Abstract
To date, most genetic analyses of phenotypes have focused on analyzing single traits or analyzing each phenotype independently. However, joint epistasis analysis of multiple complementary traits will increase statistical power and improve our understanding of the complicated genetic structure of the complex diseases. Despite their importance in uncovering the genetic structure of complex traits, the statistical methods for identifying epistasis in multiple phenotypes remains fundamentally unexplored. To fill this gap, we formulate a test for interaction between two genes in multiple quantitative trait analysis as a multiple functional regression (MFRG) in which the genotype functions (genetic variant profiles) are defined as a function of the genomic position of the genetic variants. We use large-scale simulations to calculate Type I error rates for testing interaction between two genes with multiple phenotypes and to compare the power with multivariate pairwise interaction analysis and single trait interaction analysis by a single variate functional regression model. To further evaluate performance, the MFRG for epistasis analysis is applied to five phenotypes of exome sequence data from the NHLBI’s Exome Sequencing Project (ESP) to detect pleiotropic epistasis. A total of 267 pairs of genes that formed a genetic interaction network showed significant evidence of epistasis influencing five traits. The results demonstrate that the joint interaction analysis of multiple phenotypes has a much higher power to detect interaction than the interaction analysis of a single trait and may open a new direction to fully uncovering the genetic structure of multiple phenotypes.
Author Summary
The widely used statistical methods test interaction for single phenotype. However, we often observe pleotropic genetic interaction effects. The simultaneous gene-gene (GxG) interaction analysis of multiple complementary traits will increase statistical power to detect GxG interactions. Although GxG interactions play an important role in uncovering the genetic structure of complex traits, the statistical methods for detecting GxG interactions in multiple phenotypes remains less developed owing to its potential complexity. Therefore, we extend functional regression model from single variate to multivariate for simultaneous GxG interaction analysis of multiple correlated phenotypes. Large-scale simulations are conducted to evaluate Type I error rates for testing interaction between two genes with multiple phenotypes and to compare power with traditional multivariate pair-wise interaction analysis and single trait interaction analysis by a single variate functional regression model. To further evaluate performance, the MFRG for interaction analysis is applied to five phenotypes of exome sequence data from the NHLBI’s Exome Sequencing Project (ESP) to detect pleiotropic GxG interactions. 267 pairs of genes that formed a genetic interaction network showed significant evidence of interactions influencing five traits.
Introduction
In the past several years, we have witnessed remarkable progresses in the development of methodologies for identification of epistasis that detect deviation from summation of genetic additive effects for a quantitative trait [1]. The classical approach to epistasis analysis is a single variant test. The epistasis is typically evaluated by testing interaction between a pair of variants one at a time. The classical methods for epistasis tests are originally designed to detect epistasis for common variants and are difficult applied to rare variants due to multiple testing problems and the low power to detect interaction. To overcome the critical barrier in interaction analysis for rare variants, instead of testing each pair of variants individually, group interaction tests that evaluate cumulative interaction effects of multiple genetic variants in a region or gene have recently been developed. Regression-based methods [2–8], haplotype-based methods [9–15], and machine learning-based methods [16–20] are proposed for epistasis analysis.
The classical statistical methods for interaction analysis have mainly tested association with single traits, one time analyzing one trait [21]. However, multiple phenotypes are highly correlated. More than 4.6% of the SNPs and 16.9% of the genes in previous genome-wide association studies (GWAS) are reported to be significantly associated with more than one trait [22]. These results demonstrate that genetic pleiotropic effects likely play a crucial role in the molecular basis of correlated phenotypes [23–26]. Joint epistasis analysis of multiple complementary traits will increase statistical power to unravel the interaction structure of multiple phenotypes [27, 28]. Despite their importance in understanding genetic mechanism underlying the complex diseases, the statistical methods for identifying epistasis in multiple phenotypes have been less developed [1]. The interaction analyses for multiple phenotypes have been limited to common variants in carefully controlled experimental crosses [29, 30]. Simultaneously analyzing interactions for multiple phenotypes in humans poses enormous challenges for methodologies and computations.
