Abstract
Systemic inflammation, resulting from massive release of pro-inflammatory molecules into the circulatory system, is a major risk factor for severe illness, but the precise mechanisms underlying its control are incompletely understood. We observed that prostaglandin E2 (PGE2) through its receptor EP4 is down-regulated in human systemic inflammatory disease. Mice with reduced PGE2 synthesis develop systemic inflammation, associated with translocation of gut bacteria, which can be prevented by treating with EP4 agonists. Mechanistically, we demonstrate that PGE2–EP4 signaling directly acts on type 3 innate lymphoid cells (ILCs), promoting their homeostasis and driving them to produce interleukin-22 (IL-22). Disruption of the ILC/IL-22 axis impairs PGE2–mediated inhibition of systemic inflammation. Hence, PGE2–EP4 signaling inhibits systemic inflammation through ILC/IL-22 axis–dependent protection of gut barrier dysfunction.
Systemic inflammation commonly develops from locally invasive infection, is characterized by dysregulation of the innate immune system and overproduction of pro-inflammatory cytokines and can result in severe critical illness (e.g., bacteremia, sepsis and septic shock) (1,2). Despite much research on systemic inflammation, our understanding of the precise mechanisms for its control remains incomplete and represents an unmet clinical need (1–3). Prostaglandins (PGs) are bioactive lipid mediators generated from arachidonic acid via the enzymatic activity of cyclooxygenases (COXs) (4). PGs participate in the pathogenesis of inflammatory disease (4,5) and many inflammatory conditions are treated using non-steroidal anti-inflammatory drugs (NSAIDs) that inhibit PG synthesis by blocking COXs (6). NSAID therapy is also thought to confer similar beneficial effects in treating severe inflammation, but large randomized controlled clinical trials have found that NSAIDs failed to reduce mortality in severe systemic inflammation (7,8). More importantly, NSAIDs use during evolving bacterial infection is associated with more severe critical illness (9–13). Therefore there is an imperative to define the paradoxical regulatory role of PGs in systemic inflammation (14).
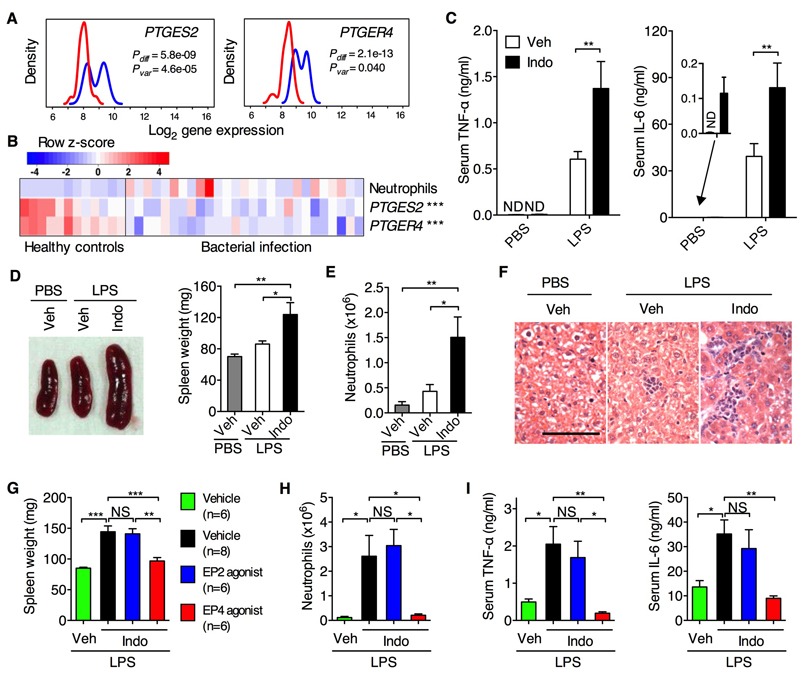
PGE2 is one of the most abundantly produced PGs and modulates immune and inflammatory responses through its receptors (EP1 – 4) (4). We performed a genome-wide gene expression analysis of whole blood samples from neonates with sepsis (15) and found that expression of PTGES2 (encoding membrane-associated PGE synthase-2) and PTGER4 (encoding EP4) were significantly diminished in the sepsis group compared with non-infected controls (Fig. 1A). The reduced expression of PTGES2 and PTGER4 was associated with increased neutrophil blood count as a marker of inflammation (Fig. 1B). Down-regulation of PTGER4 and PTGS2 (encoding COX2) was similarly observed in patients suffering from systemic inflammatory response syndrome, sepsis, septic shock or severe blunt trauma; but in contrast, expression of HPGD (encoding 15-PGDH which mediates PGE2 degradation) in these patients was up-regulated compared to non-infected controls (fig. S1). Consistent with this, blood monocytes from patients with sepsis and septic shock produced less PGE2 (16). Thus the PGE2–EP4 pathway is down-regulated in human severe systemic inflammatory disease.
