Summary
The double-edged sword nature by which IL-2 regulates autoimmunity and the unpredictable outcomes of anti-TNF therapy in autoimmunity highlight the importance for understanding how TNF regulates IL-2. Transmembrane (tm) TNF preferentially binds TNFR2 while soluble (s) TNF binds TNFR1. We have previously shown reduced IL-2 production in TNFR1−/− TNFR2−/− CD4+ T cells. Here, we generated TNFR1−/−, TNFR2−/−, or TNFR1−/− TNFR2−/− 5C.C7 TCR Il2-GFP mice and report that CD4+ T cell-intrinsic tmTNF/TNFR2 stimulates Il2 promoter activity and Il2 mRNA stability. We further utilize tmTNF Foxp3 GFP reporter mice and pharmacological TNF blockade in wild-type mice to report a tmTNF/TNFR2 interaction for Il2 expression. IL-17 is critical for host defense but its overabundance promotes autoimmunity. IL-2 represses Th17 differentiation but the role for TNFR2 in this process is not well understood. Here, we report elevated expression of TNFR2 under Th17 polarization conditions. Genetic loss-of-function experimental models as well as selective TNF blockade by etanercept and XPro™1595 in wild-type mice demonstrate that impaired tmTNF/TNFR2, but not sTNF/TNFR1, promotes Th17 differentiation in vivo and in vitro. Under Th17 polarizing conditions, elevated IL-17 production by TNFR2 KO CD4+ T cells was associated with increased STAT3 activity and decreased STAT5 activity. Increased IL-17 production in TNFR2 KO T cells was prevented by adding exogenous IL-2. We conclude that CD4+ T cell-intrinsic tmTNF/TNFR2 promotes IL-2 production that inhibits the generation of Th17 cells in a FoxP3-independent manner. Moreover, under Th17 polarizing conditions, selective blockade of CD4+ T cell-intrinsic TNFR2 appears to be sufficient to promote Th17 differentiation.
Keywords: IL-2, TNF, TNFR1, TNFR2, IL-17, NF-κ, CD4, STAT3, STAT5
Introduction
IL-2 is essential for tolerance against self-antigens as illustrated by the autoimmune phenotype of the IL-2 knockout (KO) mouse (1–3). IL-2 is produced mainly by activated CD4+ T cells. This pleotropic cytokine signals through intermediate affinity, Kd ~10−9 M, CD122 (IL-2Rβ)/CD132 (γc) and ligand-specific high affinity, Kd ~10−11 M, CD122/CD132/CD25 (IL-2Rα) receptor complexes that respond to ~1 nM and ~10 pM doses of IL-2, respectively (4) to activate downstream STAT5, MAPK, and PI3K signaling cascades (5). Consumption of IL-2 in the periphery by CD4+ effector T cells (Teffs) or T regulatory cells (Tregs) contributes to the control of Th cell differentiation and suppressor Treg function, respectively (6–9). Given the dichotomous role of IL-2 on immune regulation and the nature by which multiple cell populations compete for a common pool of IL-2, the rate and amount of IL-2 that is produced impacts the outcome of immune responses. In naïve cells, the Il2 promoter is silent but an integrated assembly of chromatin-remodeling complexes, histone modifications, and transcription factors, including AP-1, NF-κB, NFAT, and OCT-1, facilitate a rapid and transient onset of Il2 promoter activity. Il2 expression is controlled by the strength and duration of TCR signaling, co-stimulation, and rapid Il2 mRNA degradation (8,10,11). The CD28 response element (RE), located −164 to −152 bp immediately upstream of the transcriptional start site, is especially important for Il2 gene transcription and post-transcriptional regulation of Il2 mRNA stability.
Our knowledge of how different ligand-receptor interactions contribute to T cell activation and differentiation has steadily grown to include a host of co-stimulatory molecules. In addition to signal 1 through the TCR and signal 2 (co-stimulation), we and others have shown that TNF receptors also promote IL-2 production (12–14). TNF, similar to other TNF family members (e.g., LIGHT, FasL, and TRAIL), exists in membrane-bound and soluble forms. The matrix metalloprotease TNF converting enzyme (TACE) cleaves transmembrane (tm) TNF from the cell surface to generate a 17 kDa soluble (s) TNF (15). sTNF and tmTNF preferentially signal through TNF receptor type 1 (TNFR1, CD120a, p55) and TNFR2 (CD120b, p75), respectively (16,17). In contrast to the ubiquitous expression of TNFR1, TNFR2 is restricted mostly to hematopoietic cells, endothelium, microglia, and oligodendrocytes. Signaling downstream of TNFR1 and TNFR2 is distinct, yet overlapping, and is mediated by the recruitment of adaptor proteins and the activation of downstream transcription factors, including NF-κB and JNK. In contrast to TNFR2, TNFR1 contains an intracellular death domain and promotes caspase-mediated apoptosis (18, 19). Instead, TNFR2 contains intracellular TNF Receptor Associated Factor (TRAF) binding domains. We have previously associated TNFR1/TNFR2 double deficiency with impaired IL-2 production (20), but the individual contribution of each of these receptors remains undefined.
Following activation, CD4+ T cells differentiate into distinct effector subpopulations characterized by unique cytokines, transcription factors, and immune regulatory properties. CD4+ Th17 T cells are characterized by the expression of retinoic acid-related (RAR) orphan receptor (ROR)-γt and the production of two related effector cytokines, IL-17 and IL-17F. Th17 cells are essential for host protection against bacterial and fungal infections, but too much IL-17 can promote inflammation or autoimmunity (21). How TNF regulates Th17 cells is poorly understood. Given the recent interest in selective activation of TNFR2 as a therapeutic target, a better understanding of the selective roles of TNFR1 and TNFR2 on cytokine production by CD4+ T cells is needed. The objective of this study was threefold. First, determine the individual contribution of TNFR1 and TNFR2 on IL-2 expression. Second, determine whether regulation of IL-2 expression by TNFR1 or TNFR2 is CD4+ effector T cell-specific. Third, determine whether CD4+ Teff-specific ablation of TNFR2 influences Th17 cell differentiation.
To investigate the individual contribution of TNFR1 and TNFR2 on IL-2 expression, we generated 5C.C7 TCR Rag2−/− Il2-GFP reporter mice that are singly deficient for either TNFR1 or TNFR2 or doubly deficient for both TNFR1 and TNFR2 to examine IL-2 production at the level of promoter activity. We now report that an interaction of CD4+ T cell-intrinsic TNFR2 and tmTNF, but not TNFR1/sTNF, co-stimulates Il2 expression to fine tune the generation of CD4+ IL-2 producers. Although TNF has been implicated in Th17 differentiation (22, 23), not much is known about the generation of Th17 cells in response to TNFR2 signaling. Here, we show that in addition to promoting the generation of FoxP3+ Tregs, TNFR2 inhibits Th17 differentiation by promoting Il2 expression. Lastly, we show that blockade of CD4+ T cell-intrinsic TNFR2 is sufficient to promote Th17 differentiation under Th17 polarizing conditions.
Materials and Methods
Mice
All animals were bred and housed under specific pathogen-free conditions in MU facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. All experimental procedures using animals were approved by the MU Institutional Animal Care and Use Committee and were performed in accordance with the Guide for the Use and Care of Laboratory Animals. B10.A (H-2a) 5C.C7 Rag2−/− (referred to as 5C.C7) mice are specific for moth cytochrome c (MCC) aa 88–103 and pigeon cytochrome c (PCC) aa 81–104 bound to I-Ek (24). 5C.C7, 5C.C7 Il2-GFP+/+ (referred to as WT) (25), and 5C.C7 TNFR1/2 double knockout (referred to as dKO) (14, 26) mice were obtained from the National Institute of Allergy and Infectious Diseases (NIAID) contract facility (Taconic Farms, Germantown, NY). WT mice were crossed with 5C.C7 TNFR1/2 dKO mice to generate the following mouse models that are either homozygous (+/+) or heterozygous (+/−) or GFP: 5C.C7 TNFR1/2 dKO Rag2−/− Il2-GFP+/− (Il2+/−) and 5C.C7 TNFR1/2 dKO Rag2−/− Il2-GFP+/+ (Il2−/−) reporter mice (referred to as TNFR1/2 dKO); 5C.C7 TNFR1 KO Rag2−/− Il2-GFP+/− (Il2+/−) and 5C.C7 TNFR1 KO Rag2−/− Il2-GFP+/+ (Il2−/−) reporter mice (referred to as TNFR1 KO); and 5C.C7 TNFR2 KO Rag2−/− Il2-GFP+/− (Il2+/−) and 5C.C7 TNFR2 KO Rag2−/− Il2-GFP+/+ (Il2−/−) reporter mice (referred to as TNFR2 KO). Heterozygous TNFΔ1–12 mutant mice, generated by G. Kollias (27), were purchased from the Italian National Research Council European Mouse Mutant Archive (CNR-EMMA) at the International Campus in Monterotondo (CNR-EMMA Monterotondo, Rome, Italy). TNFΔ1–12 mice were generated by replacing the endogenous TNF alleles with mutant tmTNF alleles that have a deletion (Δ) of aa 1–12 to eliminate the TACE cleavage site. Lymphotoxin alpha (LTα) gene expression in homozygous TNFΔ1–12 mice is comparative to that of Tnftm/wt (27). Homozygous TNFΔ1–12 mice were backcrossed onto the C57BL/6J background for eight generations and then crossbred with C57BL/6J (H-2b) 2D2 TCR Foxp3-GFP reporter mice, carrying the Vα3.2/Vβ11 TCR transgene generated in the Kuchroo Laboratory (28). C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The Foxp3-GFP reporter mice were a gift from H. Zaghouani (University of Missouri, Columbia, MO).
Antibodies and Reagents
TCR Vβ11 (KT11), CD120α (55R-286), CD120β (TR75-89), CD26 (H194-112), CD278 (C398.4A), and CD62L (MEL-14) were purchased from BioLegend (San Diego, CA). TCRβ (H57-597), CD4 (RM4-5), CD4 (GK1.5), CD8α (Ly-2), CD120α (55R-286), CD69 (H1.2F3), CD25 (PC61), TNF (MP6-XT22), CD45Rb (16A), and CD16/CD32 (2.4 G2) were purchased from BD Biosciences (San Jose, CA). FoxP3 (FJK-16s), CD28 (37.51), and LAP (TW7-16B4) were purchased from eBioscience (San Diego, CA). Annexin V and 7-aminoactinomycin D (7-AAD) were purchased from BD Biosciences. Lyophilized recombinant mouse TNF (aa Val77-Leu233 [Accession# P06804], expressed in E. coli.) was purchased (R&D Systems, Minneapolis, MN) as a lyophilized powder, 0.2 µm filtered in PBS without a carrier protein, stored at −80°C, and reconstituted in sterile PBS with 0.01% BSA (Fraction V; Sigma-Aldrich, St. Louis, MO) as a carrier protein just prior to use. MCC aa 88–103 and myelin oligodendrocyte glycoprotein (MOG) aa 35–55 were purchased from AnaSpec Inc. (Fremont, CA). PMA, ionomycin, and cyclosporin A (CsA) were purchased from Calbiochem-EMD Millipore (Dormstadt, Germany). CFA was purchased from Sigma-Aldrich.
Pharmacological TNF Blockade
A novel dominant-negative TNF (DN-TNF) analogue XPro™1595 (Xencor, Monrovia, CA), engineered for selective sTNF inhibition, was a gift from Dr. David Szymkowski, Xencor, Inc. XPro™1595 was administered at concentrations approximately ten-fold higher than native sTNF. XPro™1595 rapidly, within minutes, neutralizes greater than 99% of sTNF while sparing tmTNF activity (29, 30). Etanercept (Enbrel®; Amgen, Thousand Oaks, CA), a decoy TNFR2 which nonselectively blocks sTNF, tmTNF, and lymphotoxin (31, 32), was purchased from a pharmacy. For in vivo TNF neutralization, groups of mice were treated with twice-weekly s.c. injections of XPro™1595 (10 mg/kg) (29, 30), etanercept (10 mg/kg) (33), or saline vehicle as control starting on the day of immunization. Entenercept has previously been shown to antagonize IL-2 production (34).
Measurement of TNF Bioactivity
The manufacturer (R&D Systems) used lysis of murine L929 cells in the presence of actinomycin D to establish an ED50 value of 0.1 ng/ml, corresponding to a specific activity of ≥ 1 × 107 units/mg. Given that sTNF is bioactive only as a homotrimer of noncovalently linked 17 kDa monomers and disintegration of the recombinant homotrimer complex is possible, we further tested the bioactivity of recombinant sTNF in our culture system. For these analyses, pooled lymph nodes (axillary, brachial, and mesenteric) and splenocytes (5 × 106 cells/ml in cRPMI) were stimulated with soluble anti-TCR-β (0.5 µg/ml) with increasing concentrations of reconstituted murine rTNF and incubated at 37°C and 5% CO2. NO, released from cells and converted to nitrite (NO2) in the culture medium, was determined using the Griess reaction as previously described (35). Aliquots (100 µl) of supernatants were incubated with an equal volume Reagent A (1% sulfanilamide in 5% phosphoric acid) plus Reagent B (0.1%, w/v, N-1-napthylethylenediamine dihydrochloride) and absorbance was measured at 543 nm using a BioTek Synergy™ 4 spectrophotometry microplate reader (BioTek; Winooski, VT). NO2 was quantified using serially diluted (0 to 100 µM) sodium nitrite (NaNO2) as a standard (36). The data are expressed as micromolar (µM) NO2 with a limit of detection of 0.78 µM.
CD4+ T cell Purification
Lymph node (LN) T cells from 5 to 16 wk old 5C.C7 Rag2−/− mice were, at times, sorted for purity of CD4+ CD45high or CD4+CD44lowCD62high (> 95%) using a MoFlo™ XDP flow cytometer (Beckman Coulter, Pasadena, CA). T cells purified from the spleens (SPLNs) of 5C.C7 mice and the lymph nodes and spleens of C57BL/6J mice were first enriched using EasySep™ CD4 negative selection (STEMCELL Technologies, Vancouver, BC, Canada), prior to MoFlo™ XDP sorting. T cells from C57BL/6J Foxp3-GFP reporter mice further sorted to exclude Foxp3-GFP positive cells. Post-sort cell viability was assessed by trypan blue exclusion. MoFlo™ XDP flow cytometer sorts typically yielded >99% purity.