Purpose of this paper is to develop a general analytic framework and novel statistical methods for simultaneous epistasis analysis of multiple correlated phenotypes. To unify the approach to epistasis analysis for both common and rare variants, we take a genome region (or gene) as a basic unit of interaction analysis and use all the information that can be accessed to collectively test interaction between all possible pairs of SNPs within two genome regions (or genes). Functional data analysis is used to reduce the dimension of next-generation sequencing data. Specifically, genetic variant profiles that will recognize information contained in the physical location of the SNP are used as a major data form. The densely typed genetic variants in a genomic region for each individual are so close that these genetic variant profiles can be treated as observed data taken from curves [8, 31]. Since standard multivariate statistical analyses often fail with functional data [32] we formulate a test for interaction between two genomic regions in multiple quantitative trait analysis as a multiple functional regression (MFRG) model [33] with scalar response. In the MFRG model the genotype functions (genetic variant profiles) are defined as a function of the genomic position of the genetic variants rather than a set of discrete genotype values and the quantitative trait is predicted by genotype functions with their interaction terms. By functional principal component analysis, the genotype functions are expanded as a few functional principal components (FPC) and the MFRG model is transformed to the classical multivariate regression model (MRG) in which FPC scores are taken as variates. Statistics are developed in this publication which can be applied to pairwise interaction tests and gene-based interaction tests for multiple phenotypes. By investigating SNP-SNP interactions or gene-gene interactions that are shared across multiple traits, pleiotropic epistasis can be studied.
To evaluate performance for multiple traits epistasis analysis, large scale simulations are used to calculate the Type I error rates of the MFRG for testing interaction between two genomic regions with multiple phenotypes and to compare power with multivariate pair-wise interaction analysis and single trait interaction analysis by functional regression (FRG) model. To further evaluate performance, the MFRG for epistasis analysis is applied to five traits: high density lipoprotein (HDL), low density lipoprotein (LDL), total cholesterol, systolic blood pressure (SBP), and diastolic blood pressure (DBP), from exome sequence data from the NHLBI’s Exome Sequencing Project (ESP) to detect pleiotropic epistasis.
Methods
Assume that n individuals are sampled. Let yik, k = 1,2,…,K, be the k-th trait values of the i-th individual. Consider two genomic regions [a1, b1] and [a2, b2]. Let xi(t) and xi(s) be genotypic functions of the i-th individual defined in the regions [a1, b1] and [a2, b2], respectively. Let yi = [yi1,…,yiK]T be the vector of the trait values measured on the i-th individual. Let t and s be a genomic position in the first and second genomic regions, respectively. Define a genotype profile xi(t) of the i-th individual as
where M and m are two alleles of the marker at the genomic position t. Recall that a regression model for interaction analysis with the k-th trait is defined as
| (1) |
where μk is an overall mean of the k-th trait, ζkd is the coefficient associated with the covariate νd, αkj is the main genetic additive effect of the j-th SNP in the first genomic region for the k-th trait, βkl is the main genetic additive effect of the l-th SNP in the second genomic region for the k-th trait, γkjl is an additive × additive interaction effect between the j-th SNP in the first genomic region and the l-th SNP in the second genomic region for the k-th trait; xij and zil are indicator variableS for the genotypes at the j-th SNP and the l-th SNP, respectively; εik, k = 1,..,K are independent and identically distributed normal variables with mean of zero and covariance matrix Σ.
Similar to the multiple regression models for interaction analysis with multiple quantitative traits, the functional regression model for a quantitative trait can be defined as
| (2) |
where α0k is an overall mean, ζkd is defined as before, αk(t) and βk(s) are genetic additive effects of two putative QTLs located at the genomic positions t and s, respectively; γk(t,s) is the interaction effect between two putative QTLs located at the genomic positions t and s for the k-th trait, k = 1,…,K, xi(t) and xi(s) are genotype profiles, and εik are independent and identically distributed normal variables with mean of zero and covariance matrix Σ. Consider covariates in the model (2) allows incorporating PCA scores for population stratification, sex, age, BMI and other biomarkers into the model.
Estimation of Interaction Effects
We assume that both phenotypes and genotype profiles are centered. The genotype profiles xi(t) and xi(s) are expanded in terms of the orthonormal basis function as:
| (3) |
where ϕj(t) and ψl(s) are sequences of the orthonormal basis functions. The more number of variants in the genes the more accurate the eigenfunction expansion. If the number of variants is less than 3 the eigenfunction expansion of the genotypic profiles is impossible. MFRG can only be used for gene with more than 3 variants.
In practice, numerical methods for the integral will be used to calculate the expansion coefficients. Substituting Eq (3) into Eq (2), we obtain (Appendix)
| (4) |
The parameters αkj, βkl and γkjl are referred to as genetic additive and additive × additive effect scores for the k-th trait. These scores can also be viewed as the expansion coefficients of the genetic effect functions with respect to orthonormal basis functions.
Then, Eq (4) can be approximated by (Appendix)
| (5) |
where and .
Therefore, we transform the original functional regression interaction model into the classical multivariate regression interaction model by eigenfunction expansions. All methods for multivariate regression interaction analysis can directly be used for solving problem (5).
