Abstract
Fluoroquinolones came into widespread use in African countries in the early 2000s, after patents for the first generation of these drugs expired. By that time, quinolone antibacterial agents had been used intensively worldwide and resistant lineages of many bacterial species had evolved. We sought to understand which Gram negative enteric pandemic lineages have been reported from Africa, as well as the nature and transmission of any indigenous resistant clones. A systematic review of articles indexed in the Medline and AJOL literature databases was conducted. We report on the findings of 43 eligible studies documenting local or pandemic fluoroquinolone-resistant enteric clones in sub-Sahara African countries. Most reports are of invasive non-typhoidal Salmonella and Escherichia coli lineages and there have been three reports of cholera outbreaks caused by fluoroquinolone-resistant Vibrio cholerae O1. Fluoroquinolone-resistant clones have also been reported from commensals and animal isolates but there are few data for non-Enterobacteriaceae and almost none for difficult-to-culture Campylobacter spp. Fluoroquinolone-resistant lineages identified in African countries were universally resistant to multiple other classes of antibacterial agents. Although as many as 972 non-duplicate articles refer to fluoroquinolone resistance in enteric bacteria from Africa, most do not report on subtypes and therefore information on the epidemiology of fluoroquinolone-resistant clones is available from only a handful of countries in the subcontinent. When resistance is reported, resistance mechanisms and lineage information is rarely investigated. Insufficient attention has been given to molecular and sequence-based methods necessary for identifying and tracking resistant clones in Africa and more research is needed in this area.
Keywords: fluoroquinolone resistance, quinolone resistance, antimicrobial resistance, Africa, Escherichia coli, Salmonella, Vibrio cholerae, Campylobacter
Introduction
Recent trends in fluoroquinolone use and resistance in africa
The antibacterial activity of nalidixic acid, a derived by-product from the synthesis of the antimicrobial chloroquine, was discovered in the 1960s. This first quinolone was the base molecule used to synthesize the even more active fluoroquinolones, which also had more favorable pharmacokinetics (Mitscher, 2005). First generation fluoroquinolones (second generation quinolones), like ciprofloxacin, became clinically available in the 1980s and were patent protected until the 2000s (Paton and Reeves, 1988). Until recently, ciprofloxacin was the most potent fluoroquinolone and highly active against virtually all bacterial enteropathogens. It was therefore widely considered to be the drug of choice for patients with enteric fever, severe gastroenteritis, cholera and salmonellosis (Brown and Reeves, 1997). These and other enteric infections are common in the sub-Saharan African sub-continent where an estimated 960–980 million people, most of them young, share the highest risk of infectious disease. Within a year of introduction of ciprofloxacin, resistance appeared and soon became commonplace (Brown and Reeves, 1997). However, 30 years since its introduction, ciprofloxacin is still the most commonly prescribed fluoroquinolone along with levofloxacin, ofloxacin, and moxifloxacin (Redgrave et al., 2014).
Being orally active, broad-spectrum, and heat-stable, the potential for fluoroquinolone application as well as misuse was high. When the efficacy of earlier orally active antibacterials such as the tetracyclines, amino penicillins, and trimethoprim-sulphonamide combinations declined due to rapidly rising antimicrobial resistance rates, fluoroquinolones were recognized as effective alternatives for common and life-threatening infections (Green and Tillotson, 1997). They quickly became among the most widely applied antimicrobials worldwide. The initial high cost of these drugs prohibited their use in the least affluent African settings. The situation changed upon ciprofloxacin patent expiry in 2003, when cheap generics entered African markets. Other fluoroquinolones, such as pefloxacin, met similar fates, and the fluoroquinolones rapidly became first-line drugs for the empiric treatment of many life-threatening infections (Orogade and Akuse, 2004).
The WHO recently highlighted fluoroquinolone resistance in Escherichia coli and related organisms as a principal public health threat (WHO, 2014). Fluoroquinolone resistance was rare worldwide in the 1980s. In Nigeria, as elsewhere, rapid escalation of fluoroquinolone use has been paralleled by a similarly rapid increase in the prevalence of resistant bacteria, including enterics (Omigie et al., 2009; Lamikanra et al., 2011; Namboodiri et al., 2011). In part because quinolone resistance evolved later but also perhaps because mechanisms conferring resistance to a wide range of drugs may have predominated before quinolone-specific mechanisms evolved, fluoroquinolone-resistant strains are typically resistant to multiple antimicrobials (Lamikanra et al., 2011; Namboodiri et al., 2011). Other factors may account for association of fluoroquinolone resistance and multiple resistance in Africa in particular. Quinolones were widely used for decades elsewhere in the world so that resistance genes and resistant clones had already evolved by the time fluoroquinolones were significantly employed in Africa. It has been hypothesized that chloroquine contributed to selective pressure for fluoroquinolone-resistant bacteria in malaria endemic settings but available data do not support this hypothesis for Nigeria (Davidson et al., 2008; Lamikanra et al., 2011). Irrespective of other factors driving fluoroquinolone resistance in enterics across Africa, introduction of fluoroquinolones into routine care in African clinics rapidly enhanced their selection and transmission in a paradigm that is instructive for future antibacterial introductions. In this paper, we review what is known about fluoroquinolone resistant enteric clones in Africa.
Fluoroquinolone resistant enteric organisms and overview of clonal expansion problem
Among enteric bacteria, fluoroquinolone resistance has predominantly been reported in the literature on E. coli (Robicsek et al., 2006), Enterobacter sp. (Robicsek et al., 2006; Cattoir et al., 2007), Citrobacter freundii (Cattoir et al., 2007) Klebsiella pneumoniae (Robicsek et al., 2006), Salmonella Typhi (Dimitrov et al., 2010; Baker et al., 2013), Salmonella enterica (Yanagi et al., 2009), Shigella flexneri (Hata et al., 2005), and Vibrio cholerae (Ismail et al., 2011). There are multiple mechanisms by which these species acquire resistance to fluoroquinolones. Resistance was initially recognized to be due to point mutations in quinolone resistance determining regions (QRDRs) of the gyrA and parC quinolone target genes and may also be attributable to other chromosomal genes that promote quinolone efflux. Secondly, plasmid mediated quinolone resistance (PMQR) can be mediated through QRDR-protective qnr alleles, the ciprofloxacin-acetylating aac(6)-Ib-cr allele or quinolone-efflux genes qepA and oqxAB (Redgrave et al., 2014; Blair et al., 2015). In general fluoroquinolone-resistant strains could have multiple genetic bases for resistance. Some accumulate multiple QRDR mutations and others carry one or more PMQR genes with one or more QRDR mutations. Sequential transition of a strain to higher level resistance is generally attributable to multiple but independent genetic events. Selective pressure from widespread fluroquinolone use appears to have allowed clones that have independently acquired a combination of natural mutation and recombination events to expand internationally (Hooper, 2003; Strahilevitz et al., 2009; Redgrave et al., 2014).
Fluoroquinolone use is an important selector of evolutionary successful resistant lineages but unlikely to be the only one (Baker et al., 2013). More research is needed to promote in-depth understanding of the drivers of emergence and persistence of resistant lineages. Bacterial clones are, by definition, descended from a common ancestor. Evolution within clones and horizontal gene transfer among them subsequently leads to diversification so that organisms of clonal origin may not always be easily discernible and clonal expansion could be difficult to distinguish from evolutionary convergence. Clone descriptions vary and only a subset of resistance mechanisms receive sufficient focus.
Examples of well-known clonal lineages include lineages of E. coli ST131, the fluoroquinolone insensitive and multi-drug resistant (MDR) Salmonella Typhimurium DT104, and the ciprofloxacin-resistant S. enterica serotype Kentucky ST198-X1-SGI1 clone. These clonal groups are marked by rapid and extensive dissemination and have been extensively tracked in many parts of the world (Threlfall, 2000; Le et al., 2013; Petty et al., 2014). However, “worldwide” or “global” studies of these and other clones frequently under-represent African isolates, and in some cases omit them altogether, pointing to the need for a systematic review of published literature on clones in Africa.
The very definition of a “resistant clone” can vary among studies causing problems in understanding the true scope of clonal emergence and subsequent expansion. Genetic studies, preferably sequence-based, are the gold standard for defining clones. Such studies include phylogenetic studies of resistant bacteria that have been characterized via multilocus sequence typing/analysis or whole genome sequencing (WGS) (Roumagnac et al., 2006; Dahiya et al., 2013; Wong et al., 2015). Sequence analysis of resistance genes or plasmids in the context of well-characterized strains can also indicate clonality but, in the absence of host background sequence, cannot robustly differentiate genetically distinct clones. Other studies characterize resistant clones by phenotypic methods such as serogrouping and resistant profiles or pulse-field gel electrophoresis (PFGE; Ke et al., 2014), which looks at genetic profile based on restriction digestion sites. Africa does have an international network to compare PFGE profiles across countries (http://www.pulsenetinternational.org/networks/africa/). However, it is known that PFGE profiles of a single clone may not be identical or chromosomal rearrangements may produce significantly different profiles within a clone (Ismail et al., 2011; Price et al., 2013). Mobile elements can be acquired in different lineages and identical phenotypic characteristics do not necessary imply genetic relatedness (Chattaway et al., 2014).
A clone definition is as good as the discovery methodology. Thus clones are often redefined when more and better information become available. Phenotypic traits—such as serogrouping, virulence, or biochemical profiling that can be acquired convergently in different lineages—cannot be accurately used to track clonal expansion. This has important implications for much of the literature from Africa where sequence-based studies are few. However, the so-called next generation and indeed third-generation sequencing technologies offer an affordable solution to sequence-based clone identification that is increasingly adopted in African studies with great potential for the future. Our review of global fluoroquinolone resistance and clonal expansion in Africa included genetic methodologies such as WGS, detection, or sequencing of fluoroquinolone resistance genes with well characterized strains as the eligible criteria.
Systematic search methods
This systematic review was conducted in accordance with the preferred reporting items of systematic reviews and meta-analyses (PRISMA) guidelines (Moher et al., 2009). Relevant English articles available in the Medline (Pubmed) and African Journals Online (AJOL) databases were retrieved by two authors using predefined search terms (Table S1). The literature search was conducted until October 2015.
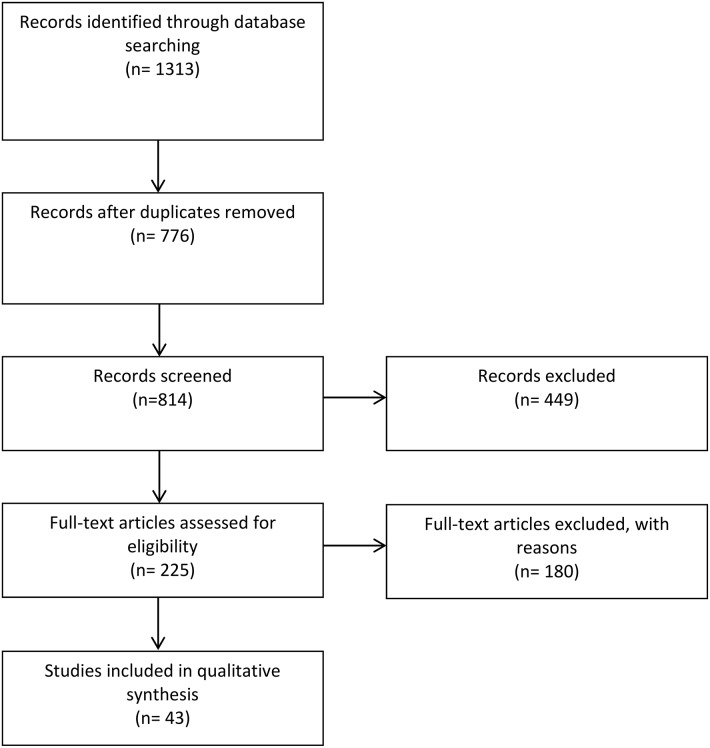
Figure 1 summarizes the study selection process. All duplicate articles were removed. Fluoroquinolone resistant organisms are defined as those having minimum inhibitory concentrations above the CLSI breakpoints (2 μg/ml for ciprofloxacin) or zone sizes below the CLSI breakpoints in standardized disk tests. Strains with MICs above the susceptible breakpoint (0.06 μg/ml and 1 μg/ml for ciprofloxacin in Salmonella and other enterics respectively) are described as fluoroquinolone non-susceptible in this review. The eligibility of published reports in this review was based primarily on polymerase chain reaction (PCR) or sequence-based detection of fluoroquinolone resistance genes and the use of at least one molecular tool for assessing clonality of the enteric pathogen including multi-locus sequencing typing (MLST), multi-locus sequencing analysis (MLSA), clone-specific PCR, or PFGE, subsequent sequencing of fluoroquinolone resistance genes, PCR amplicons, and WGS of the enteric organism. Studies where only phenotypic resistance screening methods were used were excluded.
Figure 1.
Summary of study selection process.
Results
Literature search and characteristics of the studies included in the systematic review
The systematic search of the two electronic databases yielded 1313 articles (Figure 1). After the removal of duplicate studies and assessments of titles and abstracts, 776 full-text articles were screened of which 43 studies were considered eligible for the qualitative analysis according to our inclusion criteria (three articles were not included due to inaccessibility). The majority of the articles retrieved were single center studies. Altogether, eligible studies were identified from 17 African countries, less than half of those south of the Sahara. Furthermore, 25 of these studies—almost two-thirds—were performed in just four countries: CAR, Kenya, Nigeria, and South Africa.