CD4+ T cell Activation
Naive T cells were isolated from cervical, axillary, brachial, inguinal, and mesenteric lymph nodes of 5C.C7 Rag2−/− mice. Greater than 90% of the freshly isolated cells from the LNs were CD4+ T cells. Unless otherwise indicated, no further CD4+ T cell purification was performed. Freshly isolated CD4+ T cells were washed in complete RPMI (cRPMI), collected by centrifugation, and plated at a density of 1 × 105 cells/ml in round-bottom 96-well plates (Corning, Corning, NY). Direct addition of soluble MCC88–103 to cell cultures was used to study Ag-induced responses. Alternatively, plates pre-coated with anti-TCRβ (10 µg/ml or as otherwise indicated) plus anti-CD28 or co-cultures using MCC88–103 in combination with irradiated WT splenocytes as antigen presenting cells were used. Cell cultures were maintained in RPMI-1640 (Invitrogen Life Technologies, Grand Island, NY), supplemented with 10% FCS, 2 mM L-glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin, 100 g/ml streptomycin, 40 µg/ml gentamicin, 10 mM HEPES, 1 mM sodium pyruvate, and 50 µM β-mercaptoethanol. Cultures were supplemented with anti-CD28 (37.51 ascites fluid [a gift from J. Allison, MD Anderson Cancer Center, Houston, TX] at a final dilution of 1/1000) for co-stimulation. Alternatively, cells were activated in response to PMA (10 ng/ml) and ionomycin (1 µM). Foxp3-GFP reporter and tmTNF transgenic (Tg) mice were on the H-2b–restricted C57BL/6J background and some of these mice were 2D2 TCR transgenic. T cells from these mice were isolated and purified as described above, but activated in response to plate-bound TCRβ and anti-CD28 or MOG35–55.
Cytokine Secretion
IL-2, IL-17A, IFN-γ, and TNF were quantified by mouse Ready-SET-Go!® ELISA kits (eBioscience). Cell-free culture supernatants and plasma samples were stored at −80°C until use. The absorbance (450 nm) minus background (570 nm) of the colorimetric reactions were quantified using a BioTek Synergy™ 4 spectrophotometry microplate reader (BioTek). The lower limit of sensitivity of the TNF assay was 8 pg/ml. The recognition of TNF by this assay is not interfered with by 1000X excess soluble TNFR1 or TNFR2.
Th17 Cell Differentiation
In Vivo—FACS-purified CD4+ CD62highCD44low T effector cells (1 × 106) from WT or 5C.C7 TNFR2 KO Il2-GFP+/− and Il2-GFP+/− mice were transferred into B10.A Rag2−/− mice. The host mice were then immunized with MCC88–103 (20 µg) emulsified in CFA (200 µl) as previously described (37). LNs and spleens were harvested four days after immunization. CD4+ T cells were enriched (> 90% purity) using EasySep™ CD4+ negative enrichment kits and restimulated with PMA and ionomycin for 12 h. Cell-free supernatants were collected and stored at −80°C until use. IL-17A and IFN-γ cytokine secretion were quantified by ELISA. In Vitro— Th0 polarization: Naïve CD4+ T cells (1 X 106 c/ml), purified from WT or TNFR2 KO B10.A 5C.C7 Rag2−/− Il2-GFP+/− (Il2+/−) or Il2-GFP+/+ (Il2−/−) mice were stimulated for 3 days with plate-bound α-TCRβ (H57, 5 µg/ml) and soluble α-CD28 with no added cytokines. For Th17 polarization, cells were polarized with IL-6 (50 ng/ml, PeproTech) TGF-β (2.5 ng/ml, R&D Systems), α-IFN-γ (XMG1.2, 10 µg/ml, Bio X Cell) and α-IL-4 (11B11; 10 µg/ml , Bio X Cell) in addition to the plate-bound α-TCRβ and soluble α-CD28. When indicated, IL-2 (100 U/ml, Life Technologies) was added at the beginning of stimulation. Cells were collected at 72 h for RNA, intracellular cytokine staining, flow cytometry, and ELISA.
Stability of mRNA
Naive CD4+ T cells (1 X 106) were stimulated with α-TCRβ plus plate-bound anti-CD28 for 20 h. Further Il2 transcription was blocked by addition of CsA to the cell cultures to a final concentration of 200 ng/ml without removal of the original stimulant. Cells were collected at 0, 1, 2, 4, 6, and 8 h after the CsA addition. Total RNA was isolated using the RNeasy Mini kit (Qiagen, Venlo, Limberg, Netherlands) and reverse transcribed into cDNA using Superscript® III reverse transcriptase and random hexamer primers according to the manufacturer’s recommended protocol (Invitrogen Life Technologies). The level of Il2 mRNA was determined by quantitative RT-PCR (qRT-PCR) following normalization to the T cell-specific gene, CD3δ, using the ΔΔCT method for relative quantitation with Il2 mRNA levels in unstimulated cells as the calibrator (38). TaqMan® probe and primers for Il2 and CD3δ were used (Applied Biosystems). The addition of the general inhibitor of transcription, actinomycin D (ActD), has previously been shown to stabilize Il2 mRNA degradation (39, 40). Therefore, we selected to use CsA-induced blockage of Il2 transcription (41, 42).
The exact, non-parametric Wilcoxon-Mann-Whitney test was used to determine differences in mRNA at 0 h between WT and TNFR2 KO T cells, using the ΔΔCT values from qRT-PCR. In order to determine whether Il2 mRNA degradation differed between WT and TNFRs, a generalized linear mixed model was used to model ΔΔCT values obtained from qRT-PCR for the WT and TNFR2 KO T cells (n = 4, each group). The model is of the form, ΔΔCtijk= µ + τi +Sij + γk +((τ)iγk) + εijk where i denotes the group (WT or TNFR2 KO), j denotes the mouse (j =1, 2, 3, 4, 5, 6, 7, 8), and k denotes the time (k = 0, 1, 2, 4, 6, 8). ΔΔCtijk denotes the delta delta Ct value for group i of mouse j at time k, µ is the overall mean ΔΔCT value, τi denotes the group effect, Sij denotes the effect of mouse j in group i, γk denotes the effect of time k, ((τ)iγk) denotes the interaction between group i and time k, and εijk denotes the experimental error. An F-test was used to determine differences in the degradation of Il2 mRNA between the WT and TNFR2 KO T cells.
Flow Cytometry
Cells were incubated with anti-CD16/32 prior to staining in order to prevent nonspecific Ab capture by FcRs. Cells were then stained directly with conjugated antibodies in FACS buffer (PBS containing 1% FCS, 0.05% EDTA, and 0.02% sodium azide). 7-AAD and, in some cases, Annexin V were used to exclude dead cells and quantify cell viability. FACSCalibur™ (BD Biosciences) and FlowJo v8.8.7 (FlowJo Inc., Ashland, OR) software were used.
NF-κB and AP-1 Activity
NF-κB/Rel and JNK transcriptional activity was determined by the oligonucleotide binding capability of p50, p52, RelA (p65), RelB, Rel C, c-Jun, and c-Fos. The NF-κB Family EZ-TFA® Transcription Factor Assay Chemiluminescent (EMD Millipore) and the TransAM® AP-1 DNA-binding kits (Active Motif, Carlsbad, CA) were used. Nuclear extracts were prepared, according to the manufacturer’s recommended protocols, from FACS-purified WT or TNFR2 KO CD4+ T cells after 2 and 40 h stimulation with plate-bound anti-TCRβ plus soluble anti-CD28. The Coomassie (Bradford) protein assay (Thermo Pierce, Rockford, IL) was used to quantify nuclear protein. Nuclear extracts (2.5 µg) were incubated with a biotinylated double-stranded oligonucleotide probe containing either the NF-κB/Rel consensus sequence, 5'-GGGACTTTCC-3', or the AP-1 consensus sequence, 5'-TGACGTCA-3', for AP-1 on streptavidin-coated 96-well plates. Captured complexes, including positive control NF-κB or AP-1 proteins from Raji nuclear extracts or TNF-treated HeLa whole cell extracts (supplied by manufacturers), were incubated with the primary anti-rabbit Abs, followed by horseradish peroxidase–conjugated secondary antibody and tetramethylbenzidine substrate. The absorbance of the colorimetric (450 nm) and chemiluminescence reactions were quantified using a Biotek Synergy™ 4 spectrophotometry microplate reader. All antibodies, excluding c-Rel (Santa Cruz), were provided with the EZ-TFA® and TransAM® kits.
Quantitative Real-Time Reverse-Transcription Polymerase Chain Reaction
Real-time quantification of IL-17A, IL-17F, interferon-γ (IFN-γ), forkhead box P3 (FoxP3), RAR-RORγt, RORα, aryl hydrocarbon receptor (AHR), and aryl hydrocarbon receptor nuclear translocator (ARNT) was performed using SYBR® Green PCR master mix (Applied Biosystems, Foster City, CA) and a StepOne™ Real-Time PCR System (Life Technologies). Total RNA was isolated using the RNeasy Mini Kit and reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random hexamer primers according to the manufacturer’s instructions. Purity and concentration of RNA was determined by UV-spectroscopy using a NanoDrop® 1000 Spectrophotometer (Thermo Scientific Inc.). Target gene mRNA was determined by qRT-PCR following normalization to β-actin using the ΔΔCT method for relative quantitation with target gene mRNA levels in unstimulated WT cells as the calibrator (38). PCR was performed using the following primers: The primer sets were as follows: AHR, 5′-CTGGTTGTGACAGCAGATGCCT-3′ (forward) and 5′-CGGTCTTCTGTATGGATGAGCTC-3′ (reverse); ARNT, 5′-CTCACGAAGGTCGTTCATCTGC-3′ (forward) and 5′-CCACAAAGTGAGGTTCTCCTTCC-3′ (reverse); β-actin, 5′-ATGGTGGGAATGGGTCAGAA-3′ (forward) and 5′-CCATGTCGTCCCAGTTGGTAA-3′; Foxp3, 5′-CAAGGGCTCAGAACTTCTAG-3′ (forward) and 5′-GGTTCAAGGAAGAAGAGGTG-3′ (reverse); IFN-γ, 5′-ACTGGGAAAAGGATGGTGAC-3′ (forward) and 5′-GACCTGTGGGTTGTTGACCT-3′ (reverse); IL-17A, 5′-TCCAGAAGGCCCTCAGACTA-3′ (forward) and 5′-AGCATCTTCTCGACCCTGAA-3′ (reverse); IL-17F, 5′-CCCAGGAAGACATACTTAGAAGAAA-3′ (forward) and 5′-CAACAGTAGCAAAGACTTGACCA-3′ (reverse); RORα, 5′-TTGGTCGGATGTCCAAGAAG-3′ (forward) and 5′-TGGCTGAGATGTTGTAGGTG-3′ (reverse); RORγt, 5′-CAGTCTACATGCAGAAGTGC-3′ (forward) and 5′-ATGTAAGTGTGTCTGCTCCG-3′ (reverse).
Statistical Analyses
Unless otherwise indicated, statistical analyses used GraphPad Instat 3 (GraphPad, La Jolla, CA). Differences were determined using the two-tailed Student’s t test or the non-parametric Wilcoxon-Mann-Whitney test. Significance was defined as reported.
Results
tmTNF, but not sTNF, promotes Il2 transcription during CD4+ T cell priming
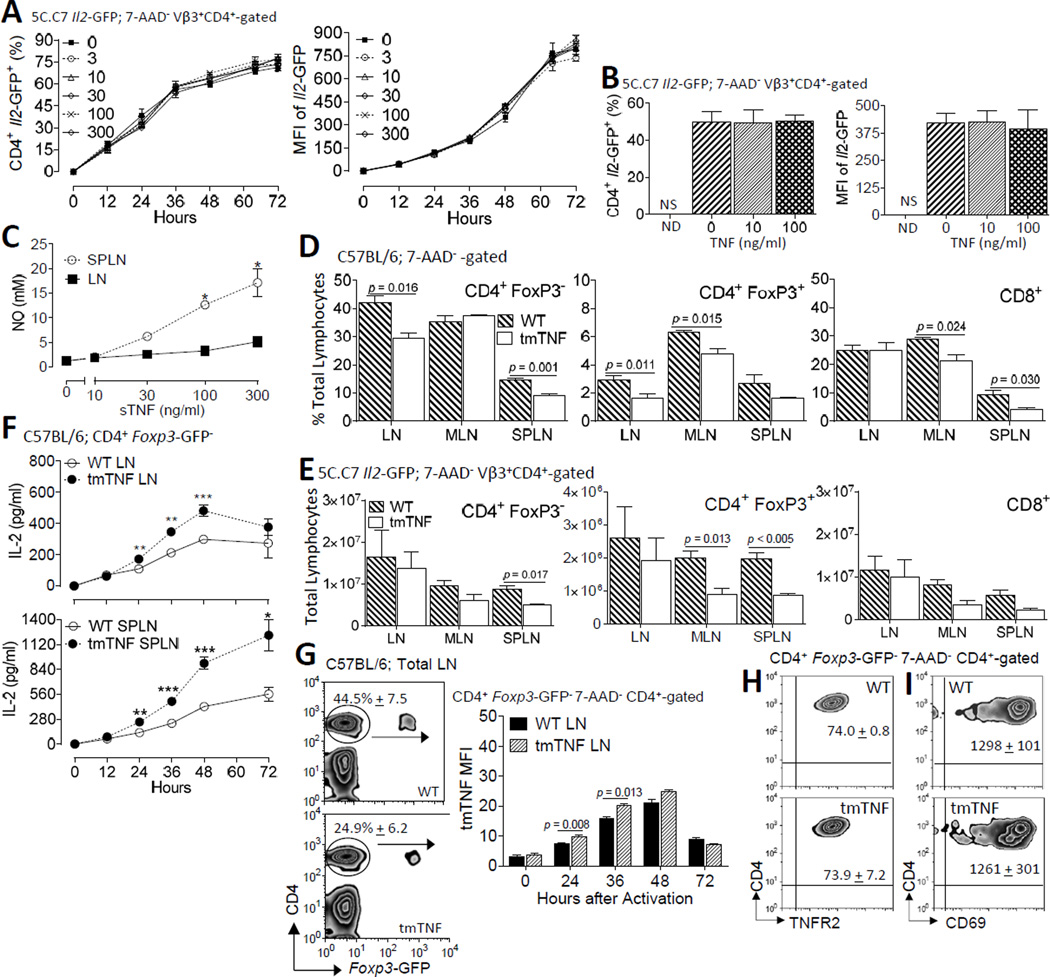
We previously showed a lower frequency of IL-2 producing CD4+ T cells in TNFR1/2 double KO mice (14). To directly study T cell-intrinsic TNFR1 and TNFR2 on Il2 transcription, we now utilize Il2-GFP reporter mice (25). These mice were originally generated by replacing the Il2 gene with a single copy of a cDNA encoding GFP and subsequently bred onto the B10.A 5C.C7 TCR Rag2−/− background (14). The gfp transcripts in these transgenic mice lack the UA-rich sequence instability elements that normally reside in the 3'-untranslated region (UTR) (43). In addition, GFP has a considerably longer half-life than Il2. Therefore, this genetic model provides an ideal system to study the role of TNFR on Il2 promoter activity. In an initial set of experiments, single cell suspensions, comprised of greater than 90% CD4+ T cells, from pooled lymph nodes isolated from homozygous Il2-GFP-reporter (IL-2−/−) mice, were activated with plate-bound anti-TCRβ plus anti-CD28 in the presence or absence of increasing concentrations (0 – 300 ng/ml) of recombinant sTNF (Fig. 1). Under these conditions, approximately 75% of all CD4+ T cells transcribed the Il2 gene as reflected by GFP expression. The addition of exogenous sTNF to the cell cultures did not influence Il2 promoter activity as determined by the time of onset of GFP expression, the frequency of IL-2 producers, and the mean fluorescence intensity (MFI) by individual IL-2 producers (Fig. 1A). Although slight shifts in the frequency and MFI of GFP+ CD4+ T cells were observed at the later times following TNF treatment, these effects were without dose-response or statistical significance (p > 0.05) (Fig. 1A). Given that CD4+ T cells up-regulate TNFR1 and TNFR2 following activation (14), we next determined whether the addition of exogenous TNF subsequent to, rather than at the time of, T cell activation was more effective in promoting Il2 promoter activity. Accordingly, the first set of experiments were repeated, except sTNF was added 24 h following T cell activation, coinciding with elevated surface TNFR2 expression on CD4+ T cells (14). Cells were collected 64 h after stimulation. Similar to the first set of experiments, no change in IL-2 production in response to sTNF was observed (Fig. 1B). Increased NO production in splenic cultures validated bioactivity of the sTNF in our culture conditions (Fig. 1C). In contrast to the splenocyte cultures, little NO was produced by lymph node cultures (Fig. 1C). These results are consistent with previous reports demonstrating NO production in spleens but not lymph nodes (44).