The standard least square estimators of B and the variance covariance matrix Σ are, respectively, given by
| (6) |
| (7) |
Denote the last JL row of the matrix (WTW)−1WT by A. Then, the estimator of the parameter γ is given by
| (8) |
The vector of the matrix γ can be written as
| (9) |
By the assumption of the variance matrix of Y, we obtain the variance matrix of vec(Y):
| (10) |
Thus, it follows from Eqs (9) and (10) that
| (11) |
Test Statistics
An essential problem in genetic interaction studies of the quantitative traits is to test the interaction between two genomic regions (or genes). Formally, we investigate the problem of testing the following hypothesis:
which is equivalent to testing the hypothesis:
Define the test statistic for testing the interaction between two genomic regions [a1, b1] and [a2, b2] with K quantitative traits as
| (12) |
Then, under the null hypothesis H0: γ = 0, TI is asymptotically distributed as a central distribution if JL components are taken in the expansion Eq (3).
Group tests often make implicit homogeneity assumptions where all putatively functional variants within the same genomic region are assumed to have the same direction of effects. However, in practice, the variants with opposite directions of effects will be simultaneously presented in the same genomic region. MFRG can efficiently use information of both risk and protective variants and allow for sign and size heterogeneity of genetic variants. In general, the trait increasing and decreasing variants will be present in different locations in the genomic region. Information of trait increasing and decreasing variants usually will be reflected in different eigenfunctions and hence will be included in different functional principal component scores. The MFRG test statistic is essentially to summarize the square of the functional principal component scores. Therefore, the opposite effects of trait increasing and decreasing variants on the phenotype will not compromise each other in the MFRG test statistics. The MFRG statistics automatically take the opposite effects of the trait increasing and decreasing variants on the phenotype into account and do not require additional computations. MFRG will take the sign and size heterogeneity of the variants into account and be less sensitive to the presence of variants with opposite directions of effect.
We can also develop likelihood ratio-based statistics for testing interaction.
Setting , we can write the model as
Under H0: γ = 0, we have the model:
The estimators will be
The likelihood for the full model and reduced model are, respectively, given by
The likelihood-ratio-based statistic for testing interaction between two genomic regions with multivariate traits is defined as
| (13) |
Under the null hypothesis H0: γ = 0, TIΛ is asymptotically distributed as a central distribution if JL components are taken in the expansion Eq (3).
Simulation Model for Type 1 Error Rate Calculation
The genetic models for simulations to calculate Type 1 error rates of the tests are briefly given below. We first assume the model with no marginal effects for all traits:
where Yi = [yi1,…,yik], μ = [μ1,…,μk], and εi is distributed as
Then, we considered the model with marginal genetic effect (additive model) at one gene:
where
where Pj is a frequency of the allele Aj, rk is a risk parameter of the k-th trait which was randomly selected from 1.1 to 1.6. The risk parameter affect the genetic effects and is used to control the contribution effort by genotype to the phenotype. The risk parameter influences the relative magnitude of the genetic effects. f0 is a baseline penetrance and set to 1 and ε are defined as before.
Finally, we consider the model with marginal genetic effects (additive model) at both genes:
where
Pj and ql are frequencies of the alleles Aj and Bl, respectively, rpk and rqk are risk parameters of the k-th trait for the SNPs in the first and second genes, respectively, and randomly selected from 1.1 to 1.6, f0 is a baseline penetrance and set to 1 and ε are defined as before.
Results
Null Distribution of Test Statistics
To examine the null distribution of test statistics, we performed a series of simulation studies to compare their empirical levels with the nominal ones. We calculated the Type I error rates for rare alleles, and common alleles. To make simulations more close to real whole exome sequencing data, we generated 50,000 datasets consisting of 1,000,000 chromosomes randomly sampled from the NHLBI’s Exome Sequencing Project (ESP) with 2,016 individuals and 18,587 genes. Each dataset included randomly selected a pair of genes from sequenced 18,587 genes. We randomly selected 20% of SNPs from each gene as causal variants. The number of sampled individuals from populations of 1,000,000 chromosomes ranged from 1,000 to 5,000. For each dataset, we repeated 5,000 simulations. We presented average type I error rates over 50,000 randomly selected pairs of genes from whole exome sequencing ESP dataset.
