Binding of IRF4 to the promoters of Il9 and Il1b was assessed by ChIP-analyses (B). For Il9 and Il1b reporter gene analyses mast cells were transfected with a plasmid coding for IRF4 (+IRF4) or with the corresponding empty vector (+mock) in combination with an Il1b (C) or Il9 (D) promoter-luciferase reporter or promoter-luciferase reporter with mutated IRF4 binding sites and stimulated for 16h with ionomycin (0.5μM). Cell lysates were prepared as outlined in materials and methods section and luminescence was measured. Mast cells transfected with promoter-luciferase vector +mock were arbitrarily set to 1. Panel (A and B) display one representative result from one out of four (A) or three (B) independent experiments. In (C) the mean of three and in (D) the mean of five independent experiments (±SD) is shown. *≤P 0.05; ***≤P 0.001 (two- tailed unpaired t-test). ns= not significant.
To examine whether IRF4 binds to the Il1b and Il9 locus thereby regulating the transcription of both genes, we performed chromatin immunoprecipitation (ChIP) analysis. As shown in Fig. 6B, inducible binding of IRF4 to the promoter regions of both genes is detectable upon stimulation of mast cells. Therefore, it can be concluded that IRF4 directly regulates the expression of Il1b and Il9 locus, respectively.
The trans-activating capacity of IRF4 for the Il1b and Il9 promoter was demonstrated with the aid of reporter gene assays and transfection of mast cells with an Il1b or Il9 reporter construct revealed an inducible promoter activity upon stimulation that was further enhanced upon ectopic expression of IRF4 (Fig. 6C and 6D). In addition, we performed in silico analyses and subsequent site-directed mutagenesis of potential IRF4 consensus binding sites to identify the putative consensus binding site of IRF4 in the Il1b and Il9 core promoters (IL-1 prom Δ−1060bp to −1043bp, and IL-9 prom Δ−243bp to −255bp upstream from translational start site). As shown in Fig. 6C and 6D, transfection of mast cells with these Il1b or Il9 promoter deletion mutants strongly impaired Il1b or Il9 promoter activity per se as well as upon ectopic expression of IRF4 indicating that these sites are essential for both Il1b and Il9 gene expression in mast cells.
Administration of sialoL as well as mast cells-specific deficiency in Irf4 or Il1r1 in vivo significantly ameliorated OVA-induced eosinophilia and AHR
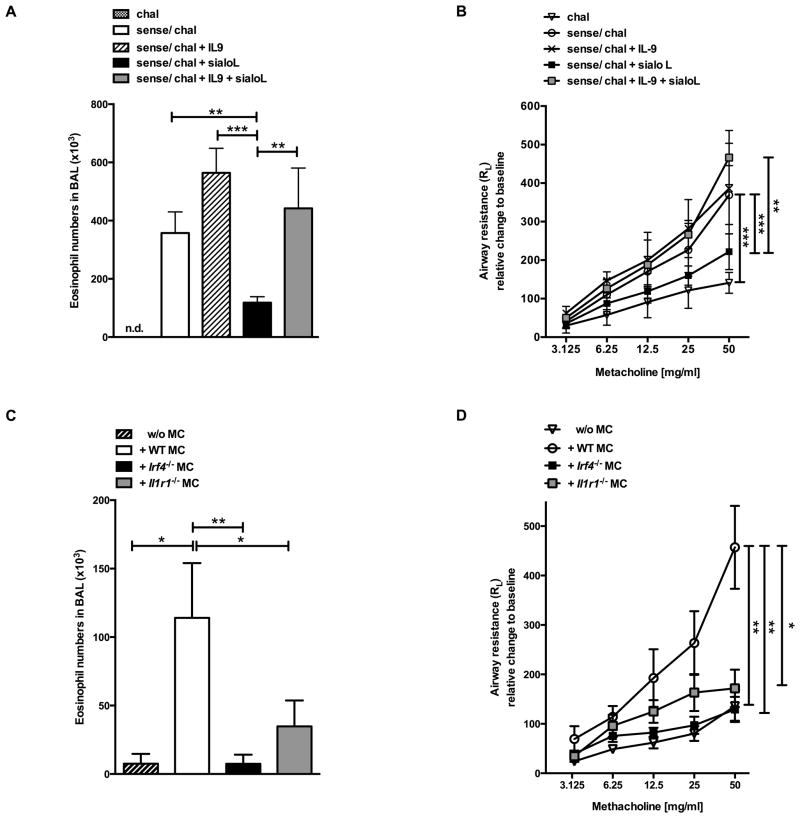
To test the in vivo relevance of the sialoL-mediated suppression of mast cell-derived IL-9 that is based on inhibition of the expression of IRF4 and IL-1β in mast cells we deployed a mast cell-dependent murine model for allergic asthma (25, 26). Major features of IL-9-induced allergic symptoms are IL-5- and eotaxin-dependent infiltration of eosinophils into the lung and an increased airway hyperresponsiveness (27). As depicted in Fig. 7A and 7B, sensitization and subsequent challenge (sense/chal) with Ovalbumin (OVA) elicits a strong influx of eosinophils into the lumen of the lungs and a considerable increase in metacholin-induced airway resistance. Addition of IL-9 (sense/chal + IL-9) further elevated the number of eosinophils and airway resistance. Notably, both asthma symptoms were substantially reduced by single administration of sialoL at the time of challenge (Fig. 7A and 7B; sense/chal + sialoL) and restored approximately to the level of the sense/chal approach upon concurrent application of IL-9 (sense/chal + IL-9 + sialoL). To further investigate the in vivo role of IRF4 and IL-1R1 signaling in mast cells, mast cell-deficient KitW-sh/W-sh mice were systemically reconstituted with mast cells derived from either WT, Irf4−/− or Il1r1−/− mice. Sensitization and challenge with OVA resulted in a robust influx of eosinophils and strong increase of methacholin-induced airway resistance when KitW-sh/W-sh mice were reconstituted with wildtype mast cells (Fig. 7C and D). These asthma symptoms were hardly detectable in the absence of mast cells in KitW-sh/W-sh mice and were significantly impaired in KitW-sh/W-sh mice that had been reconstituted with mast cells from either Irf4−/− or Il1r1−/− mice (Fig. 7C and D).
Figure 7. Administration of sialoL as well as mast cell-specific deficiency in Irf4 or Il1r1 significantly ameliorates eosinophilia and AHR.
Eosinophilia and airway hyperresponsiveness (AHR) were induced by sensitizing and challenging C57BL/6 mice with ovalbumin in the absence (sense/chal) or presence (+sialoL) of sialoL or sialoL in combination with IL-9 (+sialoL + IL-9). Mice solely challenged without sensitization (chal) served as negative control (A, B). Mast cell-deficient KitW-sh/W-sh mice were reconstituted with mast cells generated from WT, Il1r1−/− or Irf4−/− mice. Non-reconstituted KitW-sh/W-sh mice (w/o MC) served as control. 8 weeks after reconstitution mice were immunized as described in materials and methods section (C, D). Numbers of eosinophils in BAL fluid and airway resistance were determined 24h after the last challenge on day 31. Bars represent mean (±SD). * P≤0.05; **≤P 0.01; ***≤P 0.001 (unpaired t-test).
In summary, we demonstrate that the inhibition of mast cell-derived IL-9 by the tick salivary protein sialoL is based on an altered IL-1R1 signaling and a partial inhibition of IRF4 expression leading to a severely impaired self-induction of mast cell-derived IL-1β. The relevance of these findings is emphasized by the fact that mast cell-specific deficiency in Il1r1 and Irf4 prevents IL-9 production by mast cells in a murine asthma model, thereby diminishing the influx of eosinophils and the accompanying airway resistance, both cardinal features of allergic asthma.
Discussion
Adaptation of parasites to hematophagous lifecycle has led to development of parasitic immune evasion strategies that are based to a great extent on immunosuppressive agents (28). Such agents enable the survival of parasites by preventing an effective immune response and they are supposed to represent a fertile source of naturally occurring immune modulators for the development of novel drugs to treat allergic and autoimmune diseases. This discovery of novel drugs from parasites and other pathogens is called the “drugs from bugs” approach (29).