FIGURE 1. Increased IL-2 production by CD4+ T cells in response to tmTNF but not sTNF.
(A) The addition of sTNF during primary activation did not alter the frequency of IL-2 producers or the magnitude of Il2 promoter activity in Vβ3+ CD4+ 7-AAD−-gated T cells from B10.A 5C.C7 TCR Tg Il2-GFP homozygous (Il2−/−) reporter mice. Freshly isolated LN cells (1 X 106 c/ml; 200 µl/well; > 90% Vβ3+ CD4+ T cells) were stimulated with plate-bound anti-TCRβ plus soluble anti-CD28 supplemented with increasing concentrations of sTNF (0, 3, 10, 30, 100, or 300 ng/ml) for 12, 24, 36, 48, 60 and 72 h. TNF was added directly to the cell cultures at the time of activation. (B) The experiments in A were repeated, except sTNF (0, 10, or 100 ng/ml) was added to the cell cultures 24 h after the T cells were stimulated; the cells were harvested 36 h thereafter and stained for CD4 and Vβ3; 7-AAD Vβ3+ CD4+ cells were gated for analyses by flow cytometry. Values shown represent the mean ± SEM of three independent experiments in triplicate (three mice/group/experiment). Addition of sTNF did not yield a statistical significance. (C) Culture supernatants were tested for levels of NO to validate sTNF bioactivity. Values shown represent the mean + SEM of three experiments, performed in triplicate, of total splenocytes (106 c/ml; 200 µl/plate) or pooled axillary and brachial lymph nodes (106 c/ml; 200 µl/plate) from two 5C.C7 Il2-GFP+/− mice/experiment stimulated with plate-bound anti-TCRβ plus anti-CD28 in the presence or absence of increasing concentrations of sTNF (0, 3, 10, 30, 100, or 300 ng/ml) for 48 h. *p < 0.05. The percentages (D) and total numbers (E) of CD4+ FoxP3−, CD8+, and CD4+ FoxP3+ T cells from freshly isolated cervical, axillary, brachial, inguinal LN, MLN, and spleens from C57BL/6 WT and tmTNF Foxp3-GFP reporter mice were compared. 7-AAD− lymphocytes were gated for analyses by flow cytometry. (F) IL-2 secretion differs between WT and tmTNF CD4+ T cells. FACS-purified CD4+ CD45Rbhigh FoxP3− T effector cells (1 X 105 c/well) from 2D2 WT or tmTNF mice were stimulated with irradiated splenocytes (1 X 106 c/well) from WT C57BL/6J mice and cognate MOG35–55 (10 µM). IL-2 secretion from cell-free culture supernatants was quantified by ELISA at times up to 72 h. Graphs depict the results of three independent experiments performed in triplicate (mean ± SEM) **p < 0.01, ***p < 0.001. (G) In the left panel, representative staining patterns of CD4+ FoxP3− and FoxP3+ CD4+ T cells among total peripheral LNs from WT and tmTNF mice are shown with the frequencies (mean ± SEM) of CD4+ FoxP3− T cells from four mice of each strain indicated. In the right panel, viable (7-AAD−) CD4+ T cells were gated for tmTNF expression and analyzed by flow cytometry. The graph summarizes the tmTNF MFI (mean ± SEM) on CD4+ FoxP3− cells from WT and tmTNF mice before and after stimulation for the times indicated. The results from two independent experiments (three mice per experiment; cultured separately) are shown. (H) Representative staining patterns of surface TNFR2 expression on FACS-purified CD4+ FoxP3− T cells from G following stimulation for 48 h. The MFI (mean ± SEM) of TNFR2 from two independent experiments (three mice per experiment; cultured separately) are indicated. (I) Representation of CD69 expression patterns on the FACS-purified CD4+ FoxP3− T cells from G stimulated for 48 h. The MFI (mean ± SEM) of CD69 from two independent experiments (three mice per experiment; cultured separately) are shown.
Whereas tmTNF preferentially binds to TNFR2, sTNF mostly bind to TNFR1. Further, these two TNF receptors activate distinct downstream signaling cascades. To test the hypothesis that tmTNF, but not sTNF, selectively co-stimulates Il2 expression in CD4+ T cells, we crossed memTNFΔ1–12 mice, encoding an uncleavable tmTNF (27), with Foxp3-GFP mice (28) to generate tmTNF Foxp3-GFP reporter (tmTNF) mice. These tmTNF mice were then bred onto the MOG35–55-specific 2D2 TCR background to provide an Ag-specific model to assess the role of tmTNF on IL-2 production. The percentage of FoxP3− CD4+ T cells in LN (p < 0.05) and SPLN (p < 0.001) were reduced in tmTNF mice relative to age- and gender-matched WT cohorts. The frequencies of LN and mesenteric LN (MLN), but not SPLN, FoxP3+ CD4+ Tregs were also reduced in tmTNF mice compared to WT mice (p < 0.05) (Fig. 1D). In contrast to CD4+ T cells, the frequency of MLN and SPLN, but not LN, CD8+ T cells were lower in tmTNF compared to WT mice (p < 0.05) (Fig. 1D). These changes coincided with a decrease in the total number of FoxP3− CD4+ T cells in the spleen (p < 0.05), as well as, the total number of FoxP3+ CD4+ Tregs in the spleen and MLN (p < 0.05). The number of MLN or SPLN CD8+ T cells did not differ between WT and tmTNF mice (p > 0.05) (Fig. 1E). Collectively, these observations indicate that selective blockade of sTNF is sufficient to perturb T cell homeostasis, including Treg cells, in secondary lymphoid organs, but does not induce overt inflammation or spontaneous autoimmunity.
To determine whether CD4+ T cell-intrinsic tmTNF plays a regulatory role in IL-2 production, naive CD4+ CD45RbHi Foxp3-GFP− T cells from 2D2 WT or tmTNF mice were FACS-purified and activated with MOG35–55 in the presence of irradiated T cell-depleted WT C57BL/6J splenocytes. IL-2 secretion was quantified by ELISA at time points up to 72 h. CD4+ T cells from tmTNF mice produced more IL-2 (Fig. 1F) and expressed more cell surface TNF (Fig. 1G), but not TNFR2 (Fig. 1H) relative to WT CD4+ T cells. CD69 expression did not differ between WT and tmTNF CD4+ T cells (p > 0.05) to imply that increased production of IL-2 did not positively correlate with TCR signaling (Fig. 1I). The absence of sTNF secretion by tmTNF CD4+ T cells following T cell activation was confirmed by ELISA (data not shown).
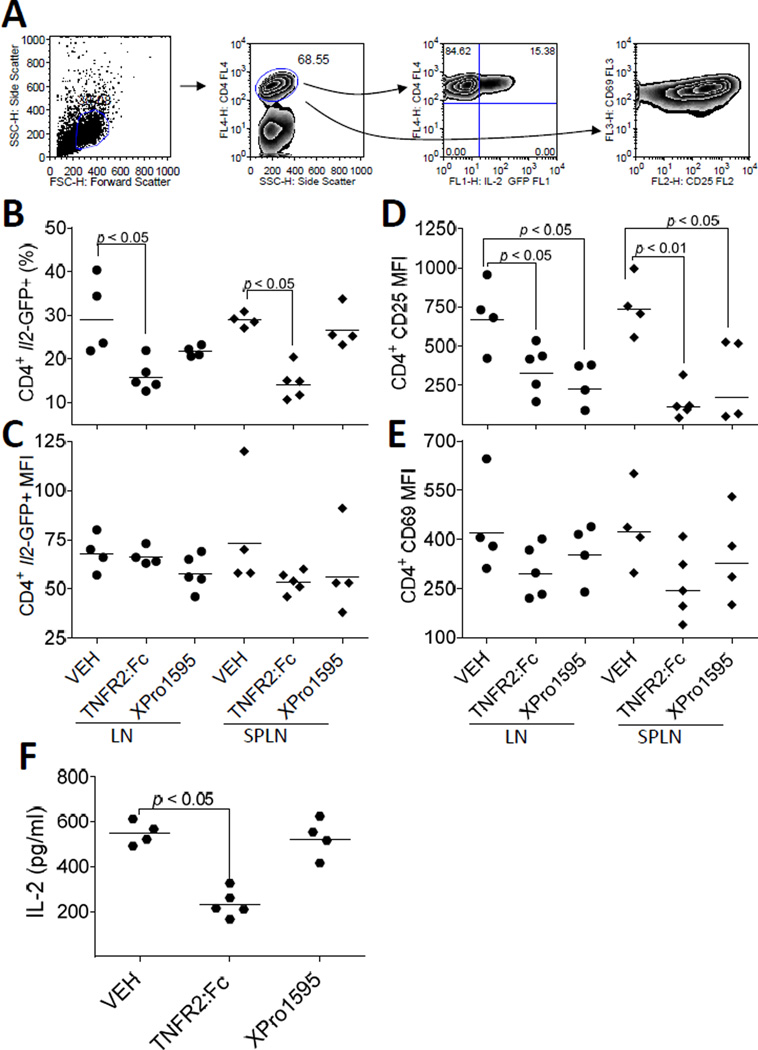
To further investigate differential regulation of IL-2 production by sTNF and tmTNF, 5C.C7 Il2-GFP+/− (Il2+/−) mice were administered (subQ) XPro™1595 (10 µg/g) or TNFR2:Fc (etanercept; 10 µg/g) prior to immunization in order to pharmacologically block sTNF or total TNF, respectively. CD4+ T cells were collected from the draining LN and spleens 24 h following immunization with MCC (300 µg) and LPS (10 µg). Viable CD4+ T cells were gated to analyze IL-2-GFP and cell surface CD69 expression by flow cytometry (Fig. 2A). The percentage of IL-2 producing CD4+ T cells was reduced following total, but not soluble, TNF blockade (p < 0.05) (Fig. 2B). Neither of the TNF blocking treatments inhibited the magnitude of IL-2 production per cell as determined by Il2-GFP MFI levels (p > 0.05) (Fig. 2C). Despite normal levels of IL-2 production, CD25 expression on CD4+-gated cells was reduced by both total TNF and sTNF-selective blockade (p < 0.05) (Fig. 2D). CD4+ T cell expression of CD69 also was unaffected by TNF blockade (p > 0.05) (Fig. 2E). Inhibition of IL-2 production by TNFR2:Fc, but not XPro™1595, was further revealed by reduced circulating levels of IL-2 (Fig. 2F). Overall, these studies suggest that tmTNF, but not sTNF, promotes IL-2 production. These observations are consistent with a model whereby tmTNF/TNFR2 provides a third signal to co-stimulate Il2 promoter activity.
FIGURE 2.
Blockade of tmTNF inhibits IL-2 production in vivo. (A) Draining LNs, spleens, and sera collected from PBS (VEH), XPro™1595 (10 µg/g, subQ), or TNFR2:Fc (10 µg/g, subQ) treated B10.A 5C.C7 Il2-GFP+/− (Il2+/−) mice 20 h following immunization with MCC88–103 and LPS (10 µg/g) associate blockade of tmTNF with impaired IL-2 production. (B - D) Each symbol represents a single mouse. Circles indicate live CD4+-gated cells isolated from the draining LNs. Diamonds indicate live CD4+-gated cells isolated from the spleens. Horizontal lines represent the geometric means. (B) The percentage of Il2-GFP-positive cells out of total live CD4+ T cells were determined by flow cytometry. (C) The MFI of Il2-GFP was determined from live GFP-positive CD4+-gated T cells shown in B. (D and E) 100% of the live LN and splenic CD4+ T cells from VEH-pretreated mice were CD25 and CD69 positive as determined by flow cytometry. Therefore, the MFI of CD25 (D) and CD69 (E) for all treatment groups were determined by gating on total live CD4+ T cells. (F) Sera was collected from the retro-orbital sinus at necropsy. The concentration of secreted IL-2 protein was determined by ELISA. Each hexagon represents an individual mouse; the mean concentrations of IL-2 (pg/ml) are indicated by horizontal lines. VEH, n = 4 mice; TNFR2:Fc, n = 5 mice, XPro™1595, n = 4 mice. Statistical significance (p < 0.05) is indicated.