The search retrieved articles focused on V. cholerae, E. coli, and other Enterobacteriaceae and Campylobacter spp. but overall, published work on fluoroquinolone-resistant enteric clones in Africa is predominantly focused on Salmonella and whole genome sequence data-based studies are almost entirely limited to this genus. One possibility is that the identification of hypervirulent invasive non-typhoidal Salmonella lineages on the continent (Okoro et al., 2012) is fueling this research. However, our reading of the literature appears to point to the enhanced focus on the molecular epidemiology of Salmonella in East and Southern Africa as being responsible for identifying this clonal expansion problem in the first place and it is probable that expanded research on other pathogens will uncover other local clones.
Many studies seeking fluoroquinolone-resistant strains found strains with intermediate susceptibility and/or strains that were resistant to the quinolones but not the fluoroquinolones (Table S2). As resistance to the fluoroquinolones typically evolves in a stepwise fashion, the wide prevalence of these intermediate strains is worrisome. While there are very few studies reporting fluoroquinolone-resistant E. coli clones, reports are increasingly using sequence-based methods to identify clones, specifically, MLST. The predominance of MLST has been helpful in placing the findings from African countries into global context and demonstrates the extension of knowledge that can come from using sequence-based tools in small studies (Tables 2, 3). There are two E. coli MLST schemes in the literature and whilst the Lacher et al. scheme (Lacher et al., 2007) has been used in African studies (Feglo et al., 2013), the more widely used (Wirth et al., 2006) scheme is more frequently applied, making comparisons among these many small studies possible. Where MLST was not applied per se, PCR markers for MLST-defined ST131 clades were used to identify members of those clones (Peirano et al., 2011, 2014; Albrechtova et al., 2012; Gqunta and Govender, 2015). Methodology of this type could be developed for and applied to other clones that are later found to be of interest in Africa (Doumith et al., 2015). A few African studies, generally older ones, have used PCR-based techniques and PFGE (Lavollay et al., 2006; Kariuki et al., 2007; Peirano et al., 2011). While these studies are the principal sources of information on distribution of FQREC soon after these strains evolved in Africa, it is difficult to connect the clones identified therein to those reported more recently.
Data from Salmonella are predominantly available from Eastern and Southern Africa, and there are two multicountry studies focused on Salmonella Kentucky ST198 (Le et al., 2011, 2013). V. cholerae studies reported from outbreaks in Western (Nigeria, Cameroon), Eastern (Kenya), and Southern Africa with one pair of studies focused on the same outbreak on either side of the Nigeria-Cameroon border (Quilici et al., 2010; Marin et al., 2013). Most E. coli data studied in this review came from single country studies, limited to CAR (Janatova et al., 2014), Ghana (Namboodiri et al., 2011; Feglo et al., 2013) Kenya (Kariuki et al., 2007; Albrechtova et al., 2012), Nigeria (Lamikanra et al., 2011; Aibinu et al., 2012a), and South Africa (Gqunta and Govender, 2015). Intercontinental studies generally included isolates from only one sub-Saharan African country (CAR or South Africa) and other non-African countries (Lavollay et al., 2006; Peirano et al., 2011, 2014). Therefore there are no data comparing FQREC isolates from different countries in sub-Saharan Africa. Additionally, multicountry or “global” studies are typically including only one country from within sub-Saharan Africa, even though it is well known that biological diversity on the African continent exceeds that in most other parts of the world.
Vibrio cholerae
Cholera, the pandemic diarrheal disease caused by toxigenic V. cholerae strains, has become increasingly problematic across Africa in the last two decades but fluoroquinolone resistance has only been recently studied (Scrascia et al., 2006, 2009; Mintz and Guerrant, 2009), Quilici et al. (2010) described the microbiology of a cholera outbreak that spanned the borders of Nigeria and Cameroon in 2009 and (Quilici et al., 2010) was due to a single V. cholerae O1 Ogawa El-tor clone. The ctxB DNA sequence from this clone was similar (only 3 SNPs) to a strain identified in India in 2007 but different from variants isolated in southern Africa, indicating a possible transmission from Asia to Africa. This clone was resistant to nalidixic acid and MICs to ciprofloxacin were 0.25–0.5 μg/ml, placing them in the susceptible category (based on general enterobacterial breakpoints), although these MICs approach the reduced susceptibility breakpoint of 1 μg/ml. Strains belonging to this clone contained mutations in the QRDR of gyrA (Ser83-Ile) and parC (Ser85-Leu) but no mutations in gyrB or parE. A more comprehensive Nigeria study looking at V. cholerae O1 strains isolated in the early 1970's and comparing them with strains of V. cholerae O1 and non-O139 strains isolated in 2009–2010 indicated that gyrA (Ser83-Ile) and parC (Ser85-Leu) mutations were not present in the 1970's or in non-O1 clinical isolates (Marin et al., 2013). Interestingly, MLVA analysis clustered the 1970's O1 El-tor strain strains with epidemic O1 El-tor strains but the strains had different ctxB types and vibrio 17th pandemic island (VSP-II) profiles indicating that they do not represent one clone (Marin et al., 2013). This can be explained by the housekeeping genes used in this analysis (pyrH, recA, and mdh) being highly conserved in V. cholerae serogroup strains. Approaches using whole genome sequence may be required to further define the inter-relatedness of these O1 strains.
The most in-depth assessment of fluoroquinolone resistant mechanisms was a South African study focused on a V. cholerae O1 Ogawa El-tor clone outbreak (Ismail et al., 2011). A selection of strains was assessed for the presence of plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrS, qnrC, and qepA) as well as Vibrio quinolone resistance determinant (qnrVC3). Nalidixic acid resistance was investigated in the presence and absence of two efflux pump inhibitors and QRDR mutations were identified by sequencing. The South Africa strains were negative for any PMQR genes and there was no evidence of active efflux conferring quinolone resistance. Nalidixic acid resistance was attributable to gyrA (Ser83-Ile) and parC (Ser85-Leu) mutations (Ismail et al., 2011). All of the recent outbreak strains described in these three studies were V. cholerae O1 El-tor (two studies also indicated serogroup Ogawa) which showed resistance to nalidixic acid but only a reduced susceptibility to ciprofloxacin (Table 1). It is highly likely that these strains are the same clone and that the same QRDR mutations are responsible for fluoroquinolone resistance. High levels of resistance to nalidixic acid or reduced susceptibility to ciprofloxacin in likely similar V. cholerae O1 clones causing epidemics in DRC and Kenya have also been shown (Mercy et al., 2014; Miwanda et al., 2015; Table 1), but a lack of standard methodology for clonal analysis prevents an understanding of clonal spread across Africa. Although ciprofloxacin has only a reduced susceptibility in these strains and continues to be used for cholera management, if additional mutations occur in these circulating clones, resistance to ciprofloxacin may develop.
Table 1.
Characterization of eligible articles that sought fluoroquinolone resistant clones in humans and animals**.
| Country | Organism | No. resistant**/No total | ID method | Study period | Study population | Genotyping tools | Resistance testing | References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WGS | MLST | ERIC-PCR | PFGE | Rep PCR | Other* | PMQR | QRDR | Efflux pump up- regulation | aac(6′)-Ib-cr | |||||||
| Angola | E. coli | 9/19 | B, S | 2009 | Stray dogs | – | – | – | √ | – | – | √ | – | – | – | Albrechtova et al., 2014a |
| Cameroon | V. cholerae O1 Ogawa El-tor | 0/9 | B, S, M | 2009 | Outbreak | – | – | – | – | – | – | – | √ | – | – | Quilici et al., 2010 |
| Central African Republic | Enterobacteriaceae | 33/121 | B, S | 2011 | Human/wildlife | – | – | – | √ | – | √ | – | – | Janatova et al., 2014 | ||
| Enterobacteriaceae | 65/65 | B | 2011–2012 | Clinical isolates (surgical site infections) | – | √ | √ | – | √ | – | √ | – | – | √ | Rafai et al., 2015 | |
| E. coli | 10/121 | B, M | 2011 | Gorillas and other wildlife | – | √ | – | – | – | – | √ | – | – | √ | Janatova et al., 2014 | |
| E. coli | 10/10 | B, M | 2000–2004 | Hospital clinical isolates | – | – | – | √ | √ | – | – | – | – | √ | Lavollay et al., 2006 | |
| S. Typhi | 0/19 | B, S | 1916–2004 | Various | √ | – | – | – | – | – | – | – | – | – | Holt et al., 2008 | |
| Cote d'Ivoire | E. coli, Enterobactericeae | 44/101 | B, S | 2012 | Human, dog, wild life | – | – | – | √ | – | – | √ | – | – | √ | Albrechtova et al., 2014b |
| DRC | Salmonella (NTS) | 0/233 | B, S | 2007–2011 | Community and Clinical isolates | – | – | – | √ | – | – | √ | √ | – | – | Lunguya et al., 2013 |
| V. cholerae | 0/1093 | B, S | 1997–2012 | Clinical Isolates | √ | – | – | – | – | MLVA | – | – | – | – | Miwanda et al., 2015 | |
| Ghana | E. coli | 23/293 | B, M | 2006–2008 | Clinical isolates from tertiary care hospitals | – | √ | – | – | – | – | √ | √ | – | – | Namboodiri et al., 2011 |
| E. coli | 28/29 | B, M | 2008–2009 | Clinical isolates: Blood, urine, sputum and wound | – | √ | – | – | – | – | – | – | – | – | Feglo et al., 2013 | |
| Ethiopia | Salmonella | 19/98 | B, S | 2000–2005 | Animal/Animal products | – | – | – | √ | – | – | – | – | – | – | Molla et al., 2007 |
| Kenya | E. coli | 66/289 | B, M | 2009 | Dogs (mainly), cats and humans | – | – | – | √ | – | – | √ | – | – | √ | Albrechtova et al., 2012 |
| E. coli | 12/17 | B, M | 2004–2005 | Clinical isolates: Urine | – | – | – | √ | – | – | √ | – | – | √ | Kariuki et al., 2007 | |
| V. cholerae | 1/76 | B, S | 2007–2010 | Clinical Isolates | – | – | – | √ | – | Ribo-typing | – | – | – | – | Mercy et al., 2014 | |
| S. Kentucky | 3/3 | B, S | 2000–2005 | Clinical Isolates | – | – | – | √ | – | √ | √ | – | – | Weill et al., 2006 | ||
| S. Typhi | 0/94 | B, S | 1988–2008 | Clinical isolates | √ | – | – | – | – | – | – | – | – | – | Kariuki et al., 2010 | |
| S. Typhi | 0/102 | B, S | 2001–2002 | Clinical isolates | – | – | – | √ | – | – | – | √ | – | – | Kariuki et al., 2004 | |
| S. Typhi | NS/55 | NS | 2001–2009 | Clinical isolates | √ | – | – | – | – | – | – | – | – | – | Wong et al., 2015 | |
| Malawi | S. Typhi | 0/112 | B, BC | 1998–2013 | Clinical isolates (blood) | √ | √ | – | – | – | – | – | – | – | – | Feasey et al., 2015 |
| S. Typhi | NS/112 | NS | 2004–2013 | Clinical isolates | √ | – | – | – | – | – | – | – | – | – | Wong et al., 2015 | |
| Nigeria | E. coli | 6/162 | B, S | 2006 | Poultry/Pigs | – | – | √ | – | – | – | √ | – | – | √ | Fortini et al., 2011 |
| E. coli | 57/114 | B, M | 2010–2011 | Human clinical isolates, bovine isolates from farms | – | √ | – | – | √ | – | √ | – | – | √ | Inwezerua et al., 2014 | |
| E. coli | 26/32 | B, M | 2011 | Humans (pregnant women) | – | √ | √ | – | – | – | √ | – | – | √ | Fortini et al., 2015 | |
| E. coli | 9/109 | B, M | 2008–2009 | Clinical isolates from tertiary care hospitals | – | ST131 and ST10 | √ | – | – | – | √ | – | – | √ | Aibinu et al., 2012a | |
| E. coli | 21/121 | B, M | 2005 | Healthy volunteers | – | √ | – | – | – | fliC -RFLP | – | √ | √ | – | Lamikanra et al., 2011 | |
| Salmonella (NTS) | 11/149 | B, S | 2009–2011 | Human/poutry/cattle/fish/vegetable | – | – | – | – | – | – | √ | – | – | – | Raufu et al., 2013 | |
| Salmonella (NTS) | 2/229 | B, S | 2010–2011 | Pigs | – | – | – | – | – | – | √ | – | – | – | Fashae and Hendriksen, 2014 | |
| S. Hiduddify | 24/130 | B, S | 2001 | Chickens/poultry meat | – | – | – | √ | – | – | – | – | – | – | Raufu et al., 2009 | |
| S. Kentucky | 197/197 | B, S | 1959–2008 | Human/Poultry/sea food/river/environment | – | √ | – | √ | – | – | √ | √ | – | √ | Le et al., 2011 | |
| S. Kentucky | 55/55 | B, S | 2007–2011 | Poultry/poultry sources | – | – | – | √ | – | – | – | – | – | – | Raufu et al., 2014 | |
| S. Kentucky | 54/70 | B, S | 1937–2013 | Humans/animals/environment | – | √ | – | √ | – | – | √ | √ | – | √ | Le et al., 2013 | |
| V. cholerae O1 and non O1 | 0/20 | B, S, M | 1971 –2010 | Outbreak, historical, environmental | – | – | – | √ | – | MLSA | – | √ | – | – | Marin et al., 2013 | |
| V. cholerae O1 Ogawa El-tor | 0/10 | B, S, M | 2009 | Outbreak | – | – | – | – | – | – | – | √ | – | – | Quilici et al., 2010 | |
| E. coli | NS/12 | B, M | 2000–2010 | Clinical isolates | – | √ | – | √ | – | Lineage PCR | – | √ | – | √ | Peirano et al., 2014 | |
| E. coli | 0/22 | B, M | 2008–2009 | Clinical isolates (Urine and pus) | – | – | – | √ | – | – | √ | – | – | √ | Peirano et al., 2011 | |
| Senegal | C. coli | 14/36 | B, M | 2000–2002 | Chicken | – | √ | – | – | – | – | – | √ | – | – | Kinana et al., 2007 |
| Salmonella (NTS) S. Typhi | 4/1623 | B, S, M | 1999–2009 | Clinical isolates | – | – | – | – | – | – | √ | √ | – | – | Harrois et al., 2014 | |
| C. jejuni | 19/46 | B, M | 2000–2002 | Chicken | – | √ | – | – | – | – | – | √ | – | – | Kinana et al., 2006 | |
| C. jejuni, C. coli | 81/181 | B, M | 2000–2003 | Poultry, Human | – | – | – | √ | – | – | – | √ | – | – | Cardinale et al., 2006 | |
| Sierra Leone | V. cholerae O1 | 0/15 | B, M, S | 2012 | Clinical isolates | – | – | – | √ | – | – | – | – | – | – | Mahmud et al., 2014 |
| South Africa | E. coli | 14/21 | B, M | 2012–2013 | Clinical isolates | – | √ | – | √ | – | – | √ | – | – | √ | Gqunta and Govender, 2015 |
| S. Typhi | 1/1 | B, S | 2010 | Clinical isolate | – | – | – | √ | – | – | √ | √ | – | – | Keddy et al., 2010 | |
| S. Typhi | 0/510 | B, S | 2003–2007 | Clinical isolates | – | – | – | √ | – | MLVA | √ | √ | – | – | Smith et al., 2010 | |
| S. Typhi | NS/41 | NS | 2004–2012 | Clinical isolates | √ | – | – | – | – | – | – | – | – | – | Wong et al., 2015 | |
| S. Kentucky | 1/1 | B, S | 2000–2005 | Clinical Isolates | – | – | – | √ | – | – | √ | √ | – | – | Weill et al., 2006 | |
| V. cholerae O1 Ogawa El-tor | 0/31 | B, S, M | 2008 | Outbreak | – | – | – | √ | – | – | – | – | – | Ismail et al., 2011 | ||
| Tanzania | S. Typhi | NS/52 | NS | 2006–2010 | Clinical isolates | √ | – | – | – | – | – | – | – | – | – | Wong et al., 2015 |
| S. Kentucky | 3/3 | B, S | 2000–2005 | Clinical Isolates | – | – | – | √ | – | √ | √ | – | – | Weill et al., 2006 | ||
KEY: ID method: B, Biochemistry; S, Serotyping; M, detection of target genes; BC, Blood culture; √, test was conducted; –, test not conducted; NS, Not stated (data not included in manuscript).