Table 1 and S1 and S2 Tables summarized the average Type I error rates of the test statistics for testing the interaction between two genes with no marginal effect and consisting of only rare variants with 5 traits, 2 traits and 10 traits, respectively, over 50,000 pairs of genes at the nominal levels α = 0.05, α = 0.01 and α = 0.001. Table 2 and S3 and S4 Tables summarized the average Type I error rates of the test statistics for testing the interaction between two genes with marginal effect at one gene consisting of only rare variants with 5 traits, 2 traits and 10 traits, respectively, over 50,000 pairs of genes at the nominal levels α = 0.05, α = 0.01 and α = 0.001. Table 3 and S5 and S6 Tables summarized the average Type I error rates of the test statistics for testing the interaction between two genes with marginal effect at both genes consisting of only rare variants with 5 traits, 2 traits and 10 traits, respectively, over 50,000 pairs of genes at the nominal levels α = 0.05, α = 0.01 and α = 0.001. For common variants, we summarized the average Type I error rates of the test statistics for testing the interaction between two genes with marginal effect at both genes consisting of only common variants with 5 traits, 2 and 10 traits, respectively, over 10 pairs of genes at the nominal levels α = 0.05, α = 0.01 and α = 0.001, in Table 4 and S7 and S8 Tables, respectively. The statistics for testing interaction between two genomic regions with only common variants have the similar Type 1 error rates in the other two scenarios: with marginal genetic effects at one gene or without marginal genetic effects at two genes. These results clearly showed that the Type I error rates of the MFRG-based test statistics for testing interaction between two genes with multiple traits and common variants with or without marginal effects were not appreciably different from the nominal α levels. For the rare variants when the sample sizes increased to 5,000, the Type 1 error rates were still not appreciably different from the nominal levels.
Table 1. Average type 1 error rates of the statistic for testing interaction between two genes with no marginal effect consisting only rare variants with 5 traits over random selected 50,000 pairs of genes from whole exome.
| Sample Size | 0.05 | 0.01 | 0.001 |
|---|---|---|---|
| 1000 | 0.0784 | 0.0188 | 0.0019 |
| 2000 | 0.0693 | 0.0097 | 0.0016 |
| 3000 | 0.0617 | 0.0135 | 0.0010 |
| 4000 | 0.0591 | 0.0126 | 0.0014 |
| 5000 | 0.0546 | 0.0095 | 0.0012 |
Table 2. Average type 1 error rates of the statistic for testing interaction between two genes with marginal effect at one gene consisting only rare variants with 5 traits over randomly selected 50,000 pairs of genes from the whole exome.
| Sample Size | 0.05 | 0.01 | 0.001 |
|---|---|---|---|
| 1000 | 0.0785 | 0.0177 | 0.0018 |
| 2000 | 0.0672 | 0.0154 | 0.0017 |
| 3000 | 0.0604 | 0.0149 | 0.0010 |
| 4000 | 0.0555 | 0.0120 | 0.0012 |
| 5000 | 0.0510 | 0.0132 | 0.0009 |
Table 3. Average type 1 error rates of the statistic for testing interaction between two genes with marginal effects at two genes consisting only rare variants with 5 traits over randomly selected 50,000 pairs of genes from the whole exome.
| Sample Size | 0.05 | 0.01 | 0.001 |
|---|---|---|---|
| 1000 | 0.0715 | 0.0152 | 0.0018 |
| 2000 | 0.0664 | 0.0133 | 0.0013 |
| 3000 | 0.0596 | 0.0105 | 0.0014 |
| 4000 | 0.0508 | 0.0098 | 0.0010 |
| 5000 | 0.0511 | 0.0106 | 0.0012 |
Table 4. Average type 1 error rates of the statistic for testing interaction between two genes with marginal effects at two genes consisting only common variants with 5 traits over randomly selected 50,000 pairs of genes from the whole exome.
| Sample Size | 0.05 | 0.01 | 0.001 |
|---|---|---|---|
| 1000 | 0.0529 | 0.0112 | 0.0011 |
| 2000 | 0.0513 | 0.0098 | 0.0013 |
| 3000 | 0.0499 | 0.0101 | 0.0009 |
| 4000 | 0.0471 | 0.0094 | 0.0010 |
| 5000 | 0.0469 | 0.0102 | 0.0008 |
Power Evaluation
To evaluate the performance of the MFRG models for interaction analysis of multiple traits, we used simulated data to estimate their power to detect interaction between two genes for two, four, five, six and ten quantitative traits. A true multiple quantitative genetic model is given as follows. Consider H pairs of quantitative trait loci (QTL) from two genes (genomic regions). Let and be two alleles at the first QTL, and and be two alleles at the second QTL, for the H pair of QTLs. Let uijkl be the genotypes of the u-th individual with and , and be its genotypic value for the m-th trait. The following multiple regression is used as a genetic model for the m-th quantitative trait:
where is a genotypic value of the h-th pair of QTLs for the m-th quantitative trait and εmu are distributed as .
Four models of interactions are considered: (1) Dominant OR Dominant, (2) Dominant AND Dominant, (3) Recessive OR Recessive and (4) Threshold model (S9 Table). We assume that the genotypes at two loci affect a complex trait. Intuitively, Dominant OR Dominant model means that presence of risk allele at least one locus will cause the phenotype variation. Dominant AND Dominant model means that only when risk alleles at both loci are present the phenotype variation can be affected. Recessive OR recessive model indicates that when both risk alleles are at least present at one locus the phenotype variation can be observed. Threshold model implies that when two risk alleles at one locus and at least one risk allele at another locus are present, the phenotype variation will be observed. Recessive AND Recessive model is excluded due to low frequency of that condition with rare variants. The risk parameter r varies from 0 to 1.