TNFR2, but not TNFR1, augments Il2 promoter activity during the priming of CD4+ T cells
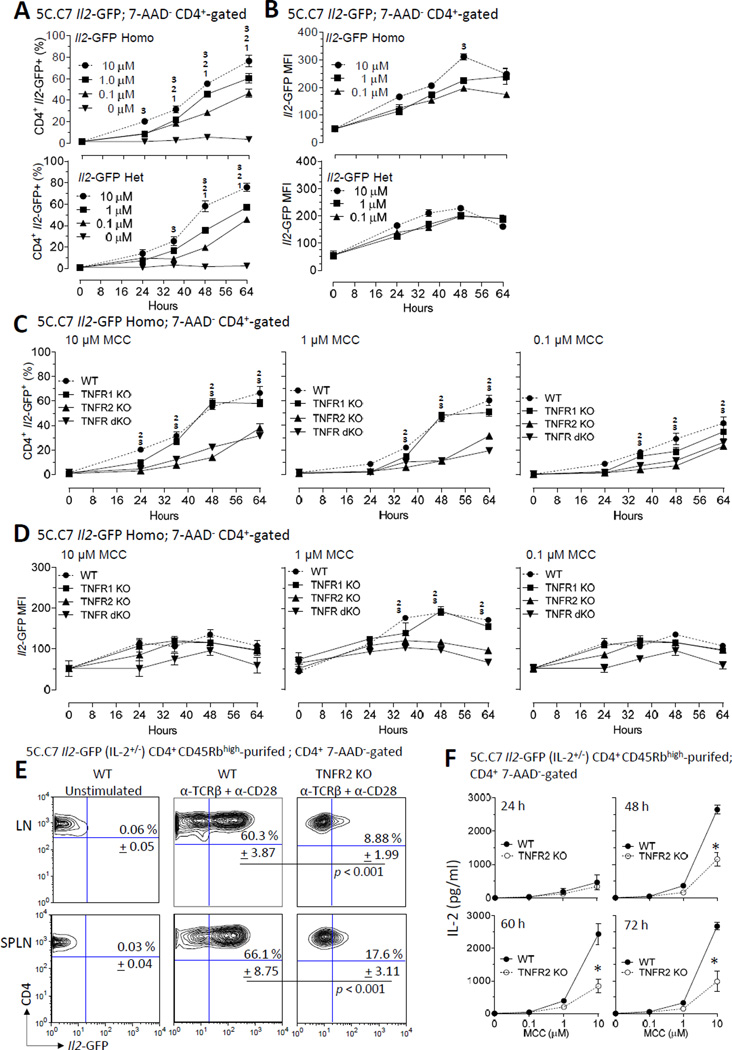
Transmembrane TNF is more effective in activating TNFR2 than sTNF (16, 17, 45). TNFR1 and TNFR2 activate distinct, albeit sometimes convergent, downstream signaling (17). Therefore, we evaluated the role for these two TNF receptors on Il2 transcription. New mouse models were first generated by crossing 5C.C7 Il2-GFP mice to 5C.C7 TNFR1/2 dKO mice to generate Ag-specific Il2-GFP mice that were deficient for either TNFR1 and/or TNFR2 as described in Materials and Methods. Lymphocytes (>90% Vβ3+ CD4+ T cells) from the lymph nodes or spleens of Il2-GFP (WT), TNFR1 KO Il2-GFP (referred to as TNFR1 KO), TNFR2 KO Il2-GFP (TNFR2 KO), or TNFR1/2 dKO Il2-GFP (referred to as TNFR dKO) mice were stimulated with increasing concentrations of MCC88–103 (Fig. 3). Mice that are homozygous for the GFP allele do not transcribe Il2 mRNA (25). Further, IL-2 signaling may indirectly impact GFP expression. Therefore, for these studies, homozygous and heterozygous Il2-GFP mice were compared. The frequency of IL-2 producers (% CD4+ Il2-GFP+) (Fig. 3A) and the magnitude of Il2 transcription (Il2-GFP MFI of the CD4+ Il2-GFP+ cells) (Fig. 3B) were quantified by flow cytometry from 7-AAD− CD4+-gated cells. These studies revealed similar Il2 promoter activity in GFP homozygous and heterozygous WT mice, suggesting IL-2 receptor signaling-independence Il2 promoter activity. The reason for the decline in GFP MFI after 48 h is not completely clear; however, cell division and GFP degradation are possible contributory factors. In comparing homozygous and heterozygous Il2-GFP cells at 48 h, the MFI of homozygous Il2-GFP producers (Fig. 3B, top panel) differed from heterozygous Il2-GFP cells (Fig. 3B, bottom panel) at the highest Ag dose (p < 0.05). Neither the absence of Il2 promoter activity in unstimulated T cells, nor the rate of onset of Il2 induction in response to T cell activation, were markedly affected by TNFR1 or TNFR2 deficiency. Moreover, consistent with the two-step quantitative biallelic model of Il2 promoter activity (14, 46), the number of IL-2 producers was equivalent for homozygous and heterozygous Il2-GFP mice. However, in contrast to heterozygous cells, the MFI of individual cells with two GFP alleles increased with strength of Ag stimulation. A combination of suboptimal T cell stimulation, delayed kinetics of GFP production (14), and cell division are possible contributors to a <2-fold increase in GFP MFI in homozygous versus heterozygous CD4+ Il2-GFP+ T cells.
FIGURE 3.
TNFR2 co-stimulates Il2 promoter activity during priming of CD4+ T cells. (A and B) The kinetics of Il2 promoter activation during the priming is dependent is antigen-concentration dependent. Naive CD4+ T cells were isolated from 5C.C7 Il2-GFP+/− (Il2+/−) or 5C.C7 Il2-GFP−/− (Il2−/−) mice and activated in vitro (2 X 105 c/well, > 90% CD4+ T cells; 96-well round bottom plate) in response to increasing concentrations of MCC88–103 (0, 0.1, 1, and 10 µM). Cells were harvested prior to (0 h) or 24, 36, 48, and 64 h after T cell activation. (A) The percentage of Il2-GF-positive cells out of total live (7-AAD−) CD4+ T cells were determined by flow cytometry. The mean + SEM of three independent experiments (three mice per group per experiment) are shown. 10.01 µM > 0 µM, p < 0.05; 21µM > 0 µM, p < 0.05; 310 µM > 0 µM, p < 0.05. (B) The MFI of Il2-GFP was determined from live GFP-positive CD4+-gated T cells shown in A. 310 µM > 0.01 µM, p < 0.05. (C and D) Genetic-deficiency of TNFR2, but not TNFR1, impairs Il2 promoter activity. Naive CD4+ T cells were obtained from TNFR1 and TNFR2 sufficient (indicated as WT), TNFR1-deficient (indicated as TNFR1 KO), TNFR2-deficient (indicated as TNFR2 KO), or TNFR1/2 dKO (indicated as TNFR dKO) 5C.C7 Il2-GFP−/− (Il2−/−) mice. Single cell suspensions (1 X 105 c/ml; 200 µl/well; > 90% CD4+ T cells) were either left unstimulated (0 µM) or stimulated with 10 µM MCC (left panel), 1 µM MCC (center panel), or 0.1µM MCC (right panel) for 24, 36, 48, and 64 h. Cells were stained for CD4 and 7-AAD and 7-AAD− CD4+ cells and gated for analyses by flow cytometry. (C) The percentage of Il2-GFP-positive cells out of total live (7-AAD−) CD4+ T cells were determined by flow cytometry. The mean + SEM of four independent experiments (three mice/group/experiment) are shown. 2TNFR2 KO < WT, p < 0.05; 3TNFR dKO < WT, p < 0.05. (D) The MFI of Il2-GFP was determined from live GFP-positive CD4+-gated T cells shown in C2TNFR2 KO < WT, p < 0.05; 3TNFR dKO < WT, p < 0.05. (E and F) CD4+ T cell-intrinsic TNFR2 co-stimulates maximal IL-2 production. Naive CD4+ CD45Rbhigh T cells were FACS-purified from LN and spleens of TNFR2-sufficient (WT) or TNFR2-deficient (TNFR2 KO) 5C.C7 Il2-GFP+/− (Il2+/−) mice and then stimulated in vitro (2 X 105 c/well; 96-well round bottom plate) with plate-bound anti-TCRβ plus anti-CD28 for 64 h. The results of three independent experiments (three mice per group per experiment) are shown. (E) The percentage of Il2-GFP-positive cells out of total live CD4+ T cells were determined by flow cytometry. The mean ± SEM of the frequency of Il2-GFP-positive cells are indicated. Statistical significance is indicated. (F) The concentration of IL-2 was determined by ELISA from tissue culture supernatants of the samples collected from E. *p < 0.05.
Comparison of TNFR sufficient (WT) CD4+ T cells with TNFR1 KO, TNFR2 KO, and TNFR1/2 dKO CD4+ T cells identified TNFR2, but not TNFR1, as a major contributor to IL-2 production (Fig. 3C). Regardless of the Ag strength, TNFR2 KO cells yielded nearly 1.8 to 1.9 –fold fewer IL-2 producers relative to WT cells (p < 0.05). Although the deficiency of TNFR1 alone resulted in a modest reduction (p > 0.05) of IL-2 producers at the early 24 h time point, regulation by TNFR1 was not evident at later time points during maximal IL-2 production (Fig. 3C). In contrast to TNF blockade in vivo, Il2-GFP was reduced in TNFR2 KO and TNFR1/2 dKO CD4+ T cells following 1 µM, but not 10 µM or 0.1 µM, MCC stimulation (Fig. 3D). Together, these studies identify TNFR2 as a regulator of Il2 transcription in CD4+ T cells.
CD4+ T cell-intrinsic TNFR2 augments IL-2 production
It is plausible that TNFR2 ligands, including TNF-α, LT-α, and PGRN, bind to TNF receptors located on non-T cells to promote IL-2 production (47–49). Therefore, we asked whether TNFR2 functions in a CD4+ T cell-intrinsic manner to regulate Il2 promoter activity. For these experiments, FACS-purified naive CD4+ T cells from LNs or spleens of WT or TNFR2 KO 5C.C7 Il2-GFP+/− mice were stimulated with anti-TCRβ and anti-CD28. The frequency of IL-2 producers (CD4+ GFP+ T cells) was similar between WT LN (60.3% ± 3.87, mean ± SEM) and WT SPLN (66.1% ± 8.75, mean ± SEM) (p > 0.05) (Fig. 3E, middle panel), but reduced in TNFR2 KO CD4+ T cells (Fig. 3E, right panel) (p < 0.001). ELISA assays further revealed reduced IL-2 protein secretion by TNFR2 KO CD4+ T cells compared to WT cells (Fig. 3F). Cell survival, as determined by 7-AAD and Annexin V, staining did not differ between WT and TNFR2 KO T cells (data not shown). Collectively, these results suggest that CD4+ T cell intrinsic TNFR2, but TNFR1, signaling contributes to maximal Il2 promoter activity.
TNF receptor signaling in CD4+ T cells: A role for NF-κB
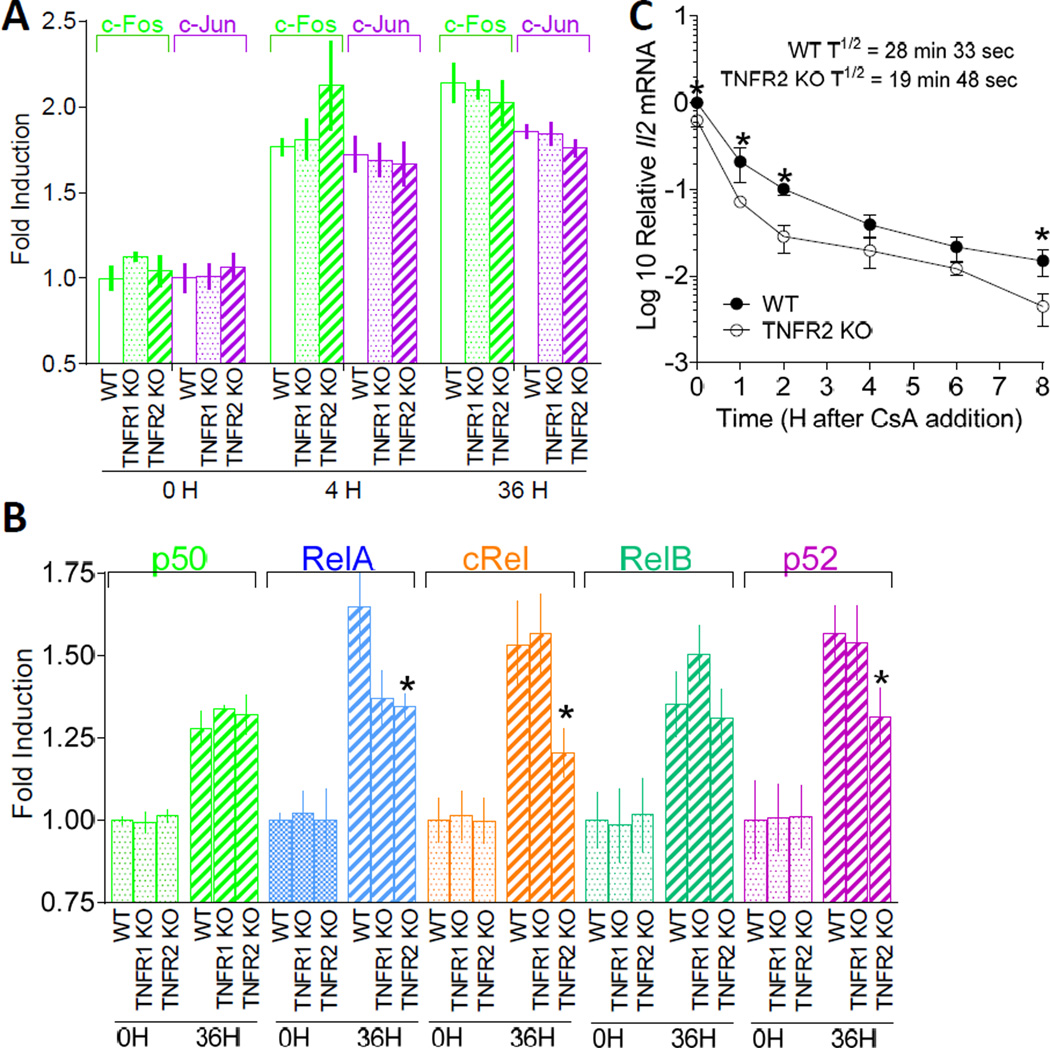
Although the integration of signaling cascades controlling Il2 induction remains enigmatic, NF-κB/Rel and JNK protein binding has been mapped to the first 300-bp minimal essential regulatory region of the Il2 promoter (14, 50–52). NF-κB and JNK cascades are activated downstream of TNFR2 activation and impaired c-Rel binding at the proximal Il2 promoter has been reported for TNFR1/2 dKO T cells (14). Therefore, we postulated that TNFR1 and TNFR2 may regulate different NF-κB/Rel and JNK signaling pathways in CD4+ T cells. To assess NF-κB/Rel activity, the binding of RelA (p65), RelB, c-Rel, p50 (NF-κB1), and p52 (NF-κB2) to the κB consensus oligonucleotide, 5'-GGGACTTTCC-3', was determined. In a similar manner, JNK/MAPK activity, was determined by recruitment of c-Fos and c-Jun to the AP-1 consensus sequence, 5'-TGACGTCA-3. For these studies, FACS-purified CD62Hi CD44Lo CD4+ T cells (purity > 96%) from 5C.C7 WT, TNFR1 KO, and TNFR2 KO mice were stimulated with anti-TCRβ plus anti-CD28 (14). Prior to T cell stimulation, constitutive NF-κB and AP-1 DNA binding did not differ between WT, TNFR1 KO, and TNFR2 KO T cells (Figs. 4A and 4B). In all cases, except for c-Rel and p52 in TNFR2 KO T cells, DNA binding increased following T cell stimulation (Fig. 4B). Although DNA binding of RelA increased following T cell stimulation in all three cell types, the magnitude of binding was substantially reduced in TNFR2 KO cells compared to WT and TNFR1 KO T cells (p < 0.05). These observations are consistent with our previous results showing impaired c-Rel binding to the proximal Il2 promoter in TNFR1/2 dKO T cells (14). Together, these results suggest that TNFR2 signaling activates canonical and noncanonical NF-κB pathways to promote Il2 induction in CD4+ T cells.