Fig. 1. PGE2–EP4 signaling controls LPS–induced systemic inflammation.
(A) Gene expression of PTGES2 and PTGER4 in whole blood samples of neonates suffering from sepsis with confirmed bacterial infection (Red, n=27) and matched non-infection controls (Blue, n=35). Line graphs display gene expression (Log2 scale) as probability density plots for both group samples. Non-parametric Wilcoxon-Rank-Sum tests (Pdiff) were used to test for differential expression of a gene between infected and control neonates. Fligner-Killeen tests (Pvar) were used to test if sepsis and control groups have substantively different inter-subject variation in gene expression levels. (B) Supervised heat map of clinical neutrophil counts and expression levels for PTGES2 and PTGER4 genes for non-infected healthy controls (n=12) and bacterially infected neonates (n=27). Colored scale bar is shown for neutrophil count or gene expression z-score transformed values, respectively. ***P<0.001 by non-parametric Spearman correlation test performed to analyze the negative association between PTGES2 (rs=−0.6111) or PTGER4 (rs=−0.6323) gene expression and blood neutrophil counts. (C to F) Serum levels of TNF-α and IL-6 (C), spleen size and weight (D), neutrophil counts in peritoneal cavity lavage (E) and liver histology (F) of WT C57BL/6 mice treated with indomethacin (Indo) or vehicle control (Veh) for 5 d followed by LPS challenge injection for another 2 (C, n=6 per group) or 24 h (D to F, n=8 per group). (G to I) Spleen weight (G), neutrophils (H) and serum levels of TNF-α and IL-6 (I) of WT C57BL/6 mice treated with indomethacin and agonists for EP2 or EP4 followed by LPS challenge for 24 (G and H) or 2 h (I). Data shown as means ± SEM are pooled from two independent experiments. Scale bar, 50 μm. *P<0.05, **P<0.01 by one-way ANOVA. NS, not significant; ND, not detected.
To understand the mechanism(s) whereby PGE2 regulates systemic inflammation, we challenged wild-type (WT) C57BL/6 mice with lipopolysaccharide (LPS) to induce systemic inflammation after pre-treatment with indomethacin, which effectively suppresses PGE2 production (17,18). Mice pre-treated with indomethacin developed an enhanced cytokine storm (e.g., tumor necrosis factor-α (TNF-α) and IL-6) (Fig. 1C) as well as other inflammatory signs in indomethacin-treated mice, e.g., splenomegaly (Fig. 1D), peritonitis characterized by accumulation of CD11b+Ly-6G+ neutrophils (Fig. 1E), and a low-grade hepatic inflammation as characterized by sinusoidal lymphocytosis and necrosis/inflammatory foci in the parenchyma (Fig. 1F). Furthermore, co-administration of an EP4 agonist almost completely diminished indomethacin-augmented systemic inflammation (Fig. 1, G to I). Thus PGE2-EP4 signaling constrains LPS-induced systemic inflammation.
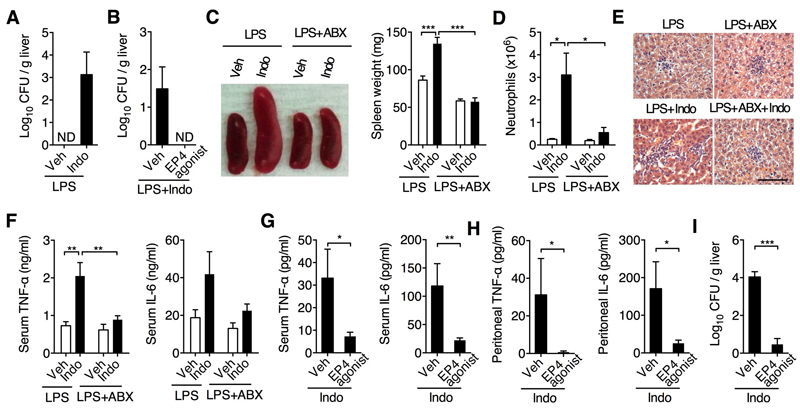
Besides the inflammatory markers described above, we also detected dissemination of gut bacteria into normally sterile tissues (e.g., liver) of indomethacin-treated, but not control, mice (Fig. 2A). The co-administration of EP4 agonist blocked the dissemination of bacteria to sterile tissues (Fig. 2B). Although pathogenic bacteria spread via the bloodstream usually cause severe systemic inflammation, gut leakage of commensal bacteria, especially in patients with damaged gut epithelium or endothelium, may also trigger systemic inflammation. We therefore tested whether these disseminated bacteria contribute to indomethacin-augmented systemic inflammation by treating mice with indomethacin and antibiotics, known to effectively deplete gut bacteria (19). Antibiotic therapy reduced indomethacin-facilitated systemic inflammation (Fig. 2, C to F). Indomethacin-dependent systemic inflammation was also observed in Rag1-/- mice (fig. S2), and this was again diminished by EP4 agonism (Fig. 2, G to I). Thus PGE2-EP4 signaling prevents systemic inflammation independently of adaptive immune cells.