Other, where specific genotypic methods are used to identify clones of that enteric pathogen.
In some cases where fluoroquinolone-resistant clones were sought but only quinolone resistant or non-susceptible isolates were identified. Details of these studies are provided in Table S2.
WGS, Whole genome sequencing; MLST, Multi-locus sequence typing; MLSA, Multi-locus sequencing analysis; MLVA, multi-locus variable-number tandem repeat analyses; PFGE, pulse field gel electrophoresis; ERIC-PCR, Enterobacterial Repetitive Intergenic Consensus – PCR; REP-PCR, repetitive element palindromic PCR; RFLP, restriction fragment length polymorphism; NTS, Nontyphoidal Salmonella, Clinical Isolates – from humans; PMQR, plasmid mediated quinolone resistance (including qnrA, qnrB, qnrS, qnrC, qepA, and qnrVC3); QRDR, quinolone resistance-determining region [including sequencing the QRDR of DNA gyrase (gyrA, gryB) and topoisomerase IV (parC, parE) genes]; efflux, efflux up regulation testing; aac(6′)-Ib-cr, aminoglycoside acetyltransferace associated with ciprofloxacin resistance; DRC, Democratic Republic of the Congo.
Campylobacter
Campylobacter jejuni is a leading enteric bacterial pathogen in developing countries particularly among children below the age of 5 years. In spite of numerous reports of C. jejuni as well as its resistance to fluoroquinolones (Coker et al., 2002) data on clones of the organisms causing human infections in Africa are non-existent. Characterization of 46 isolates from chickens in Senegal by MLST revealed seven clonal complexes which have all been associated with human disease elsewhere (Kinana et al., 2006). Clonal complex 353 was the most common and Thr-86-Ile substitution in gyrA was the major mechanism of quinolone resistance. Rather than diffusion of a single or a few clones, acquisition of resistance appears to be occurring in different lineages in response to selective pressure. Similarly, another study from Senegal, strain typing of 99 chicken and 10 human isolates of C. jejuni by PFGE that uncovered common patterns among strains circulating in human and poultry, though were no particular predominant pulsotypes (Cardinale et al., 2006). Altogether, although the data are few, there is yet to be robust evidence for expanded fluoroquinolone resistant Campylobacter clones.
Enteric and commensal Escherichia coli and Shigella
Diarrhea was the third leading cause of death in children under five in sub-Saharan Africa in 2013 (after malaria and pneumonia), accounting for 12% of the estimated 3.6 million deaths (Liu et al., 2015). In sub-Saharan Africa and south Asia, most attributable cases of moderate-to-severe childhood diarrhea in under-fives are due to the enteric bacteria E. coli and Shigella (Kotloff et al., 2013). While there was a report of a single quinolone-resistant Shigella isolate from Zaire in 1985 (Frost et al., 1985) and Shigella epidemics caused by quinolone resistant bacteria in the last century (Goma Epidemiology Group, 1995; Karas and Pillay, 1996; Vlieghe et al., 2009) we did not identify reports of fluoroquinolone-resistant clones of enteric E. coli (FQREC) in the literature before 2000. Available reports appear to suggest that most isolates were fluoroquinolone susceptible until the end of the 1990s (Wistrom et al., 1987; Felmingham and Robbins, 1992; Thornton et al., 1992; Bourgeois et al., 1993; Oyofo et al., 1997; Decousser et al., 1999; Vila et al., 1999; Ahmed et al., 2000; Okeke et al., 2000a,b; Jiang et al., 2002; Orogade and Akuse, 2004). Our findings are corroborated by other recent systematic reviews (Vlieghe et al., 2009; Leopold et al., 2014).
Reports of fluoroquinolone-resistant E. coli from cases of diarrhea and commensal reservoirs in Africa are limited to the current millennium (Aibinu et al., 2004; Lamikanra et al., 2011; Namboodiri et al., 2011; Fortini et al., 2015). One study that examined human commensal isolates from Ghana (2006–2008) found a wide range of STs associated with fluoroquinolone resistance (Namboodiri et al., 2011). However, strains belonging to the multilocus sequence type (ST) complex (specifically STs 10, 227, and 617) and the ST 156 sequence type complex (STs 156 and 1473) were detected 8 and 3 times respectively representing half of the total number (22) of FQREC detected in the study. An additional five ST10 isolates were resistant to nalidixic acid and overall, ST10 strains were computed to be overrepresented among quinolone-resistant E. coli (QREC) from Ghana (p = 0.02, Fishers exact test; Namboodiri et al., 2011). Five ST10 QREC (including four FQREC) harbored qnrS1, whilst other strains carried qepA or qnrB2 genes. The ST156 strains harbored three or more QRDR mutations, in contrast to most other strains that had 0–2 and one of the ST156 isolates also carried qepA. Interestingly, multiple qepA-bearing FQREC ST156 strains have also been reported as human commensals in the Central African Republic and Nigeria (Table 2; Janatova et al., 2014; Fortini et al., 2015). These and other lineages could be more broadly disseminated but remain unflagged because data are insufficient or unavailable. For example, qnrS1-bearing clones that have not been multilocus sequence typed (clonality was inferred by ERIC-PCR) have also been isolated from humans and pigs in Nigeria (Fortini et al., 2011, 2015).
Table 2.
Multilocus sequence types of quinolone resistant commensals reported from potential reservoirs in sub-Saharan Africa.
| City, Country | Host organism | Year | Number of independent isolates | ST | Quinolone resistance mechanisms | References |
|---|---|---|---|---|---|---|
| Accra, Ghana | Human | 2006 | 2 | E. coli (ST101) | gyrA (S83L, D87N); qnrB1 | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2006 | 1 | E. coli (ST617) | gyrA (S83L, D87N); parC (S80I); qnrS1 | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2006 | 2 | E. coli (ST648) | gyrA (S83L, D87N); parC (S80I) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2006 | 1 | E. coli (ST455) | gyrA (S83L, D87N); parC (S80I, N105S) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2006, 2007 | 2 | E. coli (ST156) | gyrA (S83L, D87N); parC (S80I, E84K), qepA | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2007 | 1 | E. coli (ST1304) | None detected | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2007 | 1 | E. coli (ST450) | gyrA (S83L, D87N) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2007, 2008 | 5 | E. coli (ST10) | gyrA (S83L, D87N); parC (S80I), qepA, qnrS1 | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2007 | 1 | E. coli (ST354) | gyrA (S83L, D87N); parC (S80I, E84G), qnrB | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST1473) | None detected | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST1496) | None detected | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 2 | E. coli (ST227) | gyrA (S83A), qnrS1 | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST410) | gyrA (S83L, D87N); parC (S80I) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST1468) | gyrA (S83L, D87N); parC (S80I) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST131) | gyrA (S83L, D87N); parC (S80I, E84V) | Namboodiri et al., 2011 |
| Accra, Ghana | Human | 2008 | 1 | E. coli (ST1470) | gyrA (S83L, D87N); parC (S80I, A108V) | Namboodiri et al., 2011 |
| Dzanga, CAR | African buffalo | 2011 | 1 | K. pneumoniae (ST1208) | oqxA | Janatova et al., 2014 |
| Dzanga, CAR | African buffalo | 2011 | 1 | K. pneumoniae (ST1209) | oqxA | Janatova et al., 2014 |
| Dzanga, CAR | Peter's Dunker | 2011 | 1 | K. pneumoniae (ST1210) | oqxA | Janatova et al., 2014 |
| Dzanga, CAR | Human | 2011 | 4 | E. coli (ST3476) | qnrS1 | Janatova et al., 2014 |
| Dzanga, CAR | Human | 2011 | 3 | E. coli (ST156) | qepA | Janatova et al., 2014 |
| Dzanga, CAR | Human | 2011 | 1 | E. coli (ST69) | oqxA | Janatova et al., 2014 |
| Dzanga, CAR | Human | 2011 | 1 | E. coli (ST218) | qnrB1 | Janatova et al., 2014 |
| Dzanga, CAR | Gorilla | 2011 | 1 | E. coli (ST424) | qepA | Janatova et al., 2014 |
| Northern Kenya | Dogs | 2012 | 3 | E. coli (ST131*) | aac(6')-Ib-cr | Janatova et al., 2014 |
| Ile-Ife, Nigeria | Human | 2005 | 9 | E. coli (ST10 complex) | gyrA (S83L), efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 2 | E. coli (ST452) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 2 | E. coli (ST517) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST494) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST156) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST521) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST448) | gyrA (S83L, D87N); parC (S80I), efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST503) | efflux | Lamikanra et al., 2011 |
| Ile-Ife, Nigeria | Human | 2005 | 1 | E. coli (ST504) | gyrA (S83L), efflux | Lamikanra et al., 2011 |
| Ibadan, Nigeria | Human | 2011 | 1 | E. coli (ST10) | qnrS1, aac(6')-Ib-cr | Fortini et al., 2015 |
| Ibadan, Nigeria | Human | 2011 | 1 | E. coli (ST156) | qepA1 | Fortini et al., 2015 |
| Ibadan, Nigeria | Human | 2011 | 1 | E. coli (ST3147) | qnrS1 | Fortini et al., 2015 |
| Senegal | Poultry | 2000–2002 | 11 | C. jejuni (ST1036) | gyrA (H81, S119, T86I, G110) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 3 | C. jejuni (ST1040) | gyrA (H81, S119, T86I) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 5 | C. jejuni (ST1039) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 3 | C. jejuni (ST1041) | gyrA (H81, S119, T86A, G110) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1081) | None detected | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST660) | None detected | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST22) | None detected | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1037) | gyrA (H81, S119, T86I) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 3 | C. jejuni (ST1038) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 4 | C. jejuni (ST52) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST824) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1359) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1211) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1370) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 1 | C. jejuni (ST1358) | gyrA (H81, S119) | Kinana et al., 2006 |
| Senegal | Poultry | 2000–2002 | 8 | C. jejuni (ST1035) | gyrA (H81, S119, G110, T86A, T86I) | Kinana et al., 2006 |
Determined by marker-based PCR not MLST.