FIGURE 4.
DNA binding activity of NF-κB/Rel proteins and stability of Il2 mRNA differs between TNFR1 KO and TNFR2 KO CD4+ T cells. (A and B) Oligonucleotide binding assays were used to determine the influence of TNFR1 and TNFR2 on NF-κB/Rel and JNK activity in CD4+ T cells. CD4+ T cells from 5C.C7 Il2-GFP+/− (Il2+/−) WT, TNFR1 KO, or TNFR2 KO mice were left unstimulated or stimulated with anti-TCRβ plus soluble anti-CD28 for 4 h or 36 h. Nuclear extracts were incubated with a biotinylated double-stranded oligonucleotide probe containing the NF-κB/Rel consensus sequence, 5'-GGGACTTTCC-3', or the AP-1 consensus sequence, 5'-TGACGTCA-3', on streptavidin-coated plates. Captured complexes were incubated with the primary anti-rabbit Abs for (A) c-Fos and c-Jun or (B) p50, RelA, c-Rel, RelB, and p52. Primary Ab capture complexes were then incubated with horseradish peroxidase–conjugated secondary anti-rabbit Ab. The c-Jun- and c-Fos-containing complexes were detected by colorimetric absorbance (450 nm) and the NF-κB/Rel complexes were detected by chemiluminescence. Results are depicted as fold increase of relative absorbance normalized to unstimulated WT controls. The data are expressed as the mean ± SEM of triplicate experiments with duplicate reactions in each experiment. (C) CD4+ T cell intrinsic TNFR2 stabilizes Il2 mRNA. Naïve CD4+ T cells were purified (> 95% Vβ3+ CD4+ T cells) from 5C.C7 Il2-GFP+/− (Il2+/−) WT or TNFR2 KO mice using EasySep™ mouse CD4+ T cell negative isolation kits (STEMCELL Technologies). Cells (1 X 106 c/well) were activated in anti-TCRβ-coated (10 µg/ml) 6-well plates in 2 ml of cRPMI containing soluble anti-CD28. After 16 h, CsA was added at a final concentration of 200 ng/ml. Cells were harvested for mRNA isolation at the time of CsA addition (0 h) and 1, 2, 4, 6, and 8 h after CsA addition. The ΔΔCT method was used for relative quantitation of steady-state mRNA. Values are expressed as log10 relative units following normalization to CD3δ using the Il2 mRNA levels in unstimulated cells as the calibrator. Determination of the half-life for mRNA decay is described in Materials and Methods. Values shown represent mean ± SEM of four independent experiments using two mice/group/experiment. *p < 0.05.
CD4+ T cell-intrinsic TNFR2 controls the stability of Il2 mRNA
In activated CD4+ T cells, transcriptional mechanisms operate in parallel with post-transcriptional mRNA stabilization to control IL-2 production. JNK pathways target 3′- and 5′- UTRs of the Il2 locus to control Il2 mRNA turnover (53). Therefore, we next determined whether TNFR2 regulates Il2 mRNA stability. For these studies, naive WT or 5C.C7 TNFR2 KO CD4+ T cells were stimulated with anti-TCRβ plus anti-CD28 for 20 h. Further transcription was blocked by the addition of CsA, a pharmacological inhibitor of the Ca2+-sensitive phosphatase, calcinuerin, and Il2 mRNA levels were determined over time by qRT-PCR. T cell activation resulted in a 1.63-fold increase in steady-state Il2 mRNA in WT, compared to TNFR2 KO, T cells prior to the addition of CsA. After blocking transcription, the decay of steady-state Il2 mRNA was more prominent in TNFR2 KO T cells (T½ = 19 min, 48 sec) then WT T cells (T½ = 28 min, 33 sec) (Fig. 4C). Overall, these results reveal that T cell-intrinsic TNFR2 signaling controls IL-2 production through transcriptional and post-transcriptional mechanisms.
Regulation of in vivo IL-2 production by TNFR2
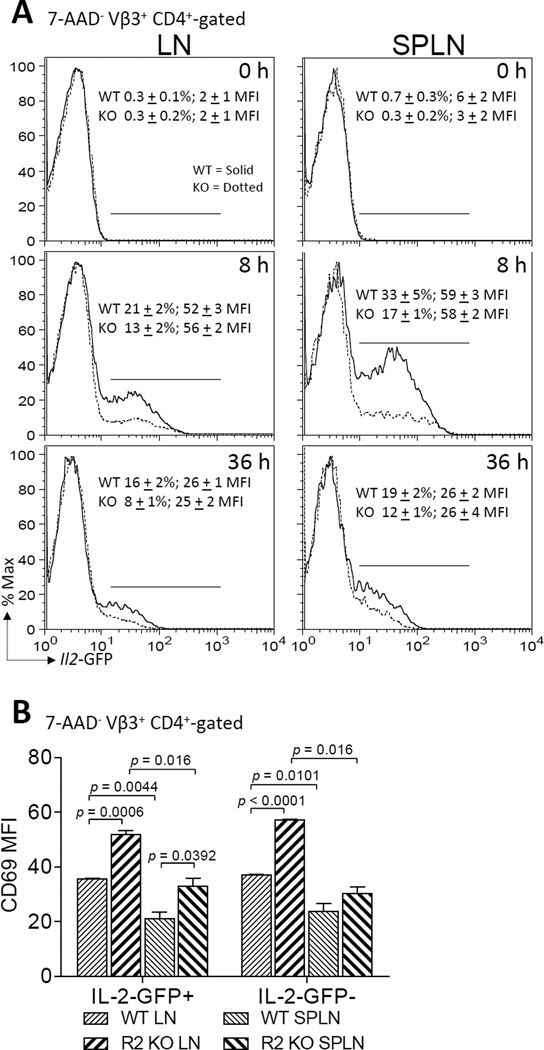
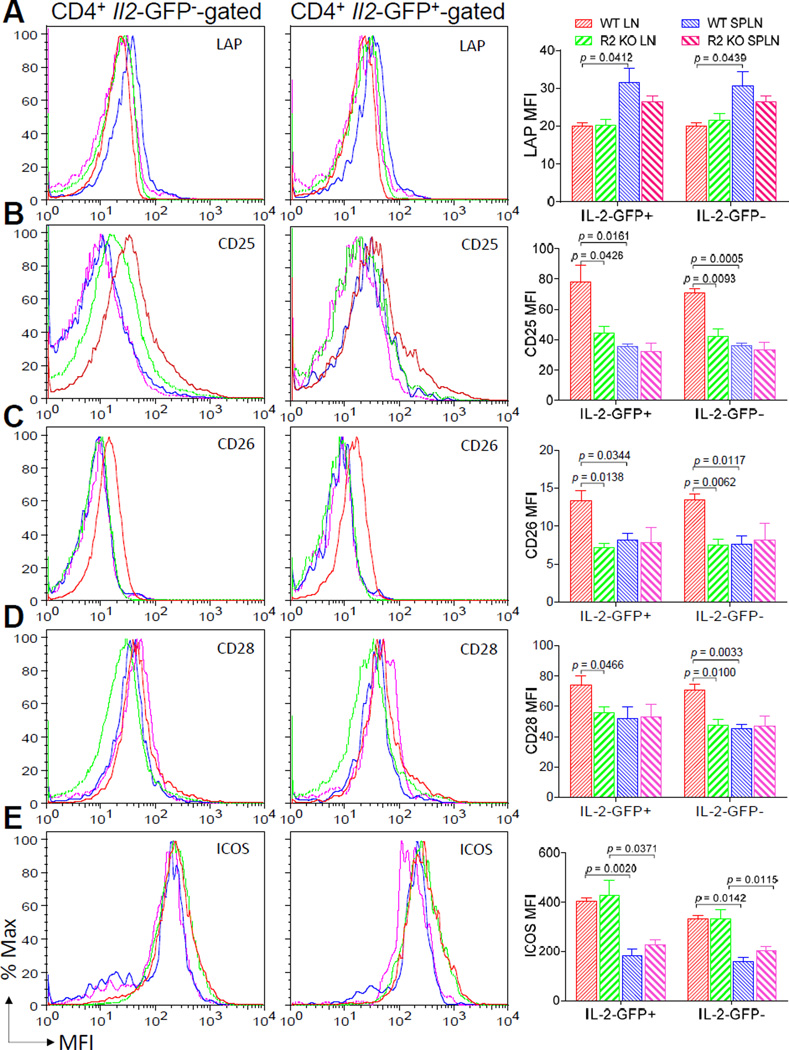
To further study regulation of Il2 promoter activity in vivo, WT or TNFR2 KO Il2-GFP+/− mice were immunized with MCC plus LPS, a potent activator of TNF. Both naive WT and TNFR2 KO CD4+ T cells required stimulation for Il2 transcription to occur (Fig. 5A). At 8 h, 33 ± 5% (mean ± SEM) of SPLN and 21 + 2% of LN CD4+ T cells from WT mice expressed GFP and this was lessened in TNFR2 KO T cells (p < 0.05). By 36 h, the frequency of Il2-GFP-expressing CD4+ T cells declined in the WT mice (19 ± 2%, SPLN; 16 ± 2%, LN) and this reduction was even greater in the TNFR 2 KO mice (12 ± 1%, SPLN; 8 ± 1%, LN) (p < 0.001). Similar to the in vitro experiments, TNFR2 deficiency did not alter the MFI of the GFP positive cells (Fig. 5A). The reduced frequency of Il2-GFP positive cells in the TNFR2 KO mice compared with WT mice did not correlate to a change in the number of CD4+ T cells (data not shown) or robustness of stimulation through the TCR as determined by CD69 expression (Fig. 5B). In fact, CD69 expression following T cell activation was enhanced in the TNFR2 KO T cells relative to WT T cells (Fig. 5B). CD69 expression did not differ (p > 0.05) between WT and TNFR2 KO CD4+ T cells prior to T cell activation (data not shown). While most commonly viewed as a stimulatory molecule that is expressed upon T cell activation, CD69 may also participate in feedback suppression of an immune response through the upregulation of transforming growth factor-β (TGF-β). TGF-β can inhibit IL-2 production (54). Furthermore, TNF antagonizes TGF-β signaling (55) and TNFR2 regulates Il2 expression in a CD4+ T cell autonomous manner. However, in our studies here, TGF-β1-latency-associated peptide (LAP) expression did not differ between TNFR2 KO and WT CD4+ T cells. These results are consistent with a T cell-intrinsic TGF-β-independent mechanism for suppression of IL-2 production in TNFR2 KO T cells. Additional surface molecules, that are also upregulated by NF-κB signaling and co-stimulate IL-2 production, were also examined (Fig. 6B – 6E). These studies revealed decreased expression of CD25, CD26, and CD28 by LN, but not splenic, TNFR2 KO T cells in comparison to WT T cells. Interestingly, elevated LAP expression by splenic T cells correlated with decreased CD25, CD26, and ICOS expression. Taken together, these data imply that TNFR2 deficiency is sufficient to disrupt Il2 promoter activity. These results further suggest possible regulatory crosstalk between T cell intrinsic TGF-β and TNFR2 on CD25, CD26, CD28, and ICOS expression in activated CD4+ T cells. It remains to be determined whether TNFR2 directly or indirectly regulates Il2 induction through modulation of these, or other, co-stimulatory molecule.
FIGURE 5.
Regulation of in vivo IL-2 production by TNFR2. 5C.C7 Il2-GFP+/− (Il2+/−) WT or TNFR2 KO mice were immunized with MCC (300 µg) plus LPS (10 µg). (A) Representative histograms depicting GFP expression in 7-AAD− CD4+-gated T cells recovered from LN (left) and SPLN (right) following no (top panels), 8 h (middle panels), and 36 h (bottom panels) immunization are shown. The first number in each panel represents the percentage (mean ± SEM) of Il2-GFP-positive cells from three separate experiments performed in triplicate. The second number in each panel indicates the MFI (mean ± SEM) of the Il2-GFP positive cells. WT CD4+ T cells are indicated by solid lines. TNFR2 KO CD4+ T cells are indicated by dashed lines. Results represent three separate experiments performed in triplicate. *p < 0.05. (B) CD4+ T cells recovered from the LN and SPLN of immunized mice in A, stained for CD69 expression. 7-AAD− CD4+ GFP− and 7-AAD− CD4+ GFP+ cells were gated for analyses. A summary of the MFI (mean ± SEM) of CD69 expression after 36 h stimulation is depicted. (C) CD4+ T cells recovered from the LN and spleens of non-immunized 5C.C7 Il2-GFP+/− (Il2+/−) WT or TNFR2 KO mice were stained for cell surface CD4 and CD69 expression and analyzed by flow cytometry. 7-AAD− CD4+ T cells were gated for analyses of CD69 expression. A representation of the MFI of CD69 expression all live CD4+ T cells is depicted.
FIGURE 6.
Expression of T cell surface co-stimulatory proteins in response to CD4+ T cell autonomous TNFR2 deficiency. CD4+ T cells recovered from the draining LNs and spleens of the immunized 5C.C7 Il2-GFP+/− (Il2+/−) WT or TNFR2 KO micedescribed in Fig. 5 were stained for cell surface CD4 and (A) LAP, (B) CD25, (C) CD26, (D) CD28, or (E) ICOS. 7-AAD− CD4+ GFP-gated (left panel) and 7-AAD− CD4+ GFP+-gated (middle panel) were analyzed for comparison. Representative flow cytometric histograms of the data are shown. The mean ± SEM of the co-stimulatory molecule MFI from three individual mice are depicted in the bar graphs (right panel). The Il2-GFP MFI for the CD4+ T cells analyzed in Fig. 6 are reported for Fig. 5.