We generated 2,000,000 chromosomes by resampling from 2,016 individuals of European origin with variants in random two genes selected from the NHLBI’s Exome Sequencing Project (ESP). Two haplotypes were randomly sampled from the population and assigned to an individual. We randomly selected 20% of the variants as causal variants. A total of 2,000 individuals for the four interaction models were sampled from the populations. A total of 1,000 simulations were repeated for the power calculation.
The power of the proposed MFRG model is compared with the single trait functional regression (SFRG) model, the multi-trait pair-wise interaction test and the regression on principal components (PCs). For SNPs genotypes in each genomic region principal component analysis (PCA) were performed. The number of principal components for each individual which can explain 80% of the total genetic variation in the genomic region will be selected as the variables. Specifically, the principal component score of the i-th individual in the first and second genomic regions are denoted by and , respectively. The regression model for detection of interaction for the m-th trait is then given by
The power of the MFRG is compared with the traditional point-wise interaction test which takes the following model:
For a pair of genes, we assume that the first gene has k1 SNPs, and the second gene has k2 SNPs, then, the total number of all possible pairs is k = k1 × k2. For each pair of SNPs, we calculated a statistic for testing pair-wise interaction Tmjpair. Finally, the maximum of Tmjpair: Tmax = max(T1,1pair,T1,2pair,…,T1,kpair,…,TM,1pair,…,TM,kpair) is computed.
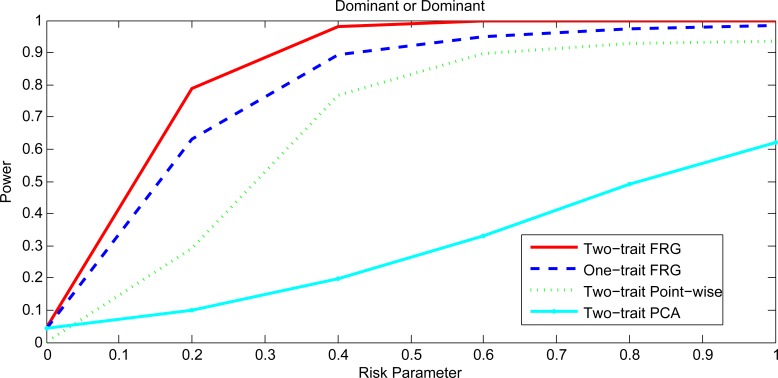
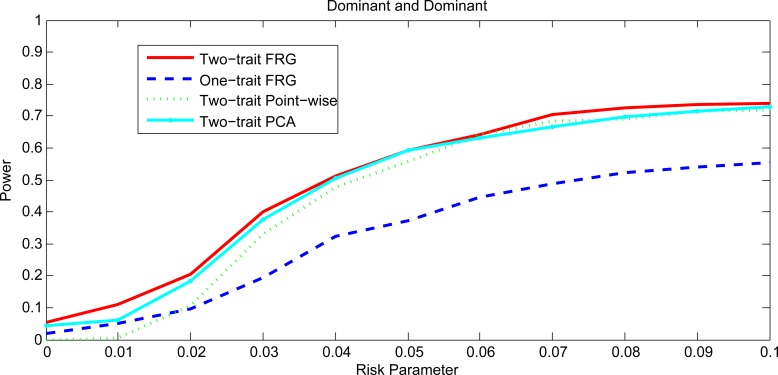
Figs 1 and 2, S1 Fig and S2 Fig plotted the power curves of the two-trait FRG, single trait FRG, two-trait regression on PCs and two-trait pair-wise interaction tests for a quantitative trait under Dominant OR Dominant, Dominant AND Dominant, Threshold, and Recessive OR Recessive models, respectively. Only two genes include rare variants. These power curves are a function of the risk parameter at the significance level α = 0.05. Permutations in the point-wise interaction tests were used to adjust for multiple testing. In all cases, the two-trait FRG had the highest power to detect epistasis. We observed two remarkable features. First, two-trait test had higher power than the one-trait test. Second, the two-trait FRG had the highest power among all two-trait tests.
Fig 1. Power curves under Dominant OR Dominant with two genes including rare variants only.
Fig 2. Power curves under Dominant AND Dominant with two genes including rare variants only.