Invasive Escherichia coli
Fluoroquinolone-resistant E. coli are a rising cause of invasive infections in Africa. The majority of reports of fluoroquinolone-resistant enterobacterial lineages only seek lineage information among ESBL-producers (Table 3; Aibinu et al., 2012a; Isendahl et al., 2012; Breurec et al., 2013; Gqunta and Govender, 2015; Rafai et al., 2015) so that the true prevalence of fluoroquinolone-resistant strains is unknown as is the existence of ESBL-negative fluoroquinolone-resistant lineages of importance. Interestingly, while the data are few and biased toward ESBL-positive lineages, there is some overlap between STs of FQREC recovered from clinical infections and those found in the commensal reservoir from humans and wild mammals (Tables 2, 3).
Table 3.
Multilocus sequence types of fluoroquinolone-resistant E. coli clinical isolates reported from sub-Saharan Africa.
| City, Country | Isolation source | Year | Number of independent isolates | ST | Reported quinolone resistance mechanisms | Other resistances | References |
|---|---|---|---|---|---|---|---|
| Kumasi, Ghana | Blood, Urine, sputum and wounds | 2008–2009 | 28 | 88 Whittam scheme (Lacher et al., 2007) | None reported | ESBLs, gentamicin, trimethoprim-sulphamethoxazole, chloramphenicol | Feglo et al., 2013 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 12 | 10 | qnrB,qnrS, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 1 | 146 | qnrB, qnrS, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 12 | 131 | qnrB, qnrS, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 5 | 156 | qnrB, qnrS, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 1 | 354 | qnrS, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Bangui, CAR | Surgical site infections | 2011–2012 | 2 | 405 | qnrB, aac(6')-Ib-cr | ESBLs | Rafai et al., 2015 |
| Cape Town, South Africa | Urine and Pus | 2008–2009 | 5 | 131* | aac(6')-Ib-cr | ESBLs | Peirano et al., 2011 |
| Ibadan, Nigeria | Urine | 2010–2011 | 1 | 2695 | qnrB, aac(6')-Ib-cr | ESBLs | Inwezerua et al., 2014 |
| Ibadan, Nigeria | Vaginal | 2010–2011 | 1 | 131 | aac(6')-Ib-cr | ESBLs | Inwezerua et al., 2014 |
| Lagos, Nigeria | Urine | 2011 | 2 | 131 | qnrB, aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Lagos, Nigeria | Urine and stool | 2011 | 2 | 617 (10 complex) | qnrB, aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Lagos, Nigeria | Urine and stool | 2011 | 1 | 295 | aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Lagos, Nigeria | Urine and stool | 2011 | 1 | 23 | aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Lagos, Nigeria | Urine and stool | 2011 | 1 | 448 | aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Lagos, Nigeria | Urine and stool | 2011 | 1 | 501 | aac(6')-Ib-cr | Aibinu et al., 2012b | |
| Port Elizabeth, South Africa | Clinical | 2012–2013 | 12 | 131* | qnrB qnrS, aac(6')-Ib-cr | ESBLs | Gqunta and Govender, 2015 |
E. coli STs are defined by the Achtman scheme unless otherwise indicated (Wirth et al., 2006).
Determined by marker-based PCR not MLST.
Kariuki et al. examined multiply resistant E. coli from urinary tract infections in Kenya (Kariuki et al., 2007). Two PFGE profiles were seen more than once. Strains belonging to these clones had identical gyrA sequences, which included Ser83Leu and Asp87Asn mutations in the QRDR regions of this gene. They also carried a plasmid-borne aac-6(Ib)-cr gene and some had qnrA and qnrB genes. Working largely with strains from other areas but including 10 isolates from Bangui, Lavollay et al. found a globally disseminated clone that carried an aac6-Ib-cr allele on its CTX-M-15 bearing plasmid and detected two isolates of this clone, as defined by REP-PCR and PFGE, among the Central African Republic isolates (Lavollay et al., 2006).
Available data suggests that pandemic lineages circulate within Africa. For example, fluoroquinolone-resistant strains belonging to the globally disseminated E. coli ST131 lineage, which is actually comprised of multiple clones, have rarely been systematically sought but there are sporadic reports that demonstrate their circulation in CAR (Rafai et al., 2015), Nigeria (Aibinu et al., 2012a; Inwezerua et al., 2014), Guinea Bissau (Isendahl et al., 2012), Tanzania (Lupindu et al., 2014), and South Africa (Peirano et al., 2011, 2014; Gqunta and Govender, 2015; Table 3). These data suggest that global dissemination may be dictated by the pandemic potential of the ST131 lineages rather than actual intercontinental transfer of specific clones.
Aibinu et al (Aibinu et al., 2012a) studied 109 E. coli isolates obtained from two tertiary care hospitals in Lagos. Of the 14 isolates that were ESBL-producers, three were ST131 E. coli. Chromosomal SNPs were not sought but two of the ST131 isolates and six other ESBL-producers (ST 10 complex—two strains, STs 23, 295, 448, and 501) harbored the aac-(6′)-lb-cr gene (Table 3). These eight strains and the pMQR-negative ST131 strain all carried the ESBL-encoding CTX-M-15 gene. One ST131 strain additionally carried a qnrB1 gene and a ST 617 strain (ST10 complex) carried qnrA1. This study demonstrated the presence of ST131, as well as other notorious E. coli clones. Many of the STs reported by Aibinu et al. were also found 2005 in the Nigerian study of Lamikanra et al. (2011) (2009 isolates from that study were not sequenced typed) as well as among 2011 isolates from Fortini et al. (2015). Feglo et al. conducted MLST on a subset of E. coli isolates from Kumasi Ghana, hypothesizing that they would find ST131 strains among them. Instead, they found that all of 29 isolates evaluated belonged to the pyleonephritis-associated lineage ST88. Thus, regional or internationally disseminated clones have been successful in African settings and that locally successful clones are not necessarily those with the greatest pandemic reputations.
Salmonella Typhi and non-typhoidal Salmonella
Salmonella causes an estimated three billion infections in humans and animals yearly (Crump et al., 2004; Coburn et al., 2007). This poses significant socio-economic challenges, especially in resource-limited settings. Classically, Salmonella can be divided into two species, S. bongori and S. enterica (Crosa et al., 1973; Schrire et al., 1987; Reeves et al., 1989). S. enterica can be further divided into six sub-groups based on carbohydrate utilization, flagellar, and lipopolysaccharide (LPS) structures (Tindall et al., 2005; Coburn et al., 2007; Grimont and Weill, 2007). Currently there are more than, 2550 serovarieties of S. enterica. S. enterica is responsible for most cases of salmonellosis in human and warm-blooded animals (Brenner et al., 2000). Based on the type of disease syndrome and host specificity, S. enterica can be broadly divided into two groups (typhoidal and non-typhoidal). “Typhoidal Salmonellae” are generally human-restricted and cause enteric fever; these include S. Typhi and S. Paratyphi. The non-typhoidal salmonellae (NTS) encompass several serovars with broad hosts including animals and humans such as S. Typhimurium, S. Enteritidis, S. Poona, and S. Isangi. (Dougan et al., 2011). In the WHO Africa sub-region, the burden of non-typhoidal gastroenteritis is estimated at 2.5 million cases and 4100 deaths per year (Majowicz et al., 2010) but this estimate is based on few data. Nontyphoidal Salmonella are also risk factors for mortality from childhood diarrhea (O'Reilly et al., 2012). NTS serovars are commonly associated with gastrointestinal disease in hosts but can sometimes cause invasive or systemic disease in humans and animals (Dougan et al., 2011; Langridge et al., 2012).
Emerging epidemiological data from across the subcontinent shows that invasive non-typhoidal Salmonella (iNTS) infection has become a major cause of bloodstream infections, overtaking typhoidal enteric fever in many places, especially among malarial and malnourished children and in HIV-positive adults (Reddy et al., 2010). Mortality rates in these vulnerable populations are reported to be as high as 30% in the absence of adequate treatment (Berkley et al., 2005; Reddy et al., 2010). Multidrug resistance of salmonellae to ampicillin, chloramphenicol and trimethoprim-sulphamethoxazole (traditional regimens, cheap, and orally active) is a global phenomenon, presenting particular salmonellosis management challenges in Africa including the epidemic of invasive S. Typhimurium ST 313. This clone consists of two closely related lineages that emerged independently ~ 55 and ~37 years ago, in close temporal association with the HIV pandemic (Kingsley et al., 2009; Okoro et al., 2012). The MDR genes of ST 313 are encoded on a Tn21-like element borne on a virulence plasmid pSLT-BT. MDR clones of S. Typhi have also emerged and spread intercontinentally and globally involving Africa countries like Kenya (Kariuki et al., 2010; Holt et al., 2011). Prevalence rates of MDR as high as 60.4, 80.7, and 94.7% have been reported from Kenya (Kariuki et al., 2010), DRC (Lunguya et al., 2013), and Malawi (Feasey et al., 2010), respectively. Increasing and dissemination of MDR clones of salmonellae warranted the current use of fluoroquinolones and third generation cephalosporins as the drugs of first choice for treatment (Kariuki and Dougan, 2014; Kariuki et al., 2015a). These newer drugs are expensive and pose significant challenges to resource poor settings. The use of these drugs, together with the use of over-the-counter and substandard antibiotics, has now selected resistance to these drugs in salmonellae (Brenner et al., 2000; Parkhill et al., 2001; McClelland et al., 2004; Berkley et al., 2005; Gordon et al., 2008; Holt et al., 2008; Kingsley et al., 2009; Majowicz et al., 2010; Reddy et al., 2010; Dougan et al., 2011; Langridge et al., 2012; Okoro et al., 2012; O'Reilly et al., 2012; Kariuki and Dougan, 2014; Kariuki et al., 2015a) Initial reports of reduced susceptibility and resistance to quinolones in NTS emerged in the mid 2000's and continue to trickle in from many sub-Saharan African countries including Nigeria (Akinyemi et al., 2007), DRC (Lunguya et al., 2013), Tanzania (associated with travel; Weill et al., 2006), Senegal (Harrois et al., 2014), and Ghana (Nielsen et al., 2012). A Mozambican study identified ~5% of isolates as being resistant to nalidixic acid; resistance to nalidixic acid is regarded as a precursor for 2nd and 3rd generation fluoroquinolone resistance (Mandomando et al., 2009). Recent reports have shown that strains of MDR S. Typhimurium ST313 primarily associated with bloodstream infections have acquired an additional incHI2 plasmid that confers additional resistance to many β-lactams including ceftriaxone. These pathovariants have been identified in Kenya (Kariuki et al., 2015b) and Malawi (Feasey et al., 2014). Other reports have shown an association with the acquisition of extended-spectrum β lactamases (ESBLs) and decreased susceptibility to fluoroquinolones in Salmonellae, the latter attributable to plasmid-borne qnr genes, although mechanism underlying the significance of this association has not been fully explored (Akinyemi et al., 2007, 2011; Harrois et al., 2014).
There are more than 20 million cases of human typhoid fever resulting in as many as 200,000 fatalities worldwide each year (Crump et al., 2004; Arjyal et al., 2011). Reports of outbreaks of MDR Typhi and Paratyphi emerged in the early 1990's in endemic countries in Asia and associated with travel to these regions in non-endemic (Parry et al., 2002; Arjyal et al., 2011). MDR in these instances was characterized by resistance to the first-line drugs of treatment at that time which included chloramphenicol, ampicillin, and cotrimoxazole. Subsequently as was in the case of NTS, fluoroquinolones were introduced as the preferred drug of treatment for enteric fever (Arjyal et al., 2011). Resistance to this drug inevitably emerged with widespread use. Isolates with full resistance or reduced resistance to nalidixic acid and second generation fluoroquinolones such as ciprofloxacin and ofloxacin now dominate in Asia and emerging as the dominant isolates associated with enteric fever in many countries in Africa (Keddy et al., 2010; Kariuki et al., 2015a; Wong et al., 2015).