SialoL is one of these immunosuppressive molecules that was characterized from tick saliva several years ago (2). Meanwhile, we demonstrated that–among other immunosuppressive activities- sialoL strongly inhibits the development of Th9 cells which produce the asthma-promoting cytokine IL-9 (4). Initially, it was reported that IL-9 production depends on IL-2, is synergistically enhanced by a combination of IL-4 and TGF-β and inhibited by IFN-γ (30). SialoL was found to inhibit the production of IL-9 and IL-2 by CD4+ T cells. Therefore, we assumed that the suppressive mechanism of sialoL is based on a reduced T cell-derived IL-2 production. However, the addition of exogenous IL-2 could not reverse the sialoL-mediated inhibition of IL-9 production suggesting a different suppressive mode of action. Th9-derived IL-9 was shown to be a major pathogenic factor in an experimental model of asthma and treatment of mice suffering from asthma with sialoL strongly reduced their asthma symptoms. Upon in vivo administration to treat asthma, sialoL can certainly have an impact on several cell types and mast cells represent prominent candidates because they were shown to produce considerable amounts of IL-9 and to induce asthmatic symptoms as well (9, 31). Therefore, it was conceivable that the sialoL-mediated inhibition of mast cell-derived IL-9 greatly contributes to the amelioration of asthmatic symptoms. Consequently, we have analyzed the influence of sialoL on mast cell-derived IL-9 in detail.
In general, we found that sialoL strongly reduced the production of IL-9 by mast cells after activation using ionomycin. This inhibitory effect of sialoL could be partially compensated for by IL-1β which represents a potent co-stimulator of mast cell-derived IL-9 production (see below and (10)). In addition, IRF4 was identified as a crucial driver of ionomycin-induced IL-9 expression by mast cells. These findings were corroborated by the fact that Il1r1−/− mast cells showed only marginal IL-9 production and Irf4−/− mast cells exhibited a strongly reduced IL-9 production that could be partially compensated for by IL-1β. By contrast, ionomycin-induced IL-6 was not affected by sialoL or IL-1β or a combination of both. Finally, comparable results could be obtained when IgE was applied as a classical mast cell activator (data not shown).
Asthma-associated eosinophilia was strongly reduced after treatment with sialoL suggesting a considerably effective suppression of endogenous IL-9 and IL-1 which both were described to have a profound eotaxin-inducing capacity preferentially by affecting airway smooth muscle and epithelial cells. This assumption was substantiated by the finding that nonresponsive mast cell-deficient mice developed clear-cut asthma-associated eosinophilia and airway hyperresponsiveness after reconstitution with wild type mast cells while the reconstitution with Irf4−/− or Il1r1−/− mast cells led to a strong reduction of the eosinophilic influx. Hence, mast cell-specific inability to produce maximal amounts of IL-9 either caused by Irf4-deficiency or IL-1 unresponsiveness is obviously a crucial condition to develop and maintain asthma symptoms in the mast cell-dependent preclinical asthma model applied herein.
Comparable to T cells, it was shown that the expression of IL-9 by mast cells could be further enhanced by NF-κB inducing stimuli like IL-1 (10) or LPS (11). In addition, a combination of IL-10 or Kit-Ligand (KL) with IL-1 strongly enhanced the production of IL-9 by mast cells (10). While stimulation of mast cells with a combination of IL-1 and IL-10 or KL increased Il9 promoter activity, only IL-10 enhanced the half-life of IL-9 mRNA suggesting post-transcriptional modifications by this cytokine. Nevertheless, IL-10 and KL exerted only a minimal effect in the absence of IL-1 thus indicating that IL-1-induced transcription factors are obviously master regulators of the Il9 locus in mast cells. The essential relevance of IL-1 was also emphasized by results which showed that an IL9-induced increase in IL-9 production by mast cells depends on endocrine IL-1β production (32). These findings prompted us to assess the role of autocrine IL-1β production when we analyzed the molecular mechanisms underlying the sialoL-mediated inhibition of mast cell-derived IL-9. The simultaneous reduction of IL-9 and IL-1β secretion in mast cells in the presence of sialoL and the fact that the addition of exogenous IL-1β could substantially revert the suppressive activity