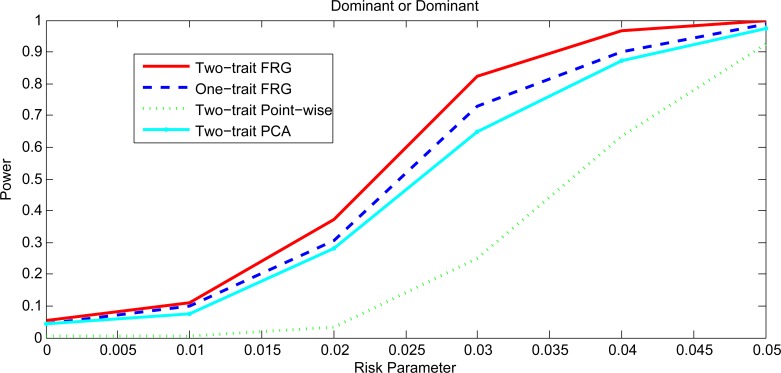
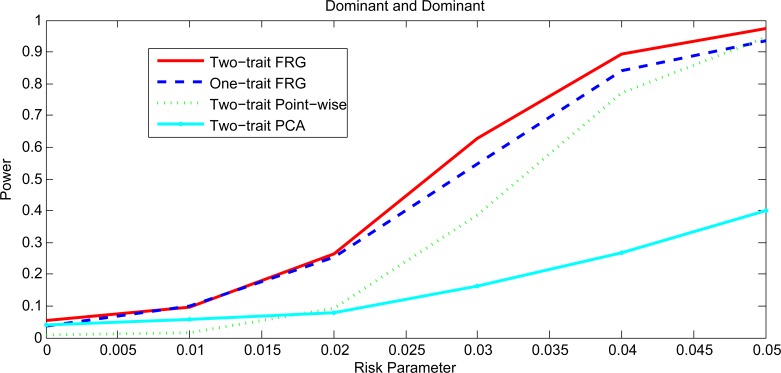
Figs 3 and 4, S3 Fig and S4 Fig plotted the power curves of the two-trait FRG, single trait FRG, two-trait regression on PCs and two-trait pair-wise interaction tests for a quantitative trait under Dominant OR Dominant, Dominant AND Dominant, Threshold and Recessive OR Recessive models, respectively. Only two genes include common variants. These power curves are a function of the risk parameter at the significance level α = 0.05. Permutations in the point-wise interaction tests were used to adjust for multiple testing. These figures showed that the power patterns of the epistasis tests for common variants were similar to that for rare variants.
Fig 3. Power curves under Dominant OR Dominant with two genes including common variants only.
Fig 4. Power curves under Dominant AND Dominant with two genes including common variants only.
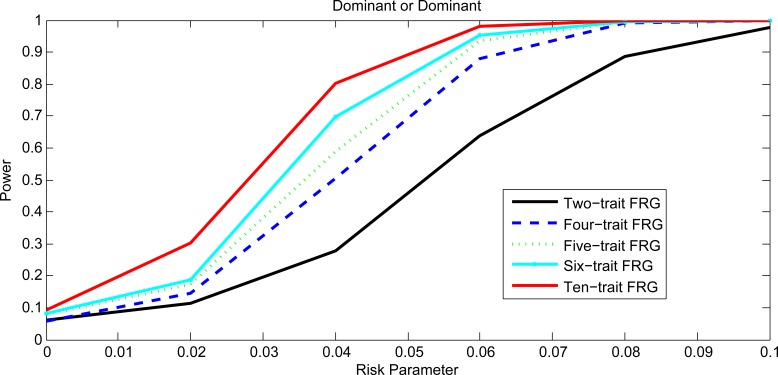
Next we investigate the impact of the number of traits on the power. Fig 5 plotted the power curves of two-trait FRG, four-trait FRG, five-trait FRG, six-trait FRG and ten-trait FRG under Dominant OR Dominant interaction model. Fig 5 showed that if the multiple phenotypes are correlated then the power of the MFRG to detect epistasis will increase as the number of phenotypes increases.
Fig 5. Power curves of MFRG with different trait number under Dominant OR Dominant.
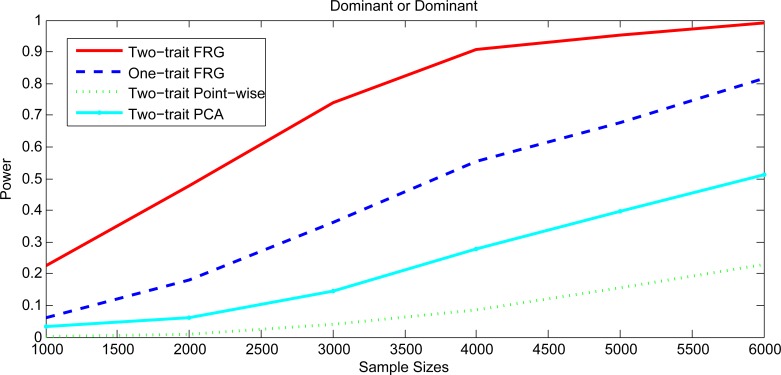
To investigate the impact of sample size on the power, we plotted Fig 6 and S5–S7 Figs showing the power of three statistics for testing the interaction between two genomic regions (or genes) with only rare variants as a function of sample sizes under four interaction models, assuming 20% of the risk rare variants and the risk parameter r = 0.05 for Dominant OR Dominant, Dominant AND Dominant, and Recessive OR Recessive, and r = 0.5 for Threshold models, respectively. Again, we observed that the power of the two-trait FRG was the highest.