The epidemiology of fluoroquinolone resistant clones in sub-Saharan Africa is still an emerging story (Kariuki et al., 2004; Feasey et al., 2015; Wong et al., 2015). A travel-associated case of ciprofloxacin resistant S. Typhi was reported in South Africa with PFGE pattern 100% identical to pattern JPPX01.0026 in the Global PulseNet Salmonella Typhi (Keddy et al., 2010). However, ciprofloxacin-resistant salmonellae appear uncommon in Africa, at least within the available literature. Quinolone resistance and decreased-susceptibility to fluoroquinolones (ciprofloxacin MIC 0.12–1.0 μg/mL) are more prevalent (Table S2). Such strains are of public health importance because there is correlation between increasing MICs to fluoroquinolones and treatment failure (Parry et al., 2011). The reported prevalence rates of decreased susceptibility to ciprofloxacin from some African countries are 0.3% (Senegal) (Harrois et al., 2014) 1.6% (Ghana) (Nielsen et al., 2012), 4.3% (DRC) (Lunguya et al., 2013), and ~5% (Mozambique) (Mandomando et al., 2009) with reports also from Nigeria (Akinyemi et al., 2007) and Tanzania (Weill et al., 2006), for NTS; and for S. Typhi: 5.3% (South Africa) (Smith et al., 2010) and 60.4% (Kenya) (Kariuki et al., 2010). Some of these prevalences are high enough to warrant shifting to other antimicrobials but there are few alternatives in some of the least affluent African settings. There is an indication that different clones of these strains could circulate in Africa (Smith et al., 2010) and herald the emergence and dissemination of fluoroquinolone resistance. With the exception of reports from specific countries most notably Kenya (Kariuki et al., 2010) Malawi (Feasey et al., 2015), and South Africa (Keddy et al., 2010; Smith et al., 2010), a systematic analyses of lineages associated with drug resistance is yet to be reported. In the absence of a clearer picture, continued first-line usage and misuse of quinolones is more than likely to contribute the acquisition of drug resistance in typhoidal Salmonellae.
Fluoroquinolone resistant salmonellae are more common in animals compared to humans. A study in Senegal identified Salmonella in almost all bovine carcasses screened. That study reviewed earlier literature demonstrating that Salmonella in animals and meat is commonplace in many African settings (Stevens et al., 2006). Recently, the evolution and global (including Africa) dissemination of fluoroquinolone resistant S. Kentucky ST 198 X1-SGI 1 clone, which has poultry as the main reservoir has occured (Le et al., 2013). This epidemic clone is widespread in poultry in Nigeria and has persisted (Le et al., 2013; Raufu et al., 2014). Using ciprofloxacin resistance as a marker Le et al. (2013) reported that the strain is also probably present in pigs in Nigeria, although the two isolates identified in that study were not sequenced typed (Fashae and Hendriksen, 2014).
Discussion
Assessing clonality and tracking expansion
Methods that have been used to identify fluoroquinolone-resistant bacterial clones in Africa vary across the continent, making between-study and cross-country comparisons difficult (Quilici et al., 2010; Ismail et al., 2011; Marin et al., 2013). For toxigenic V. cholerae for example, serotyping and biotyping—both insufficient to confirm clonality—are only occasionally performed outside clinical reference laboratories (Quilici et al., 2010; Ismail et al., 2011). Genotyping the ctxB product (Quilici et al., 2010), MLSA (Marin et al., 2013), and PFGE (Ismail et al., 2011; Marin et al., 2013) have been used to infer clonality in African studies. PFGE has been employed in multiple studies but different protocols were used and the data are therefore non-comparable. Moreover, PFGE identifies different clusters within local clones and would not be a robust method for assessing clonality across Africa (Ismail et al., 2011).
One reason why so many methods have been employed is that so many methods abound. However the robustness, replicability, and inferences that can be made from available methods differ considerably. Methods based on phenotype or PCR amplification patterns have been more widely used in the decades following the evolution of fluoroquinolone resistance but have limited replicability across laboratories and in some cases, limited resolving power (Achtman, 1996). Serotyping is very popular but has the additional downside of being dependent on very labile reagents, a concern that is of particular importance in African laboratories. To ascertain the relatedness of pathogenic bacteria, methods that are specific enough to identify clusters of related isolates and of high resolution to distinguish isolates within these groups are ideal. Many classical molecular typing methods are based on the sequences of a few small loci of interest or on electrophoretic profiles dependent on particular nucleotide patterns. These principles respectively form the basis of multilocus sequence typing (MLST) and the comparison of PFGE patterns. Assessing MLST types and PFGE patterns are the methods most commonly used to infer clonality of sporadic or epidemic cases of enteric diseases in sub-Saharan Africa. They offer much greater reproducibility and portability than PCR-based or phenotypic methods and while they were initially pricey, set-up, and consumable costs have fallen dramatically in the last two decades. The major disadvantages of these methods are that they analyze a very small portion of the bacterial genome and different parts of a genome accumulate variation at different rates and/or are horizontally inherited. Finally, whilst still utilizable in specific circumstances, these methods lack the level of resolution needed to distinguish between very closely related bacteria.
The arrival of the so-called next-generation, high-throughput sequencing technologies makes it feasible to use WGS to identify and track clones of interest. The major intrinsic challenges so far have been high cost, inadequate infrastructure to maintain sensitive equipment and lack of technical capacity. Overcoming these surmountable challenges will present the opportunity to use the best-fit methods for studying the epidemiology and transmission of infectious diseases in such high burden areas in sub-Saharan Africa. It is increasingly important to situate these resources for WGS and analysis in areas with the most need to maximize gains. In the interim, a transitional advantage of WGS is that it can be used to generate comparative data for sequence based methods with less resolution, such as MLST.
The power and utility of WGS tools is exemplified in a recent study detailing the analyses of a global collection of S. Typhi (Wong et al., 2015). The S. Typhi H58 haplotype MDR clone has been shown to rapidly expand such as is the case in Malawi in which prior to 2011, sporadic Typhoid was caused by a diversity of clades. In their study, Wong et al. used a combination of WGS and Bayesian analyses to deduce the global population structure, possible intra- and inter-continental transmission pathways and an estimate of time of emergence of the most recent common ancestor of the current H58 global pandemic. These analyses confirmed the highly clonal nature of the H58 isolates, which were very similar to each other genetically (in fact, only ~6 SNPs separates any two H58 strains). They were also able to detect potential on-going epidemics in endemic countries. The H58 clones were estimated to have emerged in the late 1980's and are associated with high levels of drug resistance and reduced susceptibility to fluoroquinolones, due to mutations in DNA gyrase subunit and topoisomerase IV genes (Wirth, 2015; Wong et al., 2015). This is consistent with epidemiological and clinical data from the regions included in the study. This kind of analysis allows identification of genomic changes and acquisitions that underpin many observable phenotypes like increased infectivity, transmissibility or fitness when compared to closely related lineages of the same species. For example, Wong et al provide evidence to show that a chromosomally borne MDR locus and loss of an IncHI1 plasmid in a vast number of examined isolates may have increased the fitness of the H58 clones (Wirth, 2015; Wong et al., 2015).
Exacerbating factors, intervention and research needs
Clonal expansion of fluoroquinolone resistance in enterics is broadly the consequence of antimicrobial pressure and bacterial and gene dissemination. Antimicrobial resistance is a “Tragedy of the Commons” evolutionary paradigm that is worsened by failure to appreciate the problem; that is the absence of a “recognition of necessity” (Hardin, 1968; Okeke, 2009). Thus, infrastructural, bacterial and human-related factors exacerbate the effects of antimicrobial pressure and bacterial exchange. Sub-par diagnostic infrastructure leads to incorrect estimation of true disease burden and veiling of significant resistant clones. The lack of national, regional and internationally coordinated antimicrobial stewardship leads to inconsistencies and non-uniformity in many reported cases; this makes it difficult to interpret data and to select truly representative studies.
Antimicrobial pressure
Antimicrobial pressure plays a large, important and potentially controllable role in the expansion of clonal resistance. Vien et al. showed that the prevalence of qnr resistance genes in gut Proteobacteria can increase following one course of antibiotic treatment (Vien le et al., 2012). In African countries, the contribution of pressure has not been measured, as antimicrobial use in humans and animals remains largely undocumented. Nonetheless, a high infectious disease burden warranting justifiable antimicrobial use, poor diagnostic test infrastructure and uptake and unregulated use of antibiotics combine to exacerbate the adverse effects that antimicrobial use has on resistance, and it's potential to select undesirable clones. While the community impact of antimicrobial use and overuse are often discussed, (Feasey et al., 2014) recently presented a case study illustrating that antimicrobial use can be associated with acquisition of fluoroquinolone resistance by a highly virulent clone of Salmonella Typhimurium in a treated patient. In that case study, recrudescence of a bloodstream infection isolate after acquisition of a resistance plasmid is the most likely explanation for death. The effects of antimicrobial pressure on the evolution of fluoroquinolone resistance extend beyond quinolone use. Evidence suggests that fluoroquinolone-resistant clones are selected and maintained by multiple antimicrobial classes, a feature that is easily explained by the fact that the successful clones in question are resistant to multiple antimicrobials. Interventions are required to prevent the over-prescription of antibiotics and preventing unsanctioned use in humans and livestock. Policies that address these issues have long been in existence but are inadequately implemented in most parts of Africa (WHO, 2001).
Antibiotic use strategies such as mixing and cycling have been a proposed tool for reducing antibiotic resistance. Such proposals are based on very few investigations, which give variable results as to their effectiveness (Brown and Nathwani, 2005; Abel zur et al., 2014). Studies with S. Typhi have shown that FQ resistance is not typically associated with fitness cost and suggests that FQ resistant strains would be naturally maintained even if fluoroquinolones use was reduced and so antibiotic cycling is unlikely to be beneficial (Baker et al., 2013). Even if mixing or cycling might be effective under some conditions, Africa currently has too few effective antimicrobial classes to effectively employ such techniques. In particular, withdrawing fluoroquinolones out of rotation limits treatment options for multiple life-threatening infections.
Dissemination
Enteric infections are principally attributable to poor sanitation, hygiene, food safety and access to potable water. Lack of such basic infrastructure contributes to endemicity in resource-poor settings and amplifies the prevalence of resistant strains in humans as well as animal and environmental reservoirs. Opportunities for bacteria to move from one colonized individual to another enhance the dissemination of resistant clones. In Africa, crowded cities and significant numbers of immunocompromised people are potential factors that could enhance the dissemination of clones. Interestingly, mathematical modeling to evaluate the impact of these risk factors found that increased pathogen transmission rates and longer durations of infectivity were even more important factors enabling spread, (Pitzer et al., 2015). Therefore, whilst unjustified antimicrobial use contributes to resistance, inadequate access to appropriate antibiotics when needed could also exacerbate the problem.
Travel has been associated with the dissemination of fluoroquinolone-resistant enteric clones in other parts of the world (Soraas et al., 2013). While there are reports pointing to export of resistant clones from African countries (Soraas et al., 2013), research does not appear to have determined whether and to what extent, resistant clones are imported into Africa. Research by Okoro et al. points to inter-country dissemination of multiply resistant non-typhoidal Salmonella in Africa but there are also very few studies documenting regional transmission as well (Okoro et al., 2012).
Clonal expansion of bacteria is often associated with selective advantage conferred by changes in bacterial genomes. These changes can be in the form the acquisition of chromosomal and/or plasmid-borne MDR determinants as well as mutations in genes targeted by antimicrobials, subsequently leading to the emergence and maintenance of fitter clones in animals and humans. This systematic review has shown that pandemic lineages within Africa are sometimes represented by different within-lineage clones than those elsewhere, pointing to bacterial factors that promote clonal expansion in select lineages. Host factors including presence of underlying conditions such immunosuppression (Gordon et al., 2007; MacLennan et al., 2008) and human migratory patterns attributable to social and economic pressures also contribute to the emergence and maintenance of MDR clones in Africa.
Surveillance
Our literature review identified very little research on fluoroquinolone-resistant enteric clones within Africa. There are however a large number of reports of fluoroquinolone resistant clones isolated in Europe and North America (Pitout et al., 2009; Soraas et al., 2013)that associate clone acquisition with travel to African countries (for example Pitout et al., 2009; Soraas et al., 2013). These “eyes of the hippopotamus” (Guerrant et al., 2005) reports point to under-detection and under reporting in Africa and the need for more local studies to estimate the true magnitude and extent of the problem. Our research also did not uncover significant information about resistance reservoirs, particularly in non-human hosts and the environment. What few animal data we did encounter however points to a large non-human reservoir of enteric organisms, many of them resistant, and the occurrence of these organism through the food chain. Studies reviewed for this paper were almost entirely small research studies on human isolates from a handful of laboratories. The location of these laboratories is the principal determinant of locations on the continent from which data are available. In order to understand resistance and track the emergence of clones, multi-center surveillance is required.
The European Antimicrobial Resistance Surveillance Network (EARS-Net) enables comparison between European countries (http://ecdc.europa.eu/en/activities/surveillance/EARS-Net/Pages/index.aspx). Recent analysis of the data has shown that an increase of fluoroquinolone consumption in Europe also increases the rate of resistance (Redgrave et al., 2014). To understand the scale of resistance in Africa, a similar model based on a regulated and coordinated surveillance system across Africa would be ideal. In contrast to the EU, where an obvious framework for coordination exists, it is less clear how to implement such a scheme across Africa. Potential frameworks include public as well as private sub-regional alliances and there are multicountry data from the Francophone Pasteur Institute Network (Breurec et al., 2013). Continental coverage cannot be obtained from either. Looking forward, a regional organization, perhaps the nascent African Society for Laboratory Medicine, or perhaps a purpose-initiated one, might be lead such an initiative. Alternatively, different laboratories and networks could feed data to a repository. As well as putting in proposed minimum reporting guidelines for research on antimicrobial resistance (Omulo et al., 2015), the surveillance system would also be expanded to include detection of clones and mechanisms of resistance that are causing resistance so that the spread can be tracked in real-time. A defined methodology for assessing clones to enable tracking would be needed so that different strains across Africa could be accurately compared and experience from Europe suggests that this is best achieved through sequence-based methods.