of sialoL indicated that the impairment of autocrine IL-1β production represents a central inhibitory mechanism of sialoL. This result was confirmed by transcriptome analysis that indicated a strong inhibition of the expression of Il9 and Il1b gene in sialoL-treated mast cells. In addition, these analyses revealed that sialoL can also inhibit the expression of the Irf4 gene suggesting that, in analogy to Th9 cells, IRF4 positively regulates mast cell-derived IL-9. Using Irf4−/− mast cells, we could demonstrate that this transcription factor is of crucial importance for the production of IL-9 in Th9 cells and mast cells as well (herein and (22). However, this does not apply to PU.1 which is a transcription factor known to substantially contribute to Il9 gene expression in Th9 cells (33). By contrast, Il9 promoter reporter analyses revealed that ectopic expression of PU.1 rather led to a silencing of the Il9 locus in mast cells (data not shown) while IRF4 strongly enhanced the activity of this reporter gene construct. Another important positive regulator of Il9 expression in mast cells is GATA-1 which was shown to be activated by p38 MAP kinase-mediated phosphorylation (12). In this context, PU.1 was found to inhibit GATA-1 function by physical interaction leading to PU.1/GATA-1 hetero dimers that prevent the binding of GATA-1 to DNA (34). Thus, PU.1 inhibits the expression of Il9 in mast cells while T cells are not negatively affected because they lack the expression of GATA-1 (35). Taken together, the regulation of the Il9 promoter in T cells and mast cells appears to share similarities but also to differ in several aspects. Nevertheless, sialoL inhibits IL-9 production of mast cells and Th9 cells indicating that it acts on molecules essentially involved in both cell types in the production of this cytokine, namely IRF4.
In light of these findings, our observation that IRF4 is decisively involved in the IL-1-dependent production of IL-9 by mast cells might be of pathophysiological importance, since it has been demonstrated that IL-9 is a central mediator in allergic asthma and decisively involved in an effective anti-melanoma immune response (36). Therapeutic approaches for the treatment of asthma on the basis of IL-9 neutralization in pre-clinical mouse models showed rather promising results (37, 38). However, neutralization of human IL-9 using the antibody MEDI-528 could not alleviate asthma symptoms in adults with uncontrolled asthma suggesting that simply blocking IL-9 in chronic asthma is not sufficient to cure such a complex disease (39). Nevertheless, the curative influence of sialoL in our preclinical asthma model is most possibly based on the manipulation of various functions in distinct cell types. Besides the inhibition of IL-9 production by Th9 cells and mast cells sialoL was also found to impair the maturation and antigen-specific activation of dendritic cells and the expansion of CD4+ T cells as well. Therefore, further comparative studies will be required to elucidate the influence of sialoL not only on the transcriptional mechanisms that are responsible for the differential regulation of the Il9 locus in mast cells and Th9 cells but also on DC functions and the proliferation of CD4+ cells in general. The resulting knowledge will presumably serve as a basis to develop innovative strategies for the treatment of asthma by exploiting the suppressive mechanisms of sialoL.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), grant DFG BO 3306/1-1, SCHM 1014/7-1, SCHM 1014/5-1, SFB 1066 project B1 (E.S.) and B8 (T.B.), SFB TR22 A16 (M.L.), GRK 1043: International Graduate School of Immunotherapy (project C4; E.S. and T.B.), Behring-Röntgen-Stiftung (to M.L.), the “Forschungszentrum Immunologie (FZI)” of the university medical center (E.S. and T.B.) and the Czech Science Foundation grant P302/11/J029 (H.L, J.K. and M.Kotsyfakis). M.K. and J.K. received financial support from the Grant Agency of the Czech Republic (GACR grant P502/12/2409). A.C.C. and J.F.A. were supported by the Intramural Research Program of the Division of Intramural Research, NIAID, National Institutes of Health. Because A.C.C. and J.F.A. aRE government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the National Institutes of Health reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. Rights can be established outside the United States subject to a government use license.
We are grateful to S. Hesse and S. Fischer for expert technical assistance.