Impaired Il2 expression in TNFR2 KO CD4+ T cells promotes IL-17 production
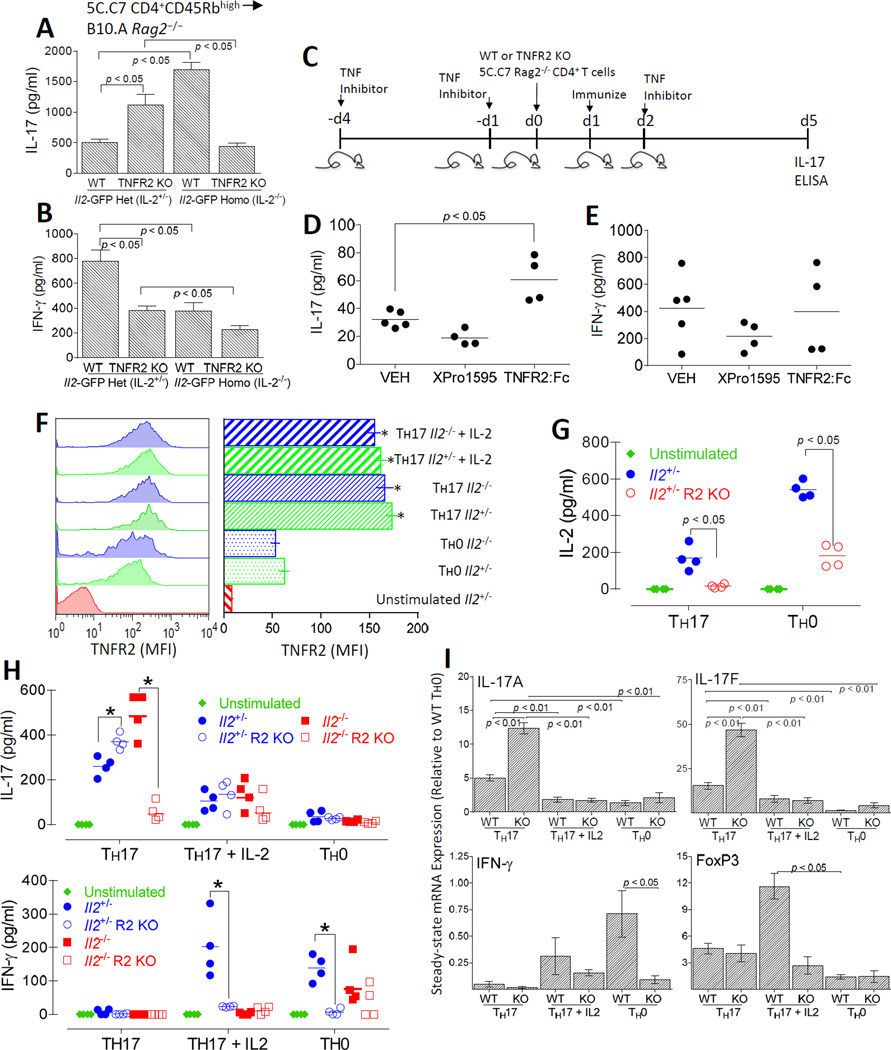
IL-2 inhibits the generation of Th17 cells, which secrete IL-17 and express the transcription factor ROR-γt (37, 56). Therefore we asked whether the deficiency of IL-2 production in TNFR2 KO CD4+ T cells was reciprocated by increased IL-17 production. For these experiments, CD4+ T cells (1 X 106) from WT or TNFR2 KO 5C.C7 TCR Il2-GFP+/− (Il2+/−) or Il2-GFP+/− (Il2+/−) donors were adoptively transferred to B10.A Rag2−/− recipient mice. Four days after immunization of the recipient mice with MCC emulsified in CFA, CD4+ T cells were isolated from draining lymph nodes and spleens and re-stimulated in vitro with PMA and ionomycin. IL-17 and IFN-γ protein secretion were determined from culture supernatants by ELISA. In support of our hypothesis, CD4+ T cells from TNFR2 Il2+/− mice had a marked increase in IL-17 production compared to WT Il2+/− cells (p < 0.05) (Fig. 7A). The increase in IL-17 production by TNFR2 KO Il2+/− T cells inversely correlated with reduced IFN-γ production compared to WT Il2+/− cells (p < 0.05) (Fig. 7B). To more clearly determine whether IL-2 was responsible for the impaired IL-17 production, we compared the differentiation of naïve CD4+ T cells from WT and TNFR2 KO Il2−/− mice. In contrast to CD4+ T cells from TNFR2 KO Il2+/− mice, T cells from TNFR2 KO Il2−/− mice did not produce increased amounts of IL-17. Instead IL-17 production by TNFR2 KO Il2−/− T cells was reduced compared to WT Il2−/− T cells (Fig. 7A). In contrast to the TNFR2 KO Il2+/− T cells, reduced IL-17 production by TNFR2 KO Il2−/− T cells was not associated with increased IFN-γ production compared to WT Il2−/− CD4+ T cells (Fig. 7B). IL-2 was hardly detectable in supernatants of IL-2+/− cells following re-stimulation and was not detected in IL-2−/− cultures (data not shown). Overall, these results suggest that tmTNF and TNFR2, but not sTNF or TNFR1, negatively regulate Th17 differentiation, at least in part, through IL-2-dependent mechanisms. To further test our hypothesis that TNFR2 co-stimulation negatively regulates Th17 differentiation, we investigated IL-17 production following TNFR2 blockade in WT mice. For these studies, B10.A Rag2−/− recipients were pretreated with TNFR2:Fc (10 µg/g) or XPro™1595 (10 µg/g) to block total TNF or only sTNF, respectively. Following adoptive transfer of naïve WT or TNFR2 KO Il2+/− CD4+ T cells, the recipients were immunized with MCC emulsified in CFA (Fig. 7C). Four days later, CD4+ T cells were isolated from the spleens and draining LNs and restimulated in vitro. As expected, TNFR2:Fc treatment augmented IL-17 production; whereas, blockade by XPro™1595 reflected an overexpression of non-cleavable TNF in transgenic mice (Fig. 7D). In contrast neither TNF blocking treatment markedly altered IFN-γ production upon re-stimulation (p > 0.05) (Fig. 7E).
FIGURE 7.
CD4+ T cell-intrinsic TNFR2 promotes Il2 expression to inhibit Th17 differentiation. B10.A Rag2−/− recipient mice were reconstituted with 1 X 106 CD4+ CD45Rbhigh cells from WT or TNFR2 KO 5C.C7 Il2-GFP+/− (Il2+/−), else WT or TNFR2 KO 5C.C7 Il2-GFP−/− (Il2−/−) donors. The recipients were then immunized with 20 µg MCC88–103 emulsified in 200 µl of CFA. CD4+ T cells from the spleens and draining lymph nodes were harvested 4 d after immunization and re-challenged with PMA + ionomycin in vitro for 12 h. IL-17A (A) and IFN-γ (B) were quantified from cell-free culture supernatants using ELISA. The results shown represent the mean ± SEM of three separate experiments performed in duplicate. (C) The experimental design to assess the role of TNF in wild-type mice using pharmacological blockade is depicted. For the results shown in D and E, B10.A Rag2−/− mice were administered (subQ) 10 µg/g XPro™1595, 10 µg/g etanercept (TNFR2:Fc), or 200 µl PBS (VEH) one day and four days prior to adoptive transfer of naïve CD4+ T cells from WT 5C.C7 Il2-GFP+/− (Il2+/−) donors. The recipients were immunized (20 µg MCC88–103 emulsified in 200 µl of CFA) the following day and received a third treatment (TNF blockade or VEH control) 48 h thereafter. CD4+ T cells were collected from the draining lymph nodes and spleens on four days following immunization and re-challenged with PMA + ionomycin in vitro for 12 h. IL-17 (D) and IFN-γ (E) were quantified from the culture supernatants by ELISA. Each symbol represents a single mouse. Horizontal lines represent the geometric means. (F – I) To determine whether enhanced production of IL-17 by TNFR2 KO CD4+ T cells could be reversed by the addition of exogenous IL-2, naive CD4+ T cells isolated from 5C.C7 Il2+/−Il2−/−, TNFR2 KO Il2+/−, or TNFR2 KO Il2−/− mice were stimulated for 4 days with plate-bound anti-TCRβ (5 µg/ml) and anti-CD28 under Th17-polarizing conditions (2.5 ng/ml TGF-β1, 50 ng/ml IL-6, 10 µg/ml α-IFN-γ, and 10 µg/ml α-IL-4) in the presence or absence of exogenous IL-2 (100 U). For comparison, CD4+ T cells were also cultured under Th0 conditions (plate-bound α-TCRβ + α-CD28). (F) Following stimulation, cells were harvested and stained for CD4, TNFR2, and 7-AAD. The MFI of cell surface TNFR2 was determined on 7AAD− CD4+-gated cells by flow cytometry. Representative flow cytometric histograms are shown (left panel) and TNFR2 MFI (mean ± SEM) of cells isolated from three independent experiments are shown (right panel). *p < 0.01, comparing Th17 or Th17 + IL-2 polarized cells to Th0 polarized counterparts. (G and H) Cell culture supernatants were collected and ELISA was performed in order to quantify IL-2 or IL-17 and IFN-γ. Each symbol represents a single mouse. Horizontal lines represent the geometric means. Diamonds indicate 7AAD− CD4+-gated from unstimulated Il2+/− mice. Circles indicate 7AAD− CD4+-gated cells isolated from WT or TNFR2 KO Il2+/− mice stimulated under Th0 or Th17 polarizing conditions. Squares indicate 7AAD− CD4+-gated cells isolated from WT or TNFR2 KO Il2−/− mice stimulated under Th0 or Th17 polarizing conditions. (I) Total RNA was extracted from gradient (Ficoll-Plaque)-purified viable cells and IL-17A, IL-17F, IFN-γ, and FoxP3 steady-state mRNA expression was determined by qRT-PCR. The ΔΔCT method was used for relative quantitation of steady-state mRNA. Values are expressed as relative expressions (fold change) following normalization to β-actin using the mRNA levels in WT Th0 cells as the calibrator. The error bars, which are visible only if they are bigger than the diameters of the symbols, indicate the SEs of the means of three independent experiments. *p < 0.05
To extend our in vivo studies, we explored a causal relationship between impaired IL-2 production and Th17 development in response toTNFR2 blockade by determining whether increased IL-17 production in TNFR2 KO CD4+ T cells was reversed with addition of exogenous IL-2. For these studies, naive CD4+ T cells isolated from 5C.C7 Il2+/−, Il2−/−, TNFR2 KO Il2+/−, or TNFR2 KO Il2−/− mice were stimulated for 4 d with plate-bound anti-TCRβ and anti-CD28 under Th17-polarizing conditions (TGF-β1, IL-6, α-IFN-γ, and α-IL-4) in the presence or absence of exogenous IL-2 (100 U). Consistent with previous reports, cell surface expression of TNFR2 was upregulated on live-gated CD4+ T cells cultured under Th0 conditions (α-TCRβ + α-CD28) but the magnitude of TNFR2 expression was markedly elevated on cells cultured under Th17 conditions (Fig. 7F). Neither genetic Il2 deficiency nor addition of exogenous IL-2 affected the frequency of CD4+ T cells that upregulated TNFR2. The MFI of TNFR2 expression was also unaffected. These results reveal that, in addition to CD4+ FoxP3 Tregs (57), TNFR2 is also highly expressed by Th17 cells. Further, the regulation of TNFR2 on Th17 cells does not appear to be influenced by T cell intrinsic or extrinsic IL-2. As expected, the amount of IL-2 produced by TNFR2 KO CD4+ T cells was greatly reduced compared to WT T cells stimulated under Th0 culture conditions. In comparison, neither WT nor TNFR2 KO T cells produced very much IL-2 when cultured under optimal Th17 conditions (Fig. 7G).
As shown in Fig. 7H, the generation of IL-17 under Th17 polarizing conditions in vitro was enhanced by deficiency of either IL-2 or TNFR2 (p < 0.05) but not by the combined deficiency of both IL-2 and TNFR2. Supplementation of the Th17 polarized cultures with IL-2 greatly reduced IL-17 production by WT Il2+/−, WT Il2−/−, and TNFR2 KO Il2+/− CD4+ T cells. In contrast to Th17 conditions, cells cultured under Th0 conditions yielded limited IL-17. Impaired IL-17 production by addition of IL-2 under Th17 polarizing conditions associated with increased IFN-γ production by WT, but not TNFR2 KO, CD4+ T cells. These in vitro results corroborate increased Th17 differentiation with TNFR2 deficiency in vivo. Consistent with these observations, when cultured under Th17 polarizing conditions in the absence of IL-2, TNFR2 KO CD4+ T cells expressed elevated steady-state Il17A and Il17F mRNA compared to WT T cells (Fig. 7I). Conversely, the addition of IL-2 during Th17 polarization suppressed Il17A and Il17F mRNA in both WT and TNFR2 KO T cells and elevated Foxp3 mRNA in WT T cells. Consistent with our earlier observation of impaired IFN-γ secretion by TNFR2 KO T cells, the induction of IFN-γ mRNA by TNFR2 KO CD4+ T cells was reduced compared to WT T cells cultured under Th0 conditions (p < 0.05). Collectively, these results suggest that TNFR2 blockade promotes Th17 differentiation independently of FoxP3 or IFN-γ.
TNFR2 targets ROR-γt expression and tyrosine phosphorylation of STAT3 and STAT5 to modulate Th17 differentiation
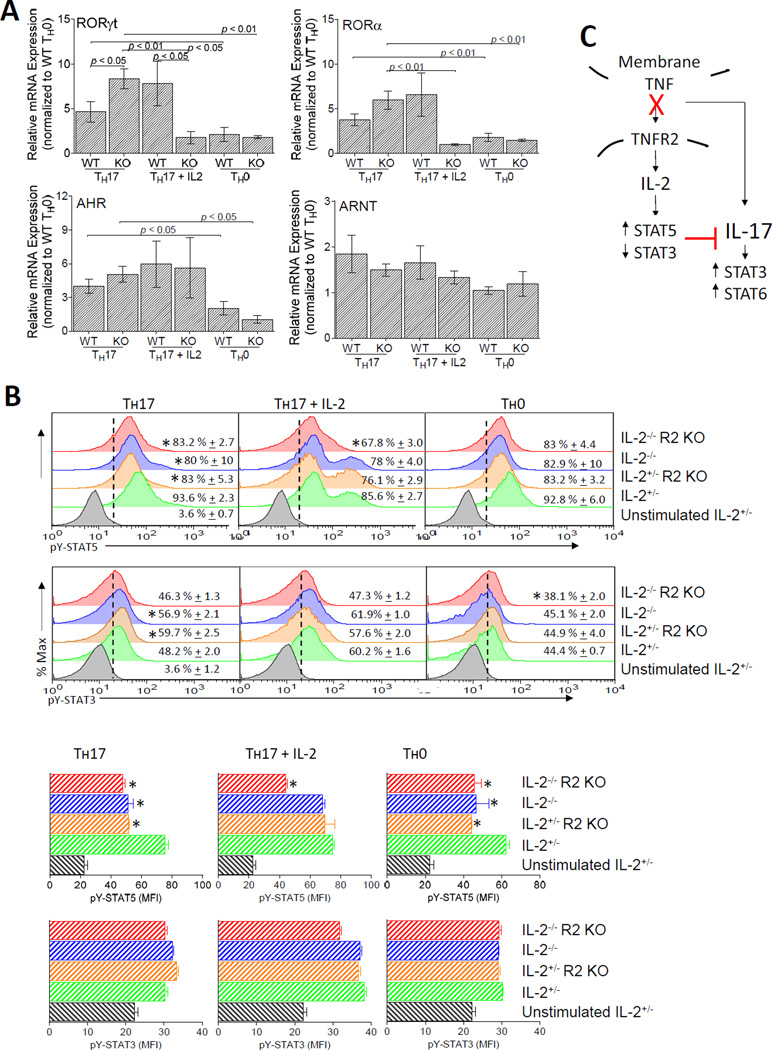
Although ROR-γt orchestrates Th17 differentiation, other transcription factors, including RORα and AHR, also regulate Il17 promoter activity (58, 59). We compared the expression of these transcription factors in WT and TNFR2 KO CD4+ T cells cultured under Th0 and Th17 polarizing conditions in the absence or presence of IL-2. The expression of Rorγt, Rora, and Ahr were elevated by Th17 conditions for both WT and TNFR2 KO T cells, and uniquely, Rorγt mRNA was more highly expressed in TNFR2 KO CD4+ T cells compared to WT T cells (Fig. 8A). In contrast, the expression of ARNT, a binding partner for AHR, was comparable between WT and TNFR2 KO CD4+ T cells, and unaffected by Th17 polarization or the addition of IL-2. IL-2 substantially inhibited Rorγt and Rora, but not Ahr, in TNFR2 KO but not WT CD4+ T cells. Collectively, these results suggest that ROR-γt may contribute to the increased IL-17 production observed in the TNFR2 KO CD4+ T cells.