A network is only as good as the data it can collate from participating countries. Implementation of such a scheme in some countries would be difficult, given the state of existing infrastructure but existing institutions in many other countries that may be well-positioned to provide a local picture of resistance problems if the necessary coordination were to occur. As has been recently reported from Ghana, it is possible to set up a national surveillance system coordinated from the ministry of health (Opintan et al., 2015). The laudable Ghanaian initiative was not without its challenges, many of them linked to resource limitations, and the problem that most of the usable data came from only a handful of participating institutions, mostly referral hospitals. However inception is the first step in building a robust surveillance system and most African countries are yet to reach that point. In low-income African settings, it is predicted that susceptibility data will save lives and costs, compared to widespread used empirical therapy without local susceptibility information (Penno et al., 2015).
In addition to microbiological infrastructure and activity, support is needed for molecular studies that will identify the underlying genetic bases for resistance, including the existence and transmission of resistant clones. Comparison of isolates from within sub-Sahara Africa to those from outside the sub-region is important as recent studies have shown that clinical, cultural practices and socio-economic factors impact on the nature of circulating MDR clones in different parts of the world. Better antibiotic stewardship at local, country, and regional levels is needed in order to monitor and standardize protocols used for testing and clinical prescription. Much of the inquiry around antimicrobial resistance in African countries occurs within academic and non-governmental sectors, a likely contributor to fragmentation and limited coverage. Government participation and lead in public health initiatives such as strengthening of existing infrastructure and development of medical and research capacity remain a critical need.
Author contributions
MC: Coordinated and collated systematic searches and references, evaluated eligible papers, performed systematic search, wrote sections of the manuscript. AA: performed systematic search, wrote sections of the manuscript, evaluated eligible papers, contributed to outlining the paper. CO, KF: performed systematic search, evaluated eligible papers, wrote sections of the manuscript. JO: evaluated eligible papers, edited sections of the manuscript and contributed to outlining the paper. IO: Initiated the project, performed systematic search, wrote sections of the manuscript, evaluated eligible papers, coordinated the writing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CO is a Branco Weiss Fellow of the Society in Science. JO is supported by Antibiotic Drug use, Monitoring, and Evaluation of Resistance in Ghana (ADMER), a DANIDA funded project in Ghana. IO is co-recipient of African Research Leader's Award MR/L00464X/1 jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and also part of the EDCTP2 programme supported by the European Union. We thank Dr. Satheesh Nair of the Gastrointestinal Bacteria Reference Unit, Public Health England for sharing his expertise on multidrug resistant clones.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00558
References
- Abel zur W. P., Kouyos R., Abel S., Viechtbauer W., Bonhoeffer S. (2014). Cycling empirical antibiotic therapy in hospitals: meta-analysis and models. PLoS Pathog. 10:e1004225. 10.1371/journal.ppat.1004225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M. (1996). A surfeit of YATMs? J. Clin. Microbiol. 34:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A. A., Osman H., Mansour A. M., Musa H. A., Ahmed A. B., Karrar Z., et al. (2000). Antimicrobial agent resistance in bacterial isolates from patients with diarrhea and urinary tract infection in the Sudan. Am. J. Trop. Med. Hyg. 63, 259–263. [PubMed] [Google Scholar]
- Aibinu I., Aednipedun E., Odugbemi T. (2004). Emergence of quinolone resistance amongst Escherichia coli strains isolated from clinical infections in some Lagos state hospitals in Nigeria. Niger. J. Health Biomed. Sci. 3, 73–78. 10.4314/njhbs.v3i2.11513 [DOI] [Google Scholar]
- Aibinu I., Odugbemi T., Koenig W., Ghebremedhin B. (2012a). Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect 18, E49–E51. 10.1111/j.1469-0691.2011.03730.x [DOI] [PubMed] [Google Scholar]
- Aibinu I., Pfeifer Y., Peters F., Ogunsola F., Adenipekun E., Odugbemi T., et al. (2012b). Emergence of bla(CTX-M-15), qnrB1 and aac(6')-Ib-cr resistance genes in Pantoea agglomerans and Enterobacter cloacae from Nigeria (sub-Saharan Africa). J. Med. Microbiol. 61, 165–167. 10.1099/jmm.0.035238-0 [DOI] [PubMed] [Google Scholar]
- Akinyemi K. O., Bamiro B. S., Coker A. O. (2007). Salmonellosis in Lagos, Nigeria: incidence of Plasmodium falciparum-associated co-infection, patterns of antimicrobial resistance, and emergence of reduced susceptibility to fluoroquinolones. J. Health Popul. Nutr. 25, 351–358. Available online at: http://www.jstor.org/stable/23499754 [PMC free article] [PubMed] [Google Scholar]
- Akinyemi K. O., Iwalokun B. A., Foli F., Oshodi K., Coker A. O. (2011). Prevalence of multiple drug resistance and screening of enterotoxin (stn) gene in Salmonella enterica serovars from water sources in Lagos, Nigeria. Public Health 125, 65–71. 10.1016/j.puhe.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Albrechtova K., Dolejska M., Cizek A., Tausova D., Klimes J., Bebora L., et al. (2012). Dogs of nomadic pastoralists in northern Kenya are reservoirs of plasmid-mediated cephalosporin- and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob. Agents Chemother. 56, 4013–4017. 10.1128/AAC.05859-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtova K., Kubelova M., Mazancova J., Dolejska M., Literak I., Cizek A. (2014a). High prevalence and variability of CTX-M-15-producing and fluoroquinolone-resistant Escherichia coli observed in stray dogs in rural Angola. Microb. Drug Resist. 20, 372–375. 10.1089/mdr.2013.0177 [DOI] [PubMed] [Google Scholar]
- Albrechtova K., Papousek I., De N. H., Pauly M., Anoh E., Mossoun A., et al. (2014b). Low rates of antimicrobial-resistant Enterobacteriaceae in wildlife in Tai National Park, Cote d'Ivoire, surrounded by villages with high prevalence of multiresistant ESBL-producing Escherichia coli in people and domestic animals. PLoS ONE 9:e113548. 10.1371/journal.pone.0113548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjyal A., Basnyat B., Koirala S., Karkey A., Dongol S., Agrawaal K. K., et al. (2011). Gatifloxacin versus chloramphenicol for uncomplicated enteric fever: an open-label, randomised, controlled trial. Lancet Infect. Dis. 11, 445–454. 10.1016/S1473-3099(11)70089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Duy P. T., Nga T. V., Dung T. T., Phat V. V., Chau T. T., et al. (2013). Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife 2:e01229. 10.7554/eLife.01229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkley J. A., Lowe B. S., Mwangi I., Williams T., Bauni E., Mwarumba S., et al. (2005). Bacteremia among children admitted to a rural hospital in Kenya. N. Engl. J. Med. 352, 39–47. 10.1056/NEJMoa040275 [DOI] [PubMed] [Google Scholar]
- Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- Bourgeois A. L., Gardiner C. H., Thornton S. A., Batchelor R. A., Burr D. H., Escamilla J., et al. (1993). Etiology of acute diarrhea among United States military personnel deployed to South America and west Africa. Am. J. Trop. Med. Hyg. 48, 243–248. [DOI] [PubMed] [Google Scholar]
- Brenner F. W., Villar R. G., Angulo F. J., Tauxe R., Swaminathan B. (2000). Salmonella nomenclature. J. Clin. Microbiol. 38, 2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breurec S., Guessennd N., Timinouni M., Le T. A., Cao V., Ngandjio A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. 10.1111/j.1469-0691.2012.03805.x [DOI] [PubMed] [Google Scholar]
- Brown E. M., Nathwani D. (2005). Antibiotic cycling or rotation: a systematic review of the evidence of efficacy. J. Antimicrob. Chemother. 55, 6–9. 10.1093/jac/dkh482 [DOI] [PubMed] [Google Scholar]
- Brown E. M., Reeves D. S. (1997). Quinolones, in Antibiotic and Chemotherapy, 7th Edn., eds O'Grady F., Lamber H. P., Finch R. G., Greenwood D. (New York, NY: Churchill Livingstone; ), 419–452. [Google Scholar]
- Cardinale E., Rose V., Perrier Gros-Claude J. D., Tall F., Rivoal K., Mead G., et al. (2006). Genetic characterization and antibiotic resistance of Campylobacter spp. isolated from poultry and humans in Senegal. J. Appl. Microbiol. 100, 209–217. 10.1111/j.1365-2672.2005.02763.x [DOI] [PubMed] [Google Scholar]
- Cattoir V., Poirel L., Rotimi V., Soussy C. J., Nordmann P. (2007). Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60, 394–397. 10.1093/jac/dkm204 [DOI] [PubMed] [Google Scholar]
- Chattaway M. A., Jenkins C., Rajendram D., Cravioto A., Talukder K. A., Dallman T., et al. (2014). Enteroaggregative Escherichia coli have evolved independently as distinct complexes within the E. coli population with varying ability to cause disease. PLoS ONE 9:e112967. 10.1371/journal.pone.0112967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Grassl G. A., Finlay B. B. (2007). Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85, 112–118. 10.1038/sj.icb.7100007 [DOI] [PubMed] [Google Scholar]
- Coker A. O., Isokpehi R. D., Thomas B. N., Amisu K. O., Obi C. L. (2002). Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8, 237–244. 10.3201/eid0803.010233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Ewing W. H., Falkow S. (1973). Molecular relationships among the Salmonelleae. J. Bacteriol. 115, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Luby S. P., Mintz E. D. (2004). The global burden of typhoid fever. Bull. World Health Organ. 82, 346–353. 10.1590/S0042-96862004000500008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya S., Kapil A., Kumar R., Das B. K., Sood S., Chaudhry R., et al. (2013). Multiple locus sequence typing of Salmonella Typhi, isolated in north India - a preliminary study. Indian J. Med Res. 137, 957–962. [PMC free article] [PubMed] [Google Scholar]
- Davidson R. J., Davis I., Willey B. M., Rizg K., Bolotin S., Porter V., et al. (2008). Antimalarial therapy selection for quinolone resistance among Escherichia coli in the absence of quinolone exposure, in tropical South America. PLoS ONE 3:e2727. 10.1371/journal.pone.0002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decousser J. W., Pfister P., Xueref X., Rakoto-Alson O., Roux J. F. (1999). [Acquired antibiotic resistance in Madagascar: first evaluation]. Med Trop. (Mars) 59, 259–265. [PubMed] [Google Scholar]
- Dimitrov T., Dashti A. A., Albaksami O., Jadaon M. M. (2010). Detection of mutations in the gyrA gene in fluoroquinolone resistance Salmonella enterica serotypes typhi and paratyphi A isolated from the Infectious Diseases Hospital, Kuwait. J. Clin. Pathol. 63, 83–87. 10.1136/jcp.2009.070664 [DOI] [PubMed] [Google Scholar]
- Dougan G., John V., Palmer S., Mastroeni P. (2011). Immunity to salmonellosis. Immunol. Rev. 240, 196–210. 10.1111/j.1600-065X.2010.00999.x [DOI] [PubMed] [Google Scholar]
- Doumith M., Day M., Ciesielczuk H., Hope R., Underwood A., Reynolds R., et al. (2015). Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 53, 160–166. 10.1128/JCM.02562-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fashae K., Hendriksen R. S. (2014). Diversity and antimicrobial susceptibility of Salmonella enterica serovars isolated from pig farms in Ibadan, Nigeria. Folia Microbiol. (Praha) 59, 69–77. 10.1007/s12223-013-0270-6 [DOI] [PubMed] [Google Scholar]
- Feasey N. A., Archer B. N., Heyderman R. S., Sooka A., Dennis B., Gordon M. A., et al. (2010). Typhoid fever and invasive nontyphoid salmonellosis, Malawi and South Africa. Emerg. Infect. Dis. 16, 1448–1451. 10.3201/eid1609.100125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey N. A., Cain A. K., Msefula C. L., Pickard D., Alaerts M., Aslett M., et al. (2014). Drug resistance in Salmonella enterica ser. Typhimurium bloodstream infection, Malawi. Emerg. Infect. Dis. 20, 1957–1959. 10.3201/eid2011.141175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey N. A., Gaskell K., Wong V., Msefula C., Selemani G., Kumwenda S., et al. (2015). Rapid emergence of multidrug resistant, h58-lineage Salmonella Typhi in Blantyre, Malawi. PLoS. Negl. Trop. Dis. 9:e0003748. 10.1371/journal.pntd.0003748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feglo P., Adu-Sarkodie Y., Ayisi L., Jain R., Spurbeck R. R., Springman A. C., et al. (2013). Emergence of a novel extended-spectrum-beta-lactamase (ESBL)-producing, fluoroquinolone-resistant clone of extraintestinal pathogenic Escherichia coli in Kumasi, Ghana. J. Clin. Microbiol. 51, 728–730. 10.1128/JCM.03006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmingham D., Robbins M. J. (1992). In vitro activity of lomefloxacin and other antimicrobials against bacterial enteritis pathogens. Diagn. Microbiol. Infect. Dis. 15, 339–343. 10.1016/0732-8893(92)90020-T [DOI] [PubMed] [Google Scholar]