Fig 6. Power curves as a function of sample sizes under Dominant OR Dominant with two genes including rare variants only.
Application to Real Data Examples
To further evaluate the performance, the MFRG for testing epistasis was applied to data from the NHLBI’s ESP Project. Five phenotypes: HDL, LDL, total cholesterol, SBP and DBP were considered with a total of 2,016 individuals of European origin from 15 different cohorts in the ESP Project. No evidence of cohort- and/or phenotype-specific effects, or other systematic biases was found [34]. Exomes from related individuals were excluded from further analysis. We took the rank-based inverse normal transformation of the phenotypes [35] as trait values. The total number of genes tested for interactions which included both common and rare variants was 18,587. The remaining annotated human genes which did not contain any SNPs in our dataset were excluded from the analysis. A P-value for declaring significant interaction after applying the Bonferroni correction for multiple tests was 2.89×10−10. Population stratification may inflate the test statistics. To reduce the inflation, the standard strategy is to adjust for population stratification via principal components. All the tests were adjusted for sex, age and population stratification via 5 principal components.
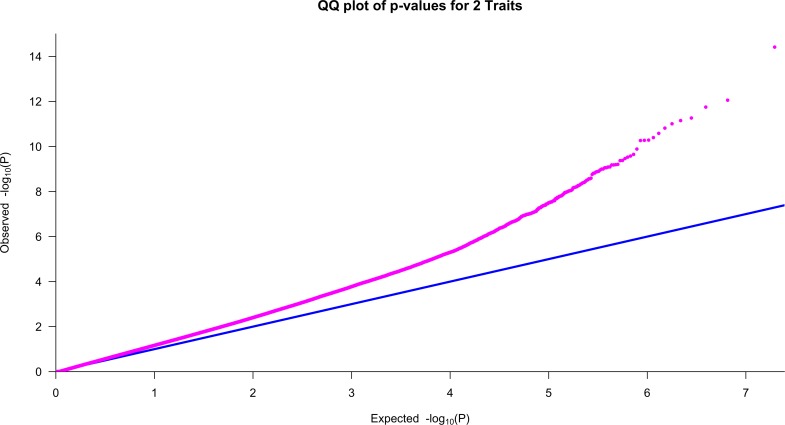
To examine the behavior of the MFRG, we plotted the QQ plot of the two-trait FRG test (Fig 7). The QQ plots showed that the false positive rate of the MFRG for detection of interaction in some degree is controlled.
Fig 7. QQ plot of the two-trait FRG test adjusted for sex, age and population stratification via five PCs.
A total of 91 pairs of genes which were derived from 85 genes showed significant evidence of epistasis with P-values < 2.7×10−10 which were calculated using the MFRG model and simultaneously analyzing interaction of inverse normally transformed HDL and LDL (S10 Table). The top 30 pairs of significantly interacted genes with HDL and LDL were listed in Table 5. In Table 5 and S10 Table, P-values for testing interactions between genes by regression on PCA and the minimum of P-values for testing all possible pairs of SNPs between two genes using standard regression model simultaneously analyzed for the HDL and LDL and P-values for testing epistasis by the FRG separately against single trait HDL or LDL were also listed.
Table 5. P-values of top 30 pairs of significantly interacted genes with HDL and LDL after adjusting for sex, age and population stratification via five PCs.