Fig. 2. PGE2 control of systemic inflammation involves gut bacterial dissemination and acts independently of adaptive immune cells.
(A) Colony forming units (CFU) present in liver homogenates from WT C57BL/6 mice (n=6) treated with indomethacin for 5 d followed by LPS challenge for another 24 h. (B) CFU present in liver homogenates from WT C57BL/6 mice treated with indomethacin plus administration of an EP4 agonist (n=6) or vehicle (n=8) for 5 d followed by LPS challenge for another 24 h. (C to F) Spleen size and weight (C), neutrophils (D), liver histology (E) and serum levels of TNF-α and IL-6 (F) of WT C57BL/6 mice (n=6 per group) treated with indomethacin and antibiotics (ABX) for 5 d followed by LPS challenge for another 24 (C to E) or 2 h (F). (G to I) TNF-α and IL-6 levels in serum (G) and peritoneal cavity lavage (H) and CFU present in liver homogenates (I) from Rag1-/- mice treated with indomethacin plus administration of an EP4 agonist or vehicle control (n=8 per group) for 4 d. Data shown as means ± SEM are pooled from two independent experiments. Scale bar, 50 μm. *P<0.05, **P<0.01, ***P<0.001 by two-way ANOVA (C, D and F) or Mann-Whitney test (G to I). ND, not detected.
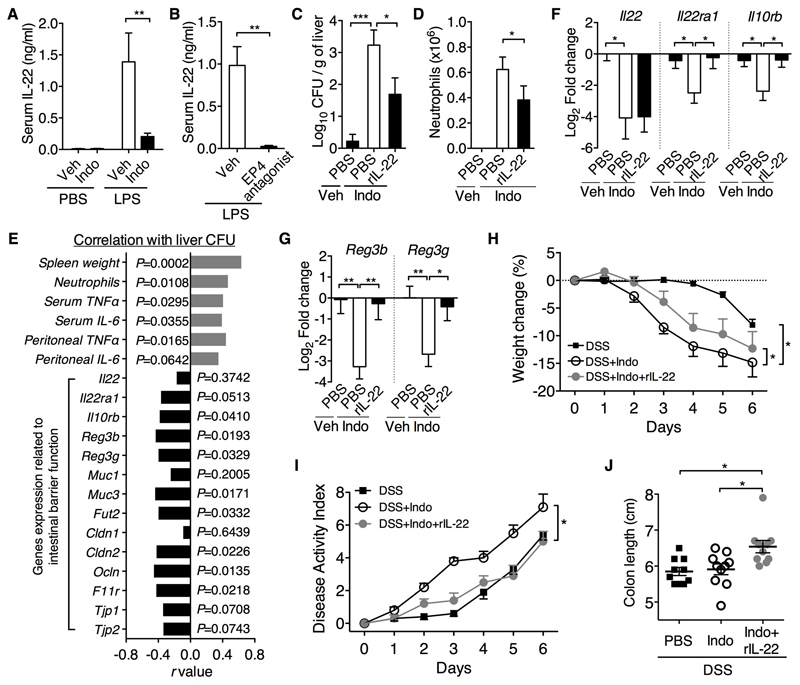
Given that IL-22 has recently been shown to inhibit inflammatory responses, particularly in the intestine (19–22), we hypothesized that IL-22 may mediate PGE2 control of systemic inflammation. To test this hypothesis, we first examined the effect of PGE2 on IL-22 production. Mice treated with indomethacin reduced LPS–induced IL-22 production (Fig. 3A), which was mimicked by an EP4 antagonist (Fig. 3B). Thus PGE2-EP4 signaling promotes IL-22 production in vivo. While LPS-induced systemic inflammation in WT mice was worsened and prevented by indomethacin and EP4 agonist, respectively (Fig. 1, G to I), both had little effects on LPS-induced inflammation in IL-22-deficient mice (ref. 23) (Fig. S3). Thus IL-22 is required for coupling PGE2-EP4 signaling-dependent control of systemic inflammation. Furthermore, co-administration of recombinant IL-22 (rIL-22) reduced indomethacin–induced bacterial dissemination, which positively correlated with reduced systemic inflammation (Fig. 3, C to E), implying that IL-22 mediates PGE2 suppression of gut bacterial dissemination and subsequent systemic inflammation.