- Fortini D., Fashae K., Garcia-Fernandez A., Villa L., Carattoli A. (2011). Plasmid-mediated quinolone resistance and beta-lactamases in Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 66, 1269–1272. 10.1093/jac/dkr085 [DOI] [PubMed] [Google Scholar]
- Fortini D., Fashae K., Villa L., Feudi C., García-Fernández A., Carattoli A. (2015). A novel plasmid carrying blaCTX-M-15 identified in commensal Escherichia coli from healthy pregnant women in Ibadan, Nigeria. J. Global Antimicrob. Resist. 3, 9–12. 10.1016/j.jgar.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Frost J. A., Willshaw G. A., Barclay E. A., Rowe B., Lemmens P., Vandepitte J. (1985). Plasmid characterization of drug-resistant Shigella dysenteriae 1 from an epidemic in Central Africa. J. Hyg. (Lond.) 94, 163–172. 10.1017/S0022172400061362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. A., Gordon S. B., Musaya L., Zijlstra E. E., Molyneux M. E., Read R. C. (2007). Primary macrophages from HIV-infected adults show dysregulated cytokine responses to Salmonella, but normal internalization and killing. AIDS 21, 2399–2408. 10.1097/QAD.0b013e3282f25107 [DOI] [PubMed] [Google Scholar]
- Gordon M. A., Graham S. M., Walsh A. L., Wilson L., Phiri A., Molyneux E., et al. (2008). Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica Serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 46, 963–969. 10.1086/529146 [DOI] [PubMed] [Google Scholar]
- Gqunta K., Govender S. (2015). Characterization of ESBL-producing Escherichia coli ST131 isolates from Port Elizabeth. Diagn. Microbiol. Infect. Dis. 81, 44–46. 10.1016/j.diagmicrobio.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Green S., Tillotson G. (1997). Use of ciprofloxacin in developing countries. Pediatr. Infect. Dis. J. 16, 150–159. 10.1097/00006454-199701000-00041 [DOI] [PubMed] [Google Scholar]
- Grimont P. A. D., Weill F. X. (2007). Antigenic Formulae of the Salmonella Serovars. Paris: Institut Pasteur. [Google Scholar]
- Goma Epidemiology Group (1995). Public health impact of Rwandan refugee crisis: what happened in Goma, Zaire, in July, 1994? Goma Epidemiology Group. Lancet 345, 339–344. 10.1016/S0140-6736(95)90338-0 [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., Oria R., Bushen O. Y., Patrick P. D., Houpt E., Lima A. A. (2005). Global impact of diarrheal diseases that are sampled by travelers: the rest of the hippopotamus. Clin. Infect. Dis 41(Suppl. 8), S524–S530. 10.1086/432946 [DOI] [PubMed] [Google Scholar]
- Hardin G. (1968). The tragedy of the commons. The population problem has no technical solution; it requires a fundamental extension in morality. Science 162, 1243–1248. [PubMed] [Google Scholar]
- Harrois D., Breurec S., Seck A., Delaune A., Le Hello S., Pardos de la Gándara M., et al. (2014). Prevalence and characterization of extended-spectrum beta-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 20, O109–O116. 10.1111/1469-0691.12339 [DOI] [PubMed] [Google Scholar]
- Hata M., Suzuki M., Matsumoto M., Takahashi M., Sato K., Ibe S., et al. (2005). Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49, 801–803. 10.1128/AAC.49.2.801-803.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Dolecek C., Chau T. T., Duy P. T., La T. T., Hoang N. V., et al. (2011). Temporal fluctuation of multidrug resistant salmonella typhi haplotypes in the mekong river delta region of Vietnam. PLoS Negl. Trop. Dis. 5:e929. 10.1371/journal.pntd.0000929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Parkhill J., Mazzoni C. J., Roumagnac P., Weill F. X., Goodhead I., et al. (2008). High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40, 987–993. 10.1038/ng.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D. (2003). Mechanisms of quinolone resistance, in Quinolone Antimicrobial Agents, 3rd Edn., eds Hooper D., Rubinstein E. (Washington DC: ASM Press; ), 41–67. [Google Scholar]
- Inwezerua C., Mendonca N., Calhau V., Domingues S., Adeleke O. E., Da Silva G. J. (2014). Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo state, Nigeria. J. Infect. Dev. Ctries 8, 774–779. 10.3855/jidc.3430 [DOI] [PubMed] [Google Scholar]
- Isendahl J., Turlej-Rogacka A., Manjuba C., Rodrigues A., Giske C. G., Naucler P. (2012). Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS ONE 7:e51981. 10.1371/journal.pone.0051981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H., Smith A. M., Sooka A., Keddy K. H. (2011). Genetic characterization of multidrug-resistant, extended-spectrum- beta-lactamase-producing Vibrio cholerae O1 outbreak strains, Mpumalanga, South Africa, 2008. J. Clin. Microbiol. 49, 2976–2979. 10.1128/JCM.00293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janatova M., Albrechtova K., Petrzelkova K. J., Dolejska M., Papousek I., Masarikova M., et al. (2014). Antimicrobial-resistant Enterobacteriaceae from humans and wildlife in Dzanga-Sangha Protected Area, Central African Republic. Vet. Microbiol. 171, 422–431. 10.1016/j.vetmic.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Jiang Z. D., Lowe B., Verenkar M. P., Ashley D., Steffen R., Tornieporth N., et al. (2002). Prevalence of enteric pathogens among international travelers with diarrhea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay). J. Infect. Dis. 185, 497–502. 10.1086/338834 [DOI] [PubMed] [Google Scholar]
- Karas J. A., Pillay D. G. (1996). Nalidixic acid-resistant Shigella dysenteriae type I in KwaZulu-Natal. S. Afr. Med. J. 86, 93. [PubMed] [Google Scholar]
- Kariuki S., Dougan G. (2014). Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann. N.Y. Acad. Sci. 1323, 43–55. 10.1111/nyas.12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Gordon M. A., Feasey N., Parry C. M. (2015a). Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 33(Suppl. 3), C21–C29. 10.1016/j.vaccine.2015.03.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Okoro C., Kiiru J., Njoroge S., Omuse G., Langridge G., et al. (2015b). Ceftriaxone-resistant Salmonella enterica serotype typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob. Agents Chemother. 59, 3133–3139. 10.1128/AAC.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Revathi G., Corkill J., Kiiru J., Mwituria J., Mirza N., et al. (2007). Escherichia coli from community-acquired urinary tract infections resistant to fluoroquinolones and extended-spectrum beta-lactams. J. Infect. Dev. Ctries 1, 257–262. [PubMed] [Google Scholar]
- Kariuki S., Revathi G., Kiiru J., Mengo D. M., Mwituria J., Muyodi J., et al. (2010). Typhoid in Kenya is associated with a dominant multidrug-resistant Salmonella enterica serovar Typhi haplotype that is also widespread in Southeast Asia. J. Clin. Microbiol. 48, 2171–2176. 10.1128/JCM.01983-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Revathi G., Muyodi J., Mwituria J., Munyalo A., Mirza S., et al. (2004). Characterization of multidrug-resistant typhoid outbreaks in Kenya. J. Clin. Microbiol. 42, 1477–1482. 10.1128/JCM.42.4.1477-1482.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke B., Sun J., He D., Li X., Liang Z., Ke C. W. (2014). Serovar distribution, antimicrobial resistance profiles, and PFGE typing of Salmonella enterica strains isolated from 2007-2012 in Guangdong, China. BMC. Infect. Dis. 14:338. 10.1186/1471-2334-14-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddy K. H., Smith A. M., Sooka A., Ismail H., Oliver S. (2010). Fluoroquinolone-resistant typhoid, South Africa. Emerg. Infect. Dis. 16, 879–880. 10.3201/eid1605.091917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinana A. D., Cardinale E., Bahsoun I., Tall F., Sire J. M., Breurec S., et al. (2007). Campylobacter coli isolates derived from chickens in Senegal: diversity, genetic exchange with Campylobacter jejuni and quinolone resistance. Res. Microbiol. 158, 138–142. 10.1016/j.resmic.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Kinana A. D., Cardinale E., Tall F., Bahsoun I., Sire J. M., Garin B., et al. (2006). Genetic diversity and quinolone resistance in Campylobacter jejuni isolates from poultry in Senegal. Appl. Environ. Microbiol. 72, 3309–3313. 10.1128/AEM.72.5.3309-3313.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley R. A., Msefula C. L., Thomson N. R., Kariuki S., Holt K. E., Gordon M. A., et al. (2009). Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19, 2279–2287. 10.1101/gr.091017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- Lacher D. W., Steinsland H., Blank T. E., Donnenberg M. S., Whittam T. S. (2007). Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189, 342–350. 10.1128/JB.01472-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamikanra A., Crowe J. L., Lijek R. S., Odetoyin B. W., Wain J., Aboderin A. O., et al. (2011). Rapid evolution of fluoroquinolone-resistant Escherichia coli in Nigeria is temporally associated with fluoroquinolone use. BMC. Infect. Dis. 11:312. 10.1186/1471-2334-11-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C., Wain J., Nair S. (2012). Invasive Salmonellosis in humans. EcoSal. Plus 5 10.1128/ecosalplus.8.6.2.2 Available online at: http://www.asmscience.org/content/journal/ecosalplus/10.1128/ecosalplus.8.6.2.2 [DOI] [PubMed]
- Lavollay M., Mamlouk K., Frank T., Akpabie A., Burghoffer B., Ben R. S., et al. (2006). Clonal dissemination of a CTX-M-15 beta-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 50, 2433–2438. 10.1128/AAC.00150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H. S., Bekhit A., Granier S. A., Barua H., Beutlich J., Zajac M., et al. (2013). The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front. Microbiol. 4:395. 10.3389/fmicb.2013.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H. S., Hendriksen R. S., Doublet B., Fisher I., Nielsen E. M., Whichard J. M., et al. (2011). International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 204, 675–684. 10.1093/infdis/jir409 [DOI] [PubMed] [Google Scholar]
- Leopold S. J., van Leth F., Tarekegn H., Schultsz C. (2014). Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. J. Antimicrob. Chemother. 69, 2337–2353. 10.1093/jac/dku176 [DOI] [PubMed] [Google Scholar]
- Liu L., Oza S., Hogan D., Perin J., Rudan I., Lawn J. E., et al. (2015). Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385, 430–440. 10.1016/S0140-6736(14)61698-6 [DOI] [PubMed] [Google Scholar]
- Lunguya O., Lejon V., Phoba M. F., Bertrand S., Vanhoof R., Glupczynski Y., et al. (2013). Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: emergence of decreased fluoroquinolone susceptibility and extended-spectrum beta lactamases. PLoS. Negl. Trop. Dis. 7:e2103. 10.1371/journal.pntd.0002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupindu A. M., Olsen J. E., Ngowi H. A., Msoffe P. L., Mtambo M. M., Scheutz F., et al. (2014). Occurrence and characterization of Shiga toxin-producing Escherichia coli O157:H7 and other non-sorbitol-fermenting E. coli in cattle and humans in urban areas of Morogoro, Tanzania. Vector Borne Zoonotic Dis. 14, 503–510. 10.1089/vbz.2013.1502 [DOI] [PubMed] [Google Scholar]
- MacLennan C. A., Gondwe E. N., Msefula C. L., Kingsley R. A., Thomson N. R., White S. A., et al. (2008). The neglected role of antibody in protection against bacteremia caused by nontyphoidal strains of Salmonella in African children. J. Clin. Invest. 118, 1553–1562. 10.1172/JCI33998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud Z. H., Islam S., Zaman R. U., Akter M., Talukder K. A., Bardhan P. K., et al. (2014). Phenotypic and genotypic characteristics of Vibrio cholerae O1 isolated from the Sierra Leone cholera outbreak in 2012. Trans. R. Soc. Trop. Med. Hyg. 108, 715–720. 10.1093/trstmh/tru137 [DOI] [PubMed] [Google Scholar]
- Majowicz S. E., Musto J., Scallan E., Angulo F. J., Kirk M., O'Brien S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- Mandomando I., Macete E., Sigauque B., Morais L., Quinto L., Sacarlal J., et al. (2009). Invasive non-typhoidal Salmonella in Mozambican children. Trop. Med. Int. Health 14, 1467–1474. 10.1111/j.1365-3156.2009.02399.x [DOI] [PubMed] [Google Scholar]
- Marin M. A., Thompson C. C., Freitas F. S., Fonseca E. L., Aboderin A. O., Zailani S. B., et al. (2013). Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS. Negl. Trop. Dis. 7:e2049. 10.1371/journal.pntd.0002049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K. E., Clifton S. W., Latreille P., Porwollik S., Sabo A., et al. (2004). Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36, 1268–1274. 10.1038/ng1470 [DOI] [PubMed] [Google Scholar]
- Mercy N., Mohamed A. A., Zipporah N., Chowdhury G., Pazhani G. P., Ramamurthy T., et al. (2014). Phenotypic and genetic characterization of Vibrio cholerae O1 isolated from various regions of Kenya between 2007 and 2010. Pan Afr. Med. J. 19, 8. 10.11604/pamj.2014.19.8.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz E. D., Guerrant R. L. (2009). A lion in our village–the unconscionable tragedy of cholera in Africa. N. Engl. J. Med. 360, 1060–1063. 10.1056/NEJMp0810559 [DOI] [PubMed] [Google Scholar]