Footnotes
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kazimírová M, Štibrániová I. Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Front Cell Infect Microbiol. 2013;3:43. doi: 10.3389/fcimb.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J Biol Chem. 2006;281:26298–26307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 3.Sa-Nunes A, Bafica A, Antonelli LR, Choi EY, Francischetti IM, Andersen JF, Shi GP, Chavakis T, Ribeiro JM, Kotsyfakis M. The immunomodulatory action of sialostatin L on dendritic cells reveals its potential to interfere with autoimmunity. J Immunol. 2009;182:7422–7429. doi: 10.4049/jimmunol.0900075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horka H, Staudt V, Klein M, Taube C, Reuter S, Dehzad N, Andersen JF, Kopecky J, Schild H, Kotsyfakis M, Hoffmann M, Gerlitzki B, Stassen M, Bopp T, Schmitt E. The Tick Salivary Protein Sialostatin L Inhibits the Th9-Derived Production of the Asthma-Promoting Cytokine IL-9 and Is Effective in the Prevention of Experimental Asthma. J Immunol. 2012;188:2669–2676. doi: 10.4049/jimmunol.1100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stassen M, Schmitt E, Bopp T. From interleukin-9 to T helper 9 cells. Ann N Y Acad Sci. 2012;1247:56–68. doi: 10.1111/j.1749-6632.2011.06351.x. [DOI] [PubMed] [Google Scholar]
- 6.Marshall JS. Mast-cell responses to pathogens. Nat Rev Immunol. 2004;4:787–799. doi: 10.1038/nri1460. [DOI] [PubMed] [Google Scholar]
- 7.Matsushima H, Yamada N, Matsue H, Shimada S. TLR3-, TLR7-, and TLR9-mediated production of proinflammatory cytokines and chemokines from murine connective tissue type skin-derived mast cells but not from bone marrow-derived mast cells. J Immunol. 2004;173:531–541. doi: 10.4049/jimmunol.173.1.531. [DOI] [PubMed] [Google Scholar]
- 8.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 9.Hultner L, Koelsch S, Stassen M, Kaspers U, Kremer JP, Mailhammer R, Moeller J, Broszeit H, Schmitt E. In activated mast cells, IL-1 up-regulates the production of several Th2-related cytokines including IL-9. J Immunol. 2000;164:5556–63. doi: 10.4049/jimmunol.164.11.5556. [DOI] [PubMed] [Google Scholar]
- 10.Stassen M, Arnold M, Hultner L, Muller C, Neudorfl C, Reineke T, Schmitt E. Murine bone marrow-derived mast cells as potent producers of IL-9: costimulatory function of IL-10 and kit ligand in the presence of IL-1. J Immunol. 2000;164:5549–5555. doi: 10.4049/jimmunol.164.11.5549. [DOI] [PubMed] [Google Scholar]
- 11.Stassen M, Muller C, Arnold M, Hultner L, Klein-Hessling S, Neudorfl C, Reineke T, Serfling E, Schmitt E. IL-9 and IL-13 production by activated mast cells is strongly enhanced in the presence of lipopolysaccharide: NF-kappa B is decisively involved in the expression of IL-9. J Immunol. 2001;166:4391–4398. doi: 10.4049/jimmunol.166.7.4391. [DOI] [PubMed] [Google Scholar]
- 12.Stassen M, Klein M, Becker M, Bopp T, Neudörfl C, Richter C, Heib V, Klein-Hessling S, Serfling E, Schild H, Schmitt E. p38 MAP kinase drives the expression of mast cell-derived IL-9 via activation of the transcription factor GATA-1. Mol Immunol. 2007;44:926–933. doi: 10.1016/j.molimm.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Mittrücker HW, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–3. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 14.Glaccum MB, Stocking KL, Charrier K, Smith JL, Willis CR, Maliszewski C, Livingston DJ, Peschon JJ, Morrissey PJ. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J Immunol. 1997;159:3364–3371. [PubMed] [Google Scholar]
- 15.Horai R, Asano M, Sudo K, Kanuka H, Suzuki M, Nishihara M, Takahashi M, Iwakura Y. Production of mice deficient in genes for interleukin (IL)-1alpha, IL-1beta, IL-1alpha/beta, and IL-1 receptor antagonist shows that IL-1beta is crucial in turpentine-induced fever development and glucocorticoid secretion. J Exp Med. 1998;187:1463–1475. doi: 10.1084/jem.187.9.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissler EN, McFarland EC, Russell ES. Analysis of pleiotropism at the dominant white-spotting (W) locus of the house mouse: a description of ten new W alleles. Genetics. 1981;97:337–361. doi: 10.1093/genetics/97.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tharp MD, Seelig LLJ, Tigelaar RE, Bergstresser PR. Conjugated avidin binds to mast cell granules. JHistochemCytochem. 