FIGURE 8.
TNFR2 targets ROR-γt expression and tyrosine phosphorylation of STAT3 and STAT5 to modulate Th17 differentiation. (A) Steady-state ROR-γt mRNA expression is elevated in TNFR2 KO CD4+ T cells compared to WT T cells. The cDNA used in these assays was obtained from the WT and TNFR2 KO Il2+/− cells stimulated under Th17-polarizing conditions in the presence or absence of exogenous IL-2 or under Th0 conditions from Figs. 7F – I. Values for steady-state ROR-γt, ROR-α, AHR, and ARNT are expressed as relative expressions (fold change) following normalization to β-actin using the mRNA levels in WT Th0 cells as the calibrator. (B) Intracellular tyrosine-phosphorylated STAT5 (upper rows) and STAT3 (lower rows) were determined for 5C.C7 +/−Il2-GFP+/− WT or TNFR2 KO CD4+ T cells left unstimulated, activated for 4 d under Th17-polarizing conditions with or without the addition of IL-2 (100 U/ml), or activated for 4 dunder Th0 conditions. IL-6 (50 ng/ml) and IL-2 (100 U/ml) were added for the final 20 min of cell culture. (Top) Representative flow cytometry histograms are shown for pY-STAT5 and pY-STAT3 using 7AAD− CD4+-gated T cells for analyses. The dotted line indicates pY-STAT3 or pY-STAT5 staining in unstimulated cells. The mean ± SEM of pY-STAT-positive cells (i.e., those to the right of the dotted line) from three independent experiments are shown. (Bottom) The mean ± SEM of the MFI of pY-STAT3- or pY-STAT5-positive CD4+ T cells from the cells acquired and analyzed above (top) are shown. *p < 0.05. (C) Overall, we conclude that tmTNF activation of TNFR2 expressed on CD4+ T cells stimulates Il2 expression and Il2 mRNA stability. During Th17 differentiation, TNFR2 is increased. When tmTNF/TNFR2 signaling is blocked or impaired, Th17 differentiation is promoted, and IL-17 activates STAT3 and STAT6
The relative balance of STAT3 and STAT5 activity is a key determinant for Il17 promoter activity. Therefore we assessed tyrosine-phosphorylation (pY) of STAT5 and STAT3. WT and TNFR2 KO T cells were stimulated under Th17-polarizing conditions with or without the addition of IL-2 and compared these results to cells stimulated under Th0 culture conditions (Fig. 8B). Under Th0 conditions, the frequency of CD4+ T cells expressing elevated levels of pY-STAT5 was unaffected, however, IL-2 and TNFR2 deficiency each reduced pY-STAT5 MFI and this was not exacerbated in TNFR2 KO Il2−/− T cells. In comparison, the frequency of cells expressing elevated levels of pY-STAT3 was reduced in the TNFR2 KO Il2−/− T cells compared to Il2+/− T cells and pY-STAT3 MFI was unaffected. Under Th17 conditions, the frequency of pY-STAT5 expressing cells and pY-STAT5 MFI was reduced by IL-2 or TNFR2 deficiency (again, this was not exacerbated in TNFR2 KO Il2−/− T cells) and this was reversed by the addition of IL-2, except for TNFR2 KO Il2−/− T cells. In comparison, under Th17 conditions, the frequency of pY-STAT3 expressing cells was increased by the absence of IL-2 or TNFR2, but not by a combined deficiency of both IL-2 and TNFR2, and this was reversed by the addition of IL-2. Together these data show altered expression of ROR-γt, STAT3, and STAT5 in TNFR2 KO CD4+ T cells.
Discussion
In this study, we show that CD4+ T cell intrinsic tmTNF/TNFR2 promotes Il2 expression to fine tune the proportion of IL-2 producing CD4+ T cells. Further, under Th17 polarizing conditions, TNFR2 blockade is sufficient to promote Th17 differentiation and this can be prevented by the addition of exogenous IL-2. Overall, we conclude that tmTNF activation of TNFR2 expressed on CD4+ T cells stimulates Il2 expression and Il2 mRNA stability. During Th17 differentiation, TNFR2 is increased. When tmTNF/TNFR2 signaling is blocked or impaired, Th17 differentiation is promoted, and IL-17 activates STAT3 and STAT6 (Fig. 8C). We propose that decreased STAT5 activity, in concert with increased STAT3 activity, is at least one mechanism by which TNFR2 blockade promotes Th17 development. Our findings are consistent with previous reports demonstrating displaced STAT3 binding at the Il17a–Il17f locus by IL-2/IL-2R signaling (56,60,61). Our findings contrast a previous report suggesting that TNF promotes Th17 differentiation (23). In these earlier studies, cell cultures were supplemented with a combination of TNF and IL-1β. The individual contribution of TNF or IL-1β were not studied. IL-1β has since been shown to promote Th17 differentiation by suppressing Socs3, an inhibitor of STAT3 (62). It remains to be determined whether IL-1β was, in fact, the key promoter of Th17 differentiation in the earlier studies.
In mice, the transcription factors ROR-γt, ROR-α, and AHR are important regulators of IL-17 producing Th17 cells. During Th17 differentiation, Rorγt expression is upregulated in TNFR2 KO T cells, but Ahr and Rorα are not. Therefore, it is not likely that either AHR or ROR-α, by themselves, cause the difference in IL-17 production between WT and TNFR2 KO T cells. CD4+ CD25+ FoxP3+ Tregs have also been demonstrated to promote Th17 differentiation through the consumption of IL-2 (37). However, Foxp3 expression is decreased in TNFR2 KO T cells. Therefore, IL-17 production in response to TNFR2 blockade appears to be Treg-independent. Overall, our observations are consistent with an increasing number of studies demonstrating immune modulation by TNF. There is little doubt that TNF plays a major role in the pathogenesis of autoimmune disorders, yet anti-TNF therapies remain unpredictable. For example, etanercept, a TNF antagonist, often triggers an unexpected onset or exacerbation of CNS demyelinating autoimmune disorders in patients with inflammatory bowel disease or rheumatoid arthritis (63). TNFR2 and IL-2 both promote the expansion of Tregs, a promising therapy for multiple disorders, including cancer, autoimmunity, and transplant rejection (11, 61, 64, 65). Therefore, strategies aimed to mimic the stimulatory actions of TNFR2 on Il2 expression while bypassing its cytotoxic effects have therapeutic potential.
Although naïve and previously-activated CD4+ cells respond very differently to TNF, little is known about TNFR1 or TNFR2 signaling cascades in these cells upon T cell activation. We now establish that TNFR2, but not TNFR1, promotes Il2 promoter activity and Il2 mRNA stability to increase IL-2 production. These observations are consistent with previous reports of impaired IL-2 production in TNFR1/2 dKO mice and reduced IL-2 production by TNFR2−/− CD8+ T cells (12–14). So, why the divergence between TNFR1 and TNFR2 signaling? One possibility is ligand specificity for receptor activation. sTNF activity is largely restricted to TNFR1 while tmTNF preferentially activates TNFR2. While our results suggest tmTNF/TNFR2 interactions, other TNFR2 ligands, including lymphotoxin-α (LT, LTα3, TNFβ) and membrane-bound LTα1β2 have been described (66, 67). In contrast to LTα1β2, which binds to a distinct receptor LTβR (68), secreted LTα3 binds to both TNFR1 and TNFR2. Although, not without controversy, the growth factor PGRN has also been proposed to bind TNFR2 (49, 69). Although TNF and TNFR2 are highly expressed by Th17 cells, not much is known about the production and function of these alternative TNFR2 ligands in Th17 cells. Therefore, it is not possible to exclude the possibility that alternative TNFR2 ligands contribute to the regulation of IL-2.
A second possibility involves receptor density. TNFR2, not TNFR1, is the predominant TNF receptor that is upregulation upon activation of naïve CD4+ T cells under Th0 and Th17 conditions (14). An alternative possibility is that TNFR1 and TNFR2 simply regulate different downstream signaling cascades, as reported in other cell types. It is not completely clear whether the failure of sTNF to modulate Il2 expression is due to low receptor density or to a signaling cascade that is distinct from tmTNF. We have considered the possibility that decreased Il2 expression in TNFR2 KO T cells is consequential to elevated TNFR1 signaling as a result of increased ligand availability as opposed to a loss of stimulatory TNFR2 signaling. However, this is in contrast to our observation that Il2 expression in TNFR2 KO T cells parallels Il2 expression in TNFR1/2 dKO T cells. If TNFR1 contributed to the silencing of the Il2 promoter, we would have expected even less Il2 expression in TNFR2 KO T cells compared to TNFR1/2 dKO T cells. Surprisingly few studies have addressed signal transduction through TNFR1 and TNFR2 in CD4+ T cells. In principal, activation of TNFR1 rapidly recruits adapter molecules TRADD, RIP1, and TRAF2 (70) to initiate a cascade of phosphorylation and ubiquitination events that activate MAPK and NFκB/Rel pathways to induce the expression of pro-inflammatory and anti-apoptotic survival genes. In contrast, TNFR2 recruits TRAF2 to activate NF-κB/Rel and MAPK pathways. Upon CD4+ T cell activation, NF-κB/Rel proteins, normally sequestered within the cytoplasm, translocate into the nucleus following the degradation of inhibitory proteins to activate canonical and noncanonical NF-κB/Rel signaling. The importance of NF-κB and MAPK in Il2 expression is well established (51). Our present data, acquired from oligonucleotide DNA binding, associate silencing of the Il2 promoter in TNFR2 KO CD4+ T cells with reduced cRel and p52 DNA binding. Similar changes were not evident in the TNFR1 KO CD4+ T cells to suggest receptor signaling specificity.
Overall, we propose that canonical and noncanonical NF-κB signaling participate in the regulation of Il2 expression by TNFR2. Our data are consistent with a requirement for cytokine-dependent degradation of cytosolic IκBβ to permit c-Rel nuclear translocation (71, 72), as well as, a requirement of c-Rel for chromatin remodeling of the Il2 locus (52). Future studies will address epigenetic modifications associated with silencing of the Il2 promoter in response to TNFR2 deficiency. Similar to other cytokines, Il2 expression is also regulated by post-transcriptional mRNA stabilization. The ability of TNFR2 to regulate Il2 stability is consistent with NF-κB- and JNK-dependent recruitment of AU-binding proteins (AUBPs) to AU-rich elements (AREs) located within the 3’ UTR, 5’ UTR, and coding region of the Il2 gene (40, 53). The precise signaling pathways by which TNFR2 regulates IL-2 expression remain important areas for future studies. Importantly, not all members of the superfamily of TNF receptors regulate IL-2 production (73), thus potentially making TNFR2 the critical signaling pathway necessary to achieve the threshold level of NF-κB activity required for Il2 promoter activity.
In characterizing the tmTNF KI FoxP3-GFP reporter mice, we observed fewer CD4+ FoxP3+ T cells in the spleen and MLN but not pooled cervical, axillary, brachial, and inguinal lymph nodes of tmTNF mice relative to WT C57BL/6 mice. These observations are consistent with previous studies suggesting that the actions of sTNF and tmTNF on immune homeostasis differ between secondary lymphoid organs (74). For example, sTNF, but not tmTNF, produced by B cells, and not T cells, is critical for secondary lymphoid organ organization (75). Moreover, B cells are the main producers of TNF in the spleen, whereas T cells generate the majority of TNF in the lymph nodes (75). Our results in the tmTNF mice suggest that sTNF plays a role in maintaining CD4+ Tregs in the spleen and MLNs. TNF has also been implicated in lymphocyte migration (74, 76), therefore dysregulated T cell trafficking may have also contributed to the observed organ-specific lymphocyte changes. Taken together, we identify CD4+ T cell intrinsic TNFR2 as an important regulator of IL-2 production to sustain IL-17 production independent of Tregs. As such, use of non-selective anti-TNF therapeutics may potentiate unwanted Th17-mediated inflammation.
Acknowledgements
We sincerely thank Drs. Habib Zaghouani for providing the Foxp3-GFP reporter mice; David Szymkowski, Xencor, Inc, for providing XPro™1595; Dr. Celso Velazquez for assistance in obtaining Etanercept; and Dr. Emily Leary for assistance with statistical analyses. We are also thank Drs. Grace Sun and Dennis Chang of the Dept. of Biochemistry for assistance with the nitric oxide assay; Andreu Garcia for technical assistance; Daniel Jackson for FACS sorting; and Sarah Dweik, Avery Day, and William Moritz for excellence with the manuscript preparation.
This work was supported by NIH R01 ES022966 (S.C.M) and a University of Missouri Richard Wallace Faculty Incentive Grant Award (S.C.M.).