- Mitscher L. A. (2005). Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem. Rev. 105, 559–592. 10.1021/cr030101q [DOI] [PubMed] [Google Scholar]
- Miwanda B., Moore S., Muyembe J. J., Nguefack-Tsague G., Kabangwa I. K., Ndjakani D. Y., et al. (2015). Antimicrobial drug resistance of Vibrio cholerae, Democratic Republic of the Congo. Emerg. Infect. Dis. 21, 847–851. 10.3201/eid2105.141233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS. Med. 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla B., Miko A., Pries K., Hildebrandt G., Kleer J., Schroeter A., et al. (2007). Class 1 integrons and resistance gene cassettes among multidrug resistant Salmonella serovars isolated from slaughter animals and foods of animal origin in Ethiopia. Acta Trop. 103, 142–149. 10.1016/j.actatropica.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Namboodiri S. S., Opintan J. A., Lijek R. S., Newman M. J., Okeke I. N. (2011). Quinolone resistance in Escherichia coli from Accra, Ghana. BMC Microbiol. 11:44. 10.1186/1471-2180-11-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M. V., Sarpong N., Krumkamp R., Dekker D., Loag W., Amemasor S., et al. (2012). Incidence and characteristics of bacteremia among children in rural Ghana. PLoS ONE 7:e44063. 10.1371/journal.pone.0044063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly C. E., Jaron P., Ochieng B., Nyaguara A., Tate J. E., Parsons M. B., et al. (2012). Risk factors for death among children less than 5 years old hospitalized with diarrhea in rural western Kenya, 2005-2007: a cohort study. PLoS Med. 9:e1001256. 10.1371/journal.pmed.1001256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke I. N. (2009). The tragedy of antimicrobial resistance: achieving a recognition of necessity. Curr. Sci. 11, 1564–1572. [Google Scholar]
- Okeke I. N., Fayinka S. T., Lamikanra A. (2000a). Antibiotic resistance in Escherichia coli from Nigerian students, 1986 1998. Emerg. Infect. Dis. 6, 393–396. 10.3201/eid0604.009913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke I. N., Lamikanra A., Czeczulin J., Dubovsky F., Kaper J. B., Nataro J. P. (2000b). Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J. Infect. Dis. 181, 252–260. 10.1086/315204 [DOI] [PubMed] [Google Scholar]
- Okoro C. K., Kingsley R. A., Connor T. R., Harris S. R., Parry C. M., Al-Mashhadani M. N., et al. (2012). Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 44, 1215–1221. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omigie O., Okoror L., Umolu P., Ikuuh G. (2009). Increasing resistance to quinolones: a four-year prospective study of urinary tract infection pathogens. Int. J. Gen. Med. 2, 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omulo S., Thumbi S. M., Njenga M. K., Call D. R. (2015). A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrob. Resist. Infect. Control 4, 1. 10.1186/s13756-014-0041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opintan J. A., Newman M. J., Arhin R. E., Donkor E. S., Gyansa-Lutterodt M., Mills-Pappoe W. (2015). Laboratory-based nationwide surveillance of antimicrobial resistance in Ghana. Infect. Drug Resist. 8, 379–389. 10.2147/IDR.S88725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orogade A. A., Akuse R. M. (2004). Changing patterns in sensitivity of causative organisms of septicaemia in children: the need for quinolones. Afr. J. Med. Sci. 33, 69–72. [PubMed] [Google Scholar]
- Oyofo B. A., Peruski L. F., Ismail T. F., el-Etr S. H., Churilla A. M., Wasfy M. O., et al. (1997). Enteropathogens associated with diarrhea among military personnel during Operation Bright Star 96, in Alexandria, Egypt. Mil. Med. 162, 396–400. [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K. D., Thomson N. R., Pickard D., Wain J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. (2002). Typhoid fever. N. Engl. J. Med. 347, 1770–1782. 10.1056/NEJMra020201 [DOI] [PubMed] [Google Scholar]
- Parry C. M., Vinh H., Chinh N. T., Wain J., Campbell J. I., Hien T. T., et al. (2011). The influence of reduced susceptibility to fluoroquinolones in Salmonella enterica serovar Typhi on the clinical response to ofloxacin therapy. PLoS. Negl. Trop. Dis. 5:e1163. 10.1371/journal.pntd.0001163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton J. H., Reeves D. S. (1988). Fluoroquinolone antibiotics. Microbiology, pharmacokinetics and clinical use. Drugs 36, 193–228. 10.2165/00003495-198836020-00004 [DOI] [PubMed] [Google Scholar]
- Peirano G., van der Bij A. K., Freeman J. L., Poirel L., Nordmann P., Costello M., et al. (2014). Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob. Agents Chemother. 58, 3762–3767. 10.1128/AAC.02428-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano G., van Greune C. H., Pitout J. D. (2011). Characteristics of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn. Microbiol. Infect. Dis. 69, 449–453. 10.1016/j.diagmicrobio.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Penno E. C., Baird S. J., Crump J. A. (2015). Cost-effectiveness of surveillance for bloodstream infections for sepsis management in low-resource settings. Am. J. Trop. Med. Hyg. 93, 850–860. 10.4269/ajtmh.15-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty N. K., Ben Zakour N. L., Stanton-Cook M., Skippington E., Totsika M., Forde B. M., et al. (2014). Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U.S.A. 111, 5694–5699. 10.1073/pnas.1322678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitout J. D., Gregson D. B., Campbell L., Laupland K. B. (2009). Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53, 2846–2851. 10.1128/AAC.00247-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzer V. E., Feasey N. A., Msefula C., Mallewa J., Kennedy N., Dube Q., et al. (2015). Mathematical modeling to assess the drivers of the recent emergence of typhoid fever in Blantyre, Malawi. Clin. Infect. Dis. 61(Suppl. 4), S251–S258. 10.1093/cid/civ710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. B., Johnson J. R., Aziz M., Clabots C., Johnston B., Tchesnokova V., et al. (2013). The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 4:e00377-13. 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilici M. L., Massenet D., Gake B., Bwalki B., Olson D. M. (2010). Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg. Infect. Dis. 16, 1804–1805. 10.3201/eid1611.100568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafai C., Frank T., Manirakiza A., Gaudeuille A., Mbecko J. R., Nghario L., et al. (2015). Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC. Microbiol. 15:15. 10.1186/s12866-015-0348-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raufu I., Bortolaia V., Svendsen C. A., Ameh J. A., Ambali A. G., Aarestrup F. M., et al. (2013). The first attempt of an active integrated laboratory-based Salmonella surveillance programme in the north-eastern region of Nigeria. J. Appl. Microbiol. 115, 1059–1067. 10.1111/jam.12304 [DOI] [PubMed] [Google Scholar]
- Raufu I., Hendriksen R. S., Ameh J. A., Aarestrup F. M. (2009). Occurrence and characterization of Salmonella Hiduddify from chickens and poultry meat in Nigeria. Foodborne. Pathog. Dis. 6, 425–430. 10.1089/fpd.2008.0150 [DOI] [PubMed] [Google Scholar]
- Raufu I. A., Fashae K., Ameh J. A., Ambali A., Ogunsola F. T., Coker A. O., et al. (2014). Persistence of fluoroquinolone-resistant Salmonella enterica serovar Kentucky from poultry and poultry sources in Nigeria. J. Infect. Dev. Ctries 8, 384–388. 10.3855/jidc.3495 [DOI] [PubMed] [Google Scholar]
- Reddy E. A., Shaw A. V., Crump J. A. (2010). Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect. Dis. 10, 417–432. 10.1016/S1473-3099(10)70072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrave L. S., Sutton S. B., Webber M. A., Piddock L. J. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445. 10.1016/j.tim.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., III. (1989). Clonal nature of Salmonella Typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A., Strahilevitz J., Sahm D. F., Jacoby G. A., Hooper D. C. (2006). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50, 2872–2874. 10.1128/AAC.01647-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumagnac P., Weill F. X., Dolecek C., Baker S., Brisse S., Chinh N. T., et al. (2006). Evolutionary history of Salmonella Typhi. Science 314, 1301–1304. 10.1126/science.1134933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrire L., Crisp S., Bear N., McStay G., Koornhof H. J., Le M. L. (1987). The prevalence of human isolates of Salmonella subspecies II in southern Africa. Epidemiol. Infect. 98, 25–31. 10.1017/S0950268800061689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrascia M., Maimone F., Mohamud K. A., Materu S. F., Grimont F., Grimont P. A., et al. (2006). Clonal relationship among Vibrio cholerae O1 El Tor strains causing the largest cholera epidemic in Kenya in the late 1990s. J. Clin. Microbiol. 44, 3401–3404. 10.1128/JCM.00611-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrascia M., Pugliese N., Maimone F., Mohamud K. A., Ali I. A., Grimont P. A., et al. (2009). Cholera in Ethiopia in the 1990 s: epidemiologic patterns, clonal analysis, and antimicrobial resistance. Int. J. Med Microbiol. 299, 367–372. 10.1016/j.ijmm.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Smith A. M., Govender N., Keddy K. H. (2010). Quinolone-resistant Salmonella Typhi in South Africa, 2003-2007. Epidemiol. Infect. 138, 86–90. 10.1017/S0950268809990331 [DOI] [PubMed] [Google Scholar]
- Soraas A., Sundsfjord A., Sandven I., Brunborg C., Jenum P. A. (2013). Risk factors for community-acquired urinary tract infections caused by ESBL-producing enterobacteriaceae–a case-control study in a low prevalence country. PLoS ONE 8:e69581. 10.1371/journal.pone.0069581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Kabore Y., Perrier-Gros-Claude J. D., Millemann Y., Brisabois A., Catteau M., et al. (2006). Prevalence and antibiotic-resistance of Salmonella isolated from beef sampled from the slaughterhouse and from retailers in Dakar (Senegal). Int. J. Food Microbiol. 110, 178–186. 10.1016/j.ijfoodmicro.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. 10.1128/CMR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton S. A., Wignall S. F., Kilpatrick M. E., Bourgeois A. L., Gardiner C., Batchelor R. A., et al. (1992). Norfloxacin compared to trimethoprim/sulfamethoxazole for the treatment of travelers' diarrhea among U.S. military personnel deployed to South America and West Africa. Mil. Med. 157, 55–58. [PubMed] [Google Scholar]
- Threlfall E. J. (2000). Epidemic Salmonella Typhimurium DT 104–a truly international multiresistant clone. J. Antimicrob. Chemother. 46, 7–10. 10.1093/jac/46.1.7 [DOI] [PubMed] [Google Scholar]
- Tindall B. J., Grimont P. A., Garrity G. M., Euzeby J. P. (2005). Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol. 55, 521–524. 10.1099/ijs.0.63580-0 [DOI] [PubMed] [Google Scholar]
- Vien le T. M., Minh N. N., Thuong T. C., Khuong H. D., Nga T. V., Thompson C., et al. (2012). The co-selection of fluoroquinolone resistance genes in the gut flora of Vietnamese children. PLoS ONE 7:e42919. 10.1371/journal.pone.0042919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila J., Vargas M., Casals C., Urassa H., Mshinda H., Schellemberg D., et al. (1999). Antimicrobial resistance of diarrheagenic Escherichia coli isolated from children under the age of 5 years from Ifakara, Tanzania. Antimicrob. Agents Chemother. 43, 3022–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe E., Phoba M. F., Tamfun J. J., Jacobs J. (2009). Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. Int. J. Antimicrob. Agents 34, 295–303. 10.1016/j.ijantimicag.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Weill F. X., Bertrand S., Guesnier F., Baucheron S., Cloeckaert A., Grimont P. A. (2006). Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg. Infect. Dis. 12, 1611–1612. 10.3201/eid1210.060589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2001). WHO Global Strategy for Cotinament of Antibiotic Resistance. Genova. [Google Scholar]
- WHO (2014). Antimicrobial Resistance: Global Report on Surveillance 2014. Genova. [Google Scholar]
- Wirth T. (2015). Massive lineage replacements and cryptic outbreaks of Salmonella Typhi in eastern and southern Africa. Nat. Genet. 47, 565–567. 10.1038/ng.3318 [DOI] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistrom J., Norrby S. R., Burman L. G., Lundholm R., Jellheden B., Englund G. (1987). Norfloxacin versus placebo for prophylaxis against travellers' diarrhoea. J. Antimicrob. Chemother. 20, 563–574. 10.1093/jac/20.4.563 [DOI] [PubMed] [Google Scholar]
- Wong V. K., Baker S., Pickard D. J., Parkhill J., Page A. J., Feasey N. A., et al. (2015). Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat. Genet. 47, 632–639. 10.1038/ng.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi D., de Vries G. C., Rahardjo D., Alimsardjono L., Wasito E. B., De I., et al. (2009). Emergence of fluoroquinolone-resistant strains of Salmonella enterica in Surabaya, Indonesia. Diagn. Microbiol. Infect. Dis 64, 422–426. 10.1016/j.diagmicrobio.2009.04.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.