1985;33:27–32. doi: 10.1177/33.1.2578142. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt E, van Brandwijk R, Van Snick J, Siebold B, Rude E. TCGF III/P40 is produced by naive murine CD4+ T cells but is not a general T cell growth factor. Eur J Immunol. 1989;19:2167–2170. doi: 10.1002/eji.1830191130. [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–4. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 22.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Liang MD, Zhang Y, McDevit D, Marecki S, Nikolajczyk BS. The interleukin-1beta gene is transcribed from a poised promoter architecture in monocytes. J Biol Chem. 2006;281:9227–9237. doi: 10.1074/jbc.M510700200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Saccani S, Shin H, Nikolajczyk BS. Dynamic protein associations define two phases of IL-1beta transcriptional activation. J Immunol. 2008;181:503–512. doi: 10.4049/jimmunol.181.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakae S, Ho LH, Yu M, Monteforte R, Iikura M, Suto H, Galli SJ. Mast cell-derived TNF contributes to airway hyperreactivity, inflammation, and T(H)2 cytokine production in an asthma model in mice. J Allergy Clin Immunol. 2007;120:48–55. doi: 10.1016/j.jaci.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 26.Williams CM, Galli SJ. Mast cells can amplify airway reactivity and features of chronic inflammation in an asthma model in mice. J Exp Med. 2000;192:455–462. doi: 10.1084/jem.192.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louahed J, Zhou Y, Maloy WL, Rani PU, Weiss C, Tomer Y, Vink A, Renauld J, Van Snick J, Nicolaides NC, Levitt RC, Haczku A. Interleukin 9 promotes influx and local maturation of eosinophils. Blood. 2001;97:1035–1042. doi: 10.1182/blood.v97.4.1035. [DOI] [PubMed] [Google Scholar]
- 28.Stibrániová I, Lahová M, Bartíková P. Immunomodulators in tick saliva and their benefits. Acta Virol. 2013;57:200–216. doi: 10.4149/av_2013_02_200. [DOI] [PubMed] [Google Scholar]
- 29.Rüter C, Hardwidge PR. “Drugs from Bugs”: bacterial effector proteins as promising biological (immune-) therapeutics. FEMS Microbiol Lett. 2014;351:126–132. doi: 10.1111/1574-6968.12333. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, Rude E. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 31.Temann UA, Geba GP, Rankin JA, Flavell RA. Expression of interleukin 9 in the lungs of transgenic mice causes airway inflammation, mast cell hyperplasia, and bronchial hyperresponsiveness. J Exp Med. 1998;188:1307–20. doi: 10.1084/jem.188.7.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiener Z, Falus A, Toth S. IL-9 increases the expression of several cytokines in activated mast cells, while the IL-9-induced IL-9 production is inhibited in mast cells of histamine-free transgenic mice. 2004. pp. 122–130. [DOI] [PubMed] [Google Scholar]
- 33.Chang H-C, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, Mckinley C, Ahyi A-N, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- 35.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in Th2 development. Molecular and Cellular Biology. 2001;21:2716–2725. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, Clark RA, Kupper TS. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearley J, Erjefalt JS, Andersson C, Benjamin E, Jones CP, Robichaud A, Pegorier S, Brewah Y, Burwell TJ, Bjermer L, Kiener PA, Kolbeck R, Lloyd CM, Coyle AJ, Humbles AA. IL-9 Governs Allergen-induced Mast Cell Numbers in the Lung and Chronic Remodeling of the Airways. Am J Respir Crit Care Med. 2010;183:865–875. doi: 10.1164/rccm.200909-1462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh CK, Raible D, Geba GP, Molfino NA. Biology of the interleukin-9 pathway and its therapeutic potential for the treatment of asthma. Inflamm Allergy Drug Targets. 2011;10:180–186. doi: 10.2174/187152811795564073. [DOI] [PubMed] [Google Scholar]
- 39.Oh CK, Leigh R, McLaurin KK, Kim K, Hultquist M, Molfino NA. A randomized, controlled trial to evaluate the effect of an anti-interleukin-9 monoclonal antibody in adults with uncontrolled asthma. Respir Res. 2013;14:93. doi: 10.1186/1465-9921-14-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.