Abbreviations used in this article
- 7-AAD
7-amino actinomycin D
- ActD
actinomycin D
- AHR
aryl hydrocarbon receptor
- APC
Allophycocyanin
- ARE
AU-rich elements
- ARNT
aryl hydrocarbon receptor nuclear translocator
- AUBP
AU-binding proteins
- cRPMI
complete RPMI
- CT
Comparative threshold
- CsA
cyclosporin A
- dKO
double knockout
- DN-TNF
dominant-negative TNF
- FoxP3
Forkhead box P3
- IBD
Inflammatory bowel disease
- KO
knockout
- LAP
latency-associated peptide
- LN
lymph node
- LTα
Lymphotoxin alpha
- MCC
moth cytochrome c
- MFI
mean fluorescence intensity
- MLN
mesenteric lymph node
- MOG
myelin oligodendrocyte glycoprotein
- NaNO2
sodium nitrite
- NO2
nitrite
- PCC
pigeon cytochrome c
- pY
tyrosine-phosphorylation
- qRT-PCR
quantitative RT-PCR
- RA
rheumatoid arthritis
- RAR
retinoic acid receptor
- RE
response element
- ROR
related orphan receptor
- SPLN
spleen
- sTNF
soluble TNF
- TACE
TNF converting enzyme
- Tg
transgenic
- tmTNF
transmembrane TNF
- TNFR
TNF Receptor
- TRAF
TNF Receptor Associated Factor
- Tregs
T regulatory cells
- UTR
untranslated region
- WT
wild type
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 2.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 4.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng G, Yu A, Malek TR. T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol Rev. 2011;241:63–76. doi: 10.1111/j.1600-065X.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao W, Lin J-X, Leonard WJ. Interleukin-2 at the Crossroads of Effector Responses, Tolerance, and Immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells AD. New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors, and epigenetic imprinting. J Immunol. 2009;182:7331–7341. doi: 10.4049/jimmunol.0803917. [DOI] [PubMed] [Google Scholar]
- 11.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim EY, Teh HS. Critical role of TNF receptor type-2 (p75) as a costimulator for IL-2 induction and T cell survival: a functional link to CD28. J Immunol. 2004;173:4500–4509. doi: 10.4049/jimmunol.173.7.4500. [DOI] [PubMed] [Google Scholar]
- 13.Kim EY, Teh HS. TNF type 2 receptor (p75) lowers the threshold of T cell activation. J Immunol. 2001;167:6812–6820. doi: 10.4049/jimmunol.167.12.6812. [DOI] [PubMed] [Google Scholar]
- 14.McKarns SC, Schwartz RH. Biphasic regulation of Il2 transcription in CD4+ T cells: roles for TNF-alpha receptor signaling and chromatin structure. J Immunol. 2008;181:1272–1281. doi: 10.4049/jimmunol.181.2.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 16.Grell M. Tumor necrosis factor (TNF) receptors in cellular signaling of soluble and membrane-expressed TNF. J Inflamm. 1995;47:8–17. [PubMed] [Google Scholar]
- 17.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 18.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 19.Hacker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 20.Hofer T, Krichevsky O, Altan-Bonnet G. Competition for IL-2 between Regulatory and Effector T Cells to Chisel Immune Responses. Front Immunol. 2012;3:268. doi: 10.3389/fimmu.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, Schluns KS, Gui J, Jetten AM, Dong C. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36:23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappu BP, Borodovsky A, Zheng TS, Yang X, Wu P, Dong X, Weng S, Browning B, Scott ML, Ma L, Su L, Tian Q, Schneider P, Flavell RA, Dong C, Burkly LC. TL1A–DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J Exp Med. 2008;205:1049–1062. doi: 10.1084/jem.20071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Seder RA, Paul WE, Davis MM, Fazekas de St Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naramura M, Hu RJ, Gu H. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 1998;9:209–216. doi: 10.1016/s1074-7613(00)80603-2. [DOI] [PubMed] [Google Scholar]
- 26.Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, Willis CR, Charrier K, Morrissey PJ, Ware CB, Mohler KM. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol. 1998;160:943–952. [PubMed] [Google Scholar]
- 27.Alexopoulou L, Kranidioti K, Xanthoulea S, Denis M, Kotanidou A, Douni E, Blackshear PJ, Kontoyiannis DL, Kollias G. Transmembrane TNF protects mutant mice against intracellular bacterial infections, chronic inflammation and autoimmunity. Eur J Immunol. 2006;36:2768–2780. doi: 10.1002/eji.200635921. [DOI] [PubMed] [Google Scholar]
- 28.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 29.Steed PM, Tansey MG, Zalevsky J, Zhukovsky EA, Desjarlais JR, Szymkowski DE, Abbott C, Carmichael D, Chan C, Cherry L, Cheung P, Chirino AJ, Chung HH, Doberstein SK, Eivazi A, Filikov AV, Gao SX, Hubert RS, Hwang M, Hyun L, Kashi S, Kim A, Kim E, Kung J, Martinez SP, Muchhal US, Nguyen DH, O’Brien C, O’Keefe D, Singer K, Vafa O, Vielmetter J, Yoder SC, Dahiyat BI. Inactivation of TNF signaling by rationally designed dominant-negative TNF variants. Science. 2003;301:1895–1898. doi: 10.1126/science.1081297. [DOI] [PubMed] [Google Scholar]
- 30.Zalevsky J, Secher T, Ezhevsky SA, Janot L, Steed PM, O’Brien C, Eivazi A, Kung J, Nguyen DH, Doberstein SK, Erard F, Ryffel B, Szymkowski DE. Dominant-negative inhibitors of soluble TNF attenuate experimental arthritis without suppressing innate immunity to infection. J Immunol. 2007;179:1872–1883. doi: 10.4049/jimmunol.179.3.1872. [DOI] [PubMed] [Google Scholar]
- 31.Goffe B, Cather JC. Etanercept: An overview. Journal of the American Academy of Dermatology. 2003;49:S105–S111. doi: 10.1016/mjd.2003.554. [DOI] [PubMed] [Google Scholar]
- 32.Victoratos P, Kollias G. Induction of autoantibody-mediated spontaneous arthritis critically depends on follicular dendritic cells. Immunity. 2009;30:130–142. doi: 10.1016/j.immuni.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Murray KM, Dahl SL. Recombinant human tumor necrosis factor receptor (p75) Fc fusion protein (TNFR:Fc) in rheumatoid arthritis. The Annals of Pharmacotherapy. 1997;31:1335–1338. doi: 10.1177/106002809703101111. [DOI] [PubMed] [Google Scholar]
- 34.Pang L, Wang L, Suo T, Hao H, Fang X, Jia J, Huang F, Tang J. Tumor necrosis factor-alpha blockade leads to decreased peripheral T cell reactivity and increased dendritic cell number in peripheral blood of patients with ankylosing spondylitis. The Journal of Rheumatology. 2008;35:2220–2228. doi: 10.3899/jrheum.080219. [DOI] [PubMed] [Google Scholar]
- 35.Cowley SC, Sedgwick JD, Elkins KL. Differential requirements by CD4+ and CD8+ T cells for soluble and membrane TNF in control of Francisella tularensis live vaccine strain intramacrophage growth. J Immunol. 2007;179:7709–7719. doi: 10.4049/jimmunol.179.11.7709. [DOI] [PubMed] [Google Scholar]
- 36.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and 15Nnitrate in biological fluids. Analytical Biochemistry. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 37.Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422–434. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Lockhart M, Miller J. Engagement of CD28 outside of the immunological synapse results in up-regulation of IL-2 mRNA stability but not IL-2 transcription. J Immunol. 2006;176:4778–4784. doi: 10.4049/jimmunol.176.8.4778. [DOI] [PubMed] [Google Scholar]
- 39.Umlauf SW, Beverly B, Lantz O, Schwartz RH. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Molecular and Cellular Biology. 1995;15:3197–3205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 41.Chen D, Rothenberg EV. Interleukin 2 transcription factors as molecular targets of cAMP inhibition: delayed inhibition kinetics and combinatorial transcription roles. J Exp Med. 1994;179:931–942. doi: 10.1084/jem.179.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emmel EA, Verweij CL, Durand DB, Higgins KM, Lacy E, Crabtree GR. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989;246:1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- 43.Ragheb JA, Deen M, Schwartz RH. CD28-Mediated regulation of mRNA stability requires sequences within the coding region of the IL-2 mRNA. J Immunol. 1999;163:120–129. [PubMed] [Google Scholar]
- 44.Xu L, Yang J, Huang Y, van der Meide PH, Levi M, Wahren B, Link H, Xiao B. Limitation of nitric oxide production: cells from lymph node and spleen exhibit distinct difference in nitric oxide production. Immunology Letters. 2000;71:177–184. doi: 10.1016/s0165-2478(00)00154-1. [DOI] [PubMed] [Google Scholar]
- 45.Shibata H, Abe Y, Yoshioka Y, Nomura T, Sato M, Kayamuro H, Kawara T, Arita S, Furuya T, Nagano K, Yoshikawa T, Kamada H, Tsunoda S, Tsutsumi Y. Generation of mouse macrophages expressing membrane-bound TNF variants with selectivity for TNFR1 or TNFR2. Cytokine. 2010;50:75–83. doi: 10.1016/j.cyto.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 46.Umlauf SW, Beverly B, Kang SM, Brorson K, Tran AC, Schwartz RH. Molecular regulation of the IL-2 gene: rheostatic control of the immune system. Immunol Rev. 1993;133:177–197. doi: 10.1111/j.1600-065x.1993.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 47.Trevejo JM, Marino MW, Philpott N, Josien R, Richards EC, Elkon KB, Falck-Pedersen E. TNF-alpha -dependent maturation of local dendritic cells is critical for activating the adaptive immune response to virus infection. Proc Natl Acad Sci U S A. 2001;98:12162–12167. doi: 10.1073/pnas.211423598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maney NJ, Reynolds G, Krippner-Heidenreich A, Hilkens CM. Dendritic Cell Maturation and Survival Are Differentially Regulated by TNFR1 and TNFR2. J Immunol. 2014;193:4914–4923. doi: 10.4049/jimmunol.1302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, Syed NM, Lai Y, Lin EA, Kong L, Su J, Yin F, Ding AH, Zanin-Zhorov A, Dustin ML, Tao J, Craft J, Yin Z, Feng JQ, Abramson SB, Yu XP, Liu CJ. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz RH. T cell clonal anergy. Curr Opin Immunol. 1997;9:351–357. doi: 10.1016/s0952-7915(97)80081-7. [DOI] [PubMed] [Google Scholar]
- 51.Rothenberg EV, Ward SB. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci U S A. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724–3731. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- 53.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes & Development. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 54.McKarns SC, Schwartz RH, Kaminski NE. Smad3 is essential for TGF-beta 1 to suppress IL-2 production and TCR-induced proliferation, but not IL-2-induced proliferation. J Immunol. 2004;172:4275–4284. doi: 10.4049/jimmunol.172.7.4275. [DOI] [PubMed] [Google Scholar]
- 55.Verrecchia F, Pessah M, Atfi A, Mauviel A. Tumor necrosis factor-alpha inhibits transforming growth factor-beta /Smad signaling in human dermal fibroblasts via AP-1 activation. The Journal of Biological Chemistry. 2000;275:30226–30231. doi: 10.1074/jbc.M005310200. [DOI] [PubMed] [Google Scholar]
- 56.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea J J. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Chen X, Subleski JJ, Kopf H, Howard OM, Mannel DN, Oppenheim JJ. Cutting edge:expression of TNFR2 defines a maximally suppressive subset of mouse CD4+CD25+FoxP3+ T regulatory cells: applicability to tumor-infiltrating T regulatory cells. J Immunol. 2008;180:6467–6471. doi: 10.4049/jimmunol.180.10.6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 59.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 60.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basu R, Whitley SK, Bhaumik S, Zindl CL, Schoeb TR, Benveniste EN, Pear WS, Hatton RD, Weaver CT. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the Th17 cell-iTreg cell balance. Nat Immunol. 2015;16:286–295. doi: 10.1038/ni.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.TNF neutralization in MS: results of a randomized, placebo-controlled multicenter study. The Lenercept Multiple Sclerosis Study Group and The University of British Columbia MS/MRI Analysis Group. Neurology. 1999;53:457–465. [PubMed] [Google Scholar]
- 64.Okubo Y, Mera T, Wang L, Faustman DL. Homogeneous expansion of human T-regulatory cells via tumor necrosis factor receptor 2. Scientific Reports. 2013;3:3153. doi: 10.1038/srep03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Wu X, Zhou Q, Howard OM, Netea MG, Oppenheim JJ. TNFR2 is critical for the stabilization of the CD4+Foxp3+ regulatory T. cell phenotype in the inflammatory environment. J Immunol. 2013;190:1076–1084. doi: 10.4049/jimmunol.1202659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sacca R, Cuff CA, Lesslauer W, Ruddle NH. Differential activities of secreted lymphotoxin-alpha3 and membrane lymphotoxin-alpha1beta2 in lymphotoxin-induced inflammation: critical role of TNF receptor 1 signaling. J Immunol. 1998;160:485–491. [PubMed] [Google Scholar]
- 67.Zhu M, Brown NK, Fu YX. Direct and indirect roles of the lymphotoxin-beta receptor pathway in central tolerance induction. Trends Immunol. 2010;31:325–331. doi: 10.1016/j.it.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crowe PD, VanArsdale TL, Walter BN, Ware CF, Hession C, Ehrenfels B, Browning JL, Din WS, Goodwin RG, Smith CA. A lymphotoxin-beta-specific receptor. Science. 1994;264:707–710. doi: 10.1126/science.8171323. [DOI] [PubMed] [Google Scholar]
- 69.Wu H, Siegel RM. Medicine. Progranulin resolves inflammation. Science. 2011;332:427–428. doi: 10.1126/science.1205992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caamano J, Hunter CA. NF-kB family of transcription factors: central regulators of innate and adaptive immune functions. Clinical Microbiology Reviews. 2002;15:414–429. doi: 10.1128/CMR.15.3.414-429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banerjee D, Liou HC, Sen R. c-Rel-dependent priming of naive T cells by inflammatory cytokines. Immunity. 2005;23:445–458. doi: 10.1016/j.immuni.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 72.Zhou XY, Yashiro-Ohtani Y, Nakahira M, Park WR, Abe R, Hamaoka T, Naramura M, Gu H, Fujiwara H. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. J Immunol. 2002;168:3847–3854. doi: 10.4049/jimmunol.168.8.3847. [DOI] [PubMed] [Google Scholar]
- 73.Gramaglia I, Jember A, Pippig SD, Weinberg AD, Killeen N, Croft M. The OX40 costimulatory receptor determines the development of CD4 memory by regulating primary clonal expansion. J Immunol. 2000;165:3043–3050. doi: 10.4049/jimmunol.165.6.3043. [DOI] [PubMed] [Google Scholar]
- 74.Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J Immunol. 2009;183:2205–2212. doi: 10.4049/jimmunol.0804324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tumanov AV, Grivennikov SI, Kruglov AA, Shebzukhov YV, Koroleva EP, Piao Y, Cui CY, Kuprash DV, Nedospasov SA. Cellular source and molecular form of TNF specify its distinct functions in organization of secondary lymphoid organs. Blood. 2010;116:3456–3464. doi: 10.1182/blood-2009-10-249177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Green DM, Trial J, Birdsall HH. TNF-a released by comigrating monocytes promotes transendothelial migration of activated lymphocytes. J Immunol. 1998;161:2481–2489. [PubMed] [Google Scholar]