Figure 3. IL-22 mediates PGE2 protection against systemic inflammation and intestinal barrier injury.
(A) Serum IL-22 levels of WT C57BL6 mice treated with indomethacin (Indo, n=6) or vehicle control (Veh, n=6) for 5 days followed by challenge with LPS or PBS for 2 h. (B) Serum IL-22 levels in WT C57BL/6 mice treated with an EP4 antagonist or vehicle (n=4 per group) in drinking water for 5 d followed by LPS challenge for another 2 h. (C to G) Rag1-/- mice were treated with vehicle (Veh, n=9) or indomethacin (Indo) plus administration of rIL-22 (n=10) or vehicle control PBS (n=10) for 4 d. (C) CFUs present in liver homogenates; (D) neutrophil counts; (E) correlation of liver CFU with systemic inflammation profile and expression profile of genes related to intestinal barrier function; (F and G) gene expression of IL-22 and its receptors (F) and antimicrobial peptides (G) in the terminal ileum. Expression was measured by real-time PCR, normalized to Gapdh gene and presented as relative expression (in Log2 Fold Change) to the vehicle control group. (H to J) Change in body weight (H), disease activity index (I) and colon length (J) of WT C57BL/6 mice (n=10 per group) treated with 2% DSS and indomethacin or vehicle control in drinking water plus administration of rIL-22 or vehicle control PBS for 6 days. Data shown as means ± SEM are pooled from two (A, H, I and J) or three (C to G) independent experiments or represent one experiment (B). *P<0.05, **P<0.01, ***P<0.001 by Mann-Whitney test (A, B and D), one-way ANOVA (C, F to J) or two-tailed Pearson correlation test (E).
As both PGE2 and IL-22 can potentially protect the gut epithelial barrier (17–22), we reasoned that inhibition of bacterial dissemination by PGE2 may act through augmenting IL-22 action on gut epithelial cells. We therefore examined gene expression related to barrier function in intestinal tissues. Indomethacin not only suppressed expression of IL-22 but also its receptors, i.e., Il22ra1 (encoding IL-22Rα1) and Il10rb (encoding IL-10Rβ), and their suppression was prevented by rIL-22 (Fig. 3F), suggesting that PGE2 may strengthen IL-22–IL-22R signaling in intestinal epithelial cells. Indeed, IL-22R–target genes such as Reg3b, Reg3g, Fut2 (24), mucins and tight junctions were similarly down-regulated by indomethacin but rescued by rIL-22 (Fig. 3G and fig. S4). Expression of genes related to IL-22R signaling and barrier function inversely correlated with gut bacterial dissemination (Fig. 3E). Given the protective role of PGE2 in acute colonic mucosal injury (17), we proposed that this protection is mediated by IL-22. Indeed, indomethacin worsened dextran sulfate sodium (DSS)-induced colitis and this was rescued by rIL-22, which elicited significantly less colitis disease activity index measured by weight loss, diarrhea and rectal bleeding (Fig. 3H-J). Our results thus indicate an important role of PGE2 in regulating the intestinal epithelial barrier function through innate IL-22 signaling.
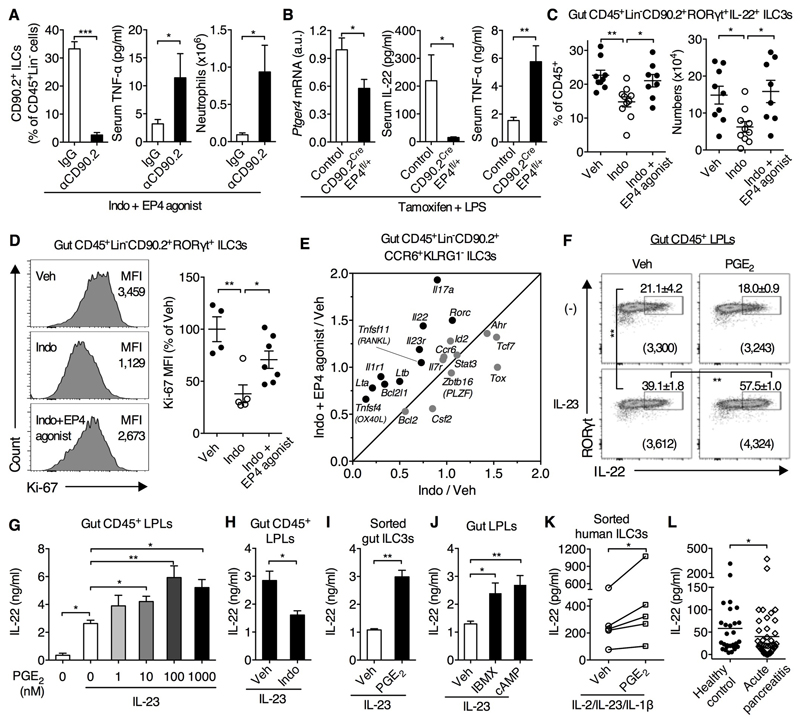
Flow cytometric analysis showed that IL-22-producing cells were CD45LowLineage(Lin)-CD90.2+RORγt(retinoid acid receptor-related orphan receptor gamma)+CCR6(CC chemokine receptor 6)+ type 3 innate lymphoid cells (ILC3s, ref. 22) in the gut and CD45+Lineage-CD90.2+CD4+ ILC3s in the spleen (fig. S5). We wondered whether these cells are involved in EP4-dependent control of systemic inflammation. Although Rag1-/- mice co-treated with indomethacin and an EP4 agonist (i.e., lacking PG signaling except through EP4) did not develop systemic inflammation (Fig. 2, G to I), depletion of CD90.2+ ILCs restored indomethacin-induced systemic inflammation in these mice (Fig. 4A). To address whether PGE2–EP4 signaling specifically in ILCs controls systemic inflammation, we crossed CD90.2-Cre mice (25) with EP4-floxed mice (26) to generate tamoxifen-induced selective down-regulation of EP4 in CD90.2+ cells (including T cells and ILCs) in heterozygous CD90.2CreEP4fl/+ mice (Fig. 4B). CD90.2CreEP4fl/+ mice produced less IL-22 in response to LPS, which was associated with augmentation of systemic inflammatory response, e.g., TNF-α production (Fig. 4B), while down-regulation of EP4 in T cells (27) affected neither IL-22 nor TNF-α production (fig. S6). These data demonstrate that PGE2-EP4 signaling controls systemic inflammation, at least in part, through the regulation of ILC3s.
Fig. 4. PGE2–EP4 signaling potentiates ILC3 homeostasis and function that contributes to control of systemic inflammatory response.
(A) Splenic CD90.2+ ILCs, serum TNF-α levels in and neutrophil counts in Rag1-/- mice co-treated with indomethacin and an EP4 agonist plus anti-CD90.2 (αCD90.2) or control IgG antibodies (n=8 per group) for 4 days. (B) Expression of Ptger4 mRNA in splenic CD90.2+ cells and serum levels of IL-22 and TNF-α in tamoxifen-treated CD90.2CreEP4fl/+ mice (n=7) and control C57BL/6 mice (n=5) at 1.5 h after LPS challenge. (C-E) Rag1-/- (C,D) or C57BL/6 (E) mice were treated with indomethacin with or without an EP4 agonist for 4-5 d. (C) Percentages and numbers of ILC3s in small intestines. Cells were ex vivo stimulated with IL-23 for 3 h. Each point represents an individual mouse. (D) Ki-67 expression in gut ILC3s. Each point represents an individual mouse. (E) Gene expression profile in gut ILC3s presented as relative expression to the vehicle control group. Data are the summary of two independent experiments with 8 mice for each group. (F) Expression of RORγt and IL-22 by Rag1-/- gut CD45+ lamina propria leukocytes (LPLs) stimulated with IL-23 and PGE2 for 4 h. Numbers in brackets represent IL-22 MFI of the RORγt+IL-22+ populations. (G to J) IL-22 production in supernatants by LPLs or ILC3s isolated from small intestines of Rag1-/- mice and then cultured with indicated conditions overnight (G, H, J) or for 3 d (I). (K) IL-22 production by human ILC3s cultured with IL-2, IL-23, IL-1β and PGE2 for 4 d (n=5 donors). (L) Plasma IL-22 levels in individuals with acute pancreatitis (AP) at the time of presentation to hospital (n=48) or from healthy individual donors (n=28). Horizon line represent the mean for each group. Data shown as means ± SEM are pooled from two or more independent experiments (A, C to E, G, J and K) or represent one (B) or two (F, H, I) independent experiments. *P<0.05, **P<0.01, ***P<0.001 by unpaired Student’s t-test (A, B, H and I), one-way ANOVA (C, D, F, G and J), ratio paired t-test (K) or Mann Whitney test (L).
Next, we investigated the impact of PGE2 on ILC3 maintenance. Indomethacin significantly diminished ILC3 numbers in the gut and spleen at the steady state (Fig. 4C and fig. S7, A to C). This reduction was prevented by an EP4 agonist and was mimicked by an EP4 antagonist (Fig. 4C and fig. S7). Consistently, ILC3s express PGE2 receptors with especially high expression levels of EP4 (fig. S8). Furthermore, indomethacin down-regulated Ki-67 expression and an EP4 agonist prevented this reduction in ILC3s (Fig. 4D), suggesting that PGE2-EP4 signaling potentiates ILC3 proliferation. PGE2-EP4 signaling also prevented ILC3 apoptosis in vitro but it had little effect on ILC3 apoptosis in vivo (Fig. S9, A and B). Moreover, down-regulation of EP4 in CD90.2+ ILCs not only decreased RORγt+ ILC3s but also weakened IL-7 responsiveness, e.g., Bcl-2 expression, in ILC3s (Fig. S9C). To further understand the effect of PGE2 on ILC3s, we measured gene expression by quantitative polymerase chain reaction in Lin-CD45lowCD90.2hiKLRG1-CCR6+ ILC3s sorted from small intestines of mice treated with indomethacin and EP4 agonist. PGE2-EP4 signaling up-regulates many ILC3 signature genes including Rorc, Il23r, Il1r1, Il22, Il17a, Lta, Ltb, Tnfsf11 and Tnfsf4 as well as Bcl2l1 (Fig. 4E).
We next asked whether and how PGE2 regulates ILC3 function. PGE2 promoted IL-23-driven IL-22 production by gut lamina propria leukocytes isolated from Rag1-/- mice in vitro (Fig. 4, F and G) and this was inhibited by indomethacin (Fig. 4H). PGE2 also promoted IL-22 production from splenic CD4+ ILC3s, which was mediated by EP2 and EP4 (fig. S10, A to C). More importantly, PGE2 increased IL-23-driven IL-22 production from highly purified CD45+Lin-CD90.2+KLRG1-CCR6+ ILC3s from intestines and CD3-CD11b-CD11c-CD4+ ILC3s from spleen or bone marrow (Fig. 4I and fig. S10D), suggesting a direct action of PGE2 on ILC3s. In addition, PGE2 also increased IL-22 production from ILC3s in response to IL-1β and IL-7 (fig. S11). PGE2 activated cyclic adenosine monophosphate signaling in ILC3s and in turn enhanced IL-22 production (Fig. 4J and fig. S12, A and B). Moreover, PGE2-augmented IL-22 production depended on the transcription factor signal transducer and activator of transcription 3, which is critical for IL-22 production by ILC3s (28), but not RORγt or Ahr (fig. S12, C to E). Altogether, these data demonstrate that PGE2–EP4 signaling promotes ILC3 homeostasis and IL-22 production through promoting ILC3 proliferation and enhancing cytokine (e.g., IL-7, IL-23 and IL-1) responsiveness.
Finally, we questioned whether PGE2 promotes IL-22 production from human ILC3s. To answer this, we sorted human Lin-CD161+CD127+CD117+CRTH2- ILC3s from periphery blood of healthy donors and cultured them with IL-2, IL-23 and IL-1β to induce IL-22 production. Addition of PGE2 similarly up-regulated IL-22 production from sorted human ILC3s (Fig. 4K). Moreover, expression of the human IL22 gene in healthy individuals infused with a bacterial endotoxin (ref. 29) positively correlated with expression of PTGS2 and PTGES (encoding mPGES-1) (fig. S13); two inducible enzymes that mediate PGE2 synthesis (4). Acute pancreatitis (AP) is a sterile initiator of systemic inflammation that results in multiple organ dysfunction where gut barrier injury is central to the pathogenesis (30). Given that IL-22 was protective in an animal model of AP (31), we measured IL-22 levels in patients with AP. IL-22 concentrations in plasma were lower in patients with AP compared to healthy controls at the time of admission to hospital (Fig. 4L), further confirming that the reduction of IL-22 signaling is associated with development of systemic inflammation.
In summary, we identify a physiological role of endogenous PGE2–EP4 signaling in activating the ILC3/IL-22 axis, which functionally contributes to crosstalk between the innate immune system, gut epithelium and microflora, and subsequently constrains systemic inflammation (fig. S14). These findings provide valuable insight towards understanding how inactivation of COXs may be harmful in severe bacterial infection and inflammation (32–34) and advance a crucial cellular and molecular mechanism for a scenario in which maintaining or augmenting PGE2 signaling, e.g., by inhibiting the PGE2-degrading enzyme 15-PGDH, protects intestinal barrier injury and potentiates its repair and control of systemic inflammation (35, 36).
Supplementary Material
www.sciencemag.org/content/xxx/xxxx/xxx/suppl/DC1
Materials and Methods
Figs. S1 to S14
References (37–55)
Acknowledgments
We thank J. Allen for critically reading and editing the manuscript; P.J. Brophy and D. Mahad for CD90.2(Thy1.2)-Cre mice; J.C. Renauld for IL-22-deficient mice; J. Pollard, P.J. Spence and J. Thompson for Rag1-/- mice; T. Walsh and J. Rennie for human blood samples; Y.X. Fu, X.G. Guo, D. Ruckerl, T. Kendall, J. Hu, Q. Huang, G.T. Ho for invaluable advice, excellent technical assistance and/or discussion; and S. Johnston and W. Ramsay for cell sorting and analysis. We thank the Wellcome Trust Clinical Research Facility staff and the Department of Surgery, NHS Lothian. IL-22-deficient mice are available from Jean-Ludwig Institute For Cancer Research under a material transfer agreement with Jean-Christophe Renauld. This work was supported in part by the University of Edinburgh start-up funding (C.Yao.), Wellcome Trust Institutional Strategic Support Fund (C.Yao, J.P.I., S.M.A), Medical Research Council (MRC) UK (J.P.I., S.M.A., R.D., A.G.R.), NIH (DK37097) and a VA Merit (1BX000616 to R.M.B.), Health Foundation/Academy of Medical Sciences (D.J.M), University of Edinburgh/GlaxoSmithKline DPAC collaboration (D.J.M.), Wellcome Trust Investigator Award (R.M.M., Ref 106122) and Rainin Trust (R.M.M. 13H6), Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Agency (S.N.), and EU FP7 IAPP project ClouDx-I, Chief Scientists Office (ETM202) and BBSRC (BB/K091121/1 to P.G.). The data presented in this manuscript are tabulated in the main paper and the supplementary materials. The authors declare no financial conflicts of interest.
References and Notes
- 1.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;3481:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 2.LaRosa SP, Opal SM. Immune aspects of sepsis and hope for new therapeutics. Curr Infect Dis Rep. 2012;14:474–483. doi: 10.1007/s11908-012-0276-2. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J, Opal S, Calandra T. Sepsis studies need new direction. Lancet Infect Dis. 2012;12:503–505. doi: 10.1016/S1473-3099(12)70136-6. [DOI] [PubMed] [Google Scholar]
- 4.Hirata T, Narumiya S. Prostanoids as regulators of innate and adaptive immunity. Adv Immunol. 2012;116:143–174. doi: 10.1016/B978-0-12-394300-2.00005-3. [DOI] [PubMed] [Google Scholar]
- 5.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day RO. Non-steroidal anti-inflammatory drugs (NSAIDs) BMJ. 2013;346:f3195. doi: 10.1136/bmj.f3195. [DOI] [PubMed] [Google Scholar]
- 7.Bernard GR, et al. The effects of ibuprofen on the physiology and survival of patients with sepsis. The ibuprofen study group. N Engl J Med. 1997;336:912–918. doi: 10.1056/NEJM199703273361303. [DOI] [PubMed] [Google Scholar]
- 8.Haupt MT, Jastremski MS, Clemmer TP, Metz CA, Goris GB. Effect of ibuprofen in patients with severe sepsis: a randomized, double-blind, multicenter study. The ibuprofen study group. Crit Care Med. 1991;19:1339–1347. doi: 10.1097/00003246-199111000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Aronoff DM. Cyclooxygenase inhibition in sepsis: is there life after death? Mediat Inflamm. 2012;2012:696897. doi: 10.1155/2012/696897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen DP. Manifold beneficial effects of acetyl salicylic acid and non-steroidal anti-inflammatory drugs on sepsis. Intensive Care Med. 2012;38:1249–1257. doi: 10.1007/s00134-012-2570-8. [DOI] [PubMed] [Google Scholar]
- 11.Stevens DL. Could nonsteroidal anti-inflammatory drugs (NSAIDs) enhance the progression of bacterial infections to toxic shock syndrome? Clin Infect Dis. 1995;21:977–980. doi: 10.1093/clinids/21.4.977. [DOI] [PubMed] [Google Scholar]
- 12.Legras A, et al. A multicentre case-control study of nonsteroidal anti-inflammatory drugs as a risk factor for severe sepsis and septic shock. Crit Care. 2009;13:R43. doi: 10.1186/cc7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BH, et al. Association of body temperature and antipyretic treatments with mortality of critically ill patients with and without sepsis: multi-centered prospective observational study. Crit Care. 2012;16:R33. doi: 10.1186/cc11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullerton JN, O’Brien AJ, Gilroy DW. Lipid mediators in immune dysfunction after severe inflammation. Trends Immunol. 2014;35:12–21. doi: 10.1016/j.it.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith CL, et al. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun. 2014;5:4649. doi: 10.1038/ncomms5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruegel M, et al. Sepsis-associated changes of the arachidonic acid metabolism and their diagnostic potential in septic patients. Crit Care Med. 2012;40:1478–1486. doi: 10.1097/CCM.0b013e3182416f05. [DOI] [PubMed] [Google Scholar]
- 17.Kabashima K, et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J Clin Invest. 2002;109:883–893. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinen T, et al. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat Commun. 2011;2:190. doi: 10.1038/ncomms1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 22.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: A new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreymborg K, et al. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J Immunol. 2007;179:8098–8104. doi: 10.4049/jimmunol.179.12.8098. [DOI] [PubMed] [Google Scholar]
- 24.Goto Y, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345:1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zonta B, et al. A critical Role for Neurofascin in regulating action potential Initiation through maintenance of the Axon Initial Segment. Neuron. 2011;69:945–956. doi: 10.1016/j.neuron.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider A, et al. Generation of a conditional allele of the mouse prostaglandin EP4 receptor. Genesis. 2004;40:7–14. doi: 10.1002/gene.20048. [DOI] [PubMed] [Google Scholar]
- 27.Yao C, et al. Prostaglandin E2 promotes Th1 differentiation via synergistic amplification of IL-12 signalling by cAMP and PI3-kinase. Nat Commun. 2013;4:1685. doi: 10.1038/ncomms2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 2014;40:25–39. doi: 10.1016/j.immuni.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 30.Fishman JE, et al. The intestinal mucus layer is a critical component of the gut barrier that is damaged during acute pancreatitis. Shock. 2014;42:264–270. doi: 10.1097/SHK.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng D, et al. Interleukin-22 ameliorates cerulein-induced pancreatitis in mice by inhibiting the autophagic pathway. Int J Biol Sci. 2012;8:249–257. doi: 10.7150/ijbs.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang B, et al. Platelets protect from septic shock by inhibiting macrophage-dependent inflammation via the cyclooxygenase 1 signalling pathway. Nat Commun. 2013;4:2657. doi: 10.1038/ncomms3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fredenburgh LE, et al. Cyclooxygenase-2 deficiency leads to intestinal barrier dysfunction and increased mortality during polymicrobial sepsis. J Immunol. 2011;187:5255–5267. doi: 10.4049/jimmunol.1101186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton SM, Bayer CR, Stevens DL, Bryant AE. Effects of selective and nonselective nonsteroidal anti-inflammatory drugs on antibiotic efficacy of experimental group A streptococcal myonecrosis. J Infect Dis. 2014;209:1429–1435. doi: 10.1093/infdis/jit594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Inhibition of the prostaglandin-degrading enzyme 15-PGDH potentiates tissue regeneration. Science. 2015;348:aaa2340. doi: 10.1126/science.aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Németh K, et al. Bone marrow stromal cells attenuate sepsis via prostaglandin E2-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med. 2009;15:42–49. doi: 10.1038/nm.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.