| Gene 1 | Chr | Gene 2 | Chr | P-values | ||||
|---|---|---|---|---|---|---|---|---|
| Two Traits | HDL | LDL | ||||||
| MFRG | Pair-wise | PCA | FRG | FRG | ||||
| (minimum) | ||||||||
| SHPK | 17 | ST20 | 15 | 1.42E-19 | 4.30E-04 | 3.03E-02 | 2.36E-08 | 6.48E-11 |
| STK3 | 8 | CSMD1 | 8 | 5.98E-16 | 5.58E-04 | 1.76E-01 | 2.82E-07 | 6.06E-05 |
| ST20 | 15 | FRMD5 | 15 | 6.97E-15 | 7.14E-04 | 2.13E-03 | 9.36E-07 | 4.03E-07 |
| C5orf64 | 5 | PSMD1 | 2 | 9.81E-15 | 1.21E-05 | 2.64E-01 | 1.01E-06 | 3.75E-07 |
| ST20 | 15 | PDE4DIP | 1 | 3.65E-14 | 3.42E-06 | 2.18E-01 | 1.88E-03 | 1.95E-08 |
| SHPK | 17 | CSMD1 | 8 | 2.44E-13 | 2.37E-04 | 4.91E-05 | 1.26E-03 | 2.93E-04 |
| C5orf64 | 5 | SPRY1 | 4 | 2.64E-13 | 1.21E-05 | 2.94E-03 | 1.12E-06 | 1.67E-07 |
| NARG2 | 15 | CSMD1 | 8 | 3.77E-13 | 8.07E-05 | 4.74E-02 | 2.24E-03 | 1.11E-04 |
| SIGLEC7 | 19 | NBPF1 | 1 | 4.03E-13 | 6.34E-04 | 2.64E-02 | 5.04E-06 | 5.90E-07 |
| SHPK | 17 | NRG1 | 8 | 5.33E-13 | 3.91E-04 | 3.73E-03 | 2.48E-04 | 1.58E-07 |
| PLTP | 20 | NBPF1 | 1 | 8.35E-13 | 2.03E-03 | 2.03E-01 | 6.77E-06 | 6.34E-05 |
| DIAPH3-AS1 | 13 | SPRY1 | 4 | 1.06E-12 | 1.35E-05 | 5.48E-04 | 1.35E-04 | 1.34E-06 |
| MPG | 16 | NBPF1 | 1 | 1.11E-12 | 3.77E-03 | 5.56E-01 | 3.34E-04 | 2.18E-07 |
| FRMD5 | 15 | SLC8A3 | 14 | 2.92E-12 | 7.85E-05 | 9.79E-04 | 2.99E-03 | 1.67E-08 |
| DIAPH3-AS1 | 13 | PPRC1 | 10 | 3.57E-12 | 9.90E-04 | 7.71E-01 | 3.33E-05 | 2.01E-06 |
| DIAPH3-AS1 | 13 | STK3 | 8 | 3.57E-12 | 3.64E-03 | 4.74E-01 | 1.82E-06 | 5.95E-05 |
| CD300A | 17 | CSMD1 | 8 | 3.89E-12 | 3.19E-04 | 2.59E-01 | 3.65E-04 | 1.71E-04 |
| RNF40 | 16 | DIAPH3-AS1 | 13 | 4.09E-12 | 3.39E-03 | 2.68E-01 | 5.05E-05 | 1.37E-07 |
| CGB2 | 19 | CSMD1 | 8 | 4.38E-12 | 1.82E-04 | 3.19E-04 | 1.87E-05 | 5.23E-06 |
| SHPK | 17 | RYR3 | 15 | 4.47E-12 | 3.25E-04 | 2.47E-05 | 1.14E-02 | 7.29E-05 |
| FRMD5 | 15 | C5orf64 | 5 | 4.75E-12 | 1.42E-02 | 3.65E-01 | 8.14E-06 | 4.60E-05 |
| PPM1A | 14 | CSMD1 | 8 | 4.77E-12 | 6.84E-05 | 2.37E-03 | 1.09E-06 | 2.46E-05 |
| CSMD1 | 8 | ZBTB47 | 3 | 5.53E-12 | 4.02E-06 | 2.71E-04 | 9.85E-07 | 1.78E-03 |
| ST20 | 15 | PSMD1 | 2 | 5.75E-12 | 1.06E-05 | 9.13E-02 | 4.41E-06 | 4.93E-06 |
| CSMD1 | 8 | KIF3A | 5 | 6.56E-12 | 9.52E-04 | 2.35E-02 | 7.04E-03 | 1.16E-02 |
| TRIM22 | 11 | SORCS2 | 4 | 6.62E-12 | 1.05E-04 | 2.01E-01 | 2.98E-07 | 1.52E-05 |
| CREBBP | 16 | CSMD1 | 8 | 7.56E-12 | 2.64E-05 | 2.77E-12 | 2.78E-03 | 3.16E-03 |
| ADRA1B | 5 | PSMD1 | 2 | 9.05E-12 | 1.08E-05 | 5.86E-01 | 1.70E-05 | 1.04E-05 |
| TRIM22 | 11 | STK3 | 8 | 1.17E-11 | 3.46E-04 | 5.43E-02 | 1.48E-06 | 6.03E-05 |
| DIAPH3-AS1 | 13 | SH2B3 | 12 | 1.25E-11 | 5.62E-04 | 3.94E-01 | 5.91E-06 | 6.44E-05 |
Several remarkable features from these results were observed. First, we observed that although pairs of genes showed no strong evidence of interactions influencing individual trait HDL or LDL, they indeed demonstrated significant interactions if interactions were simultaneously analyzed for correlated HDL and LDL. Second, the MFRG often had a much smaller P-value to detect interaction than regression on the PCA and the minimum of P-values of pair-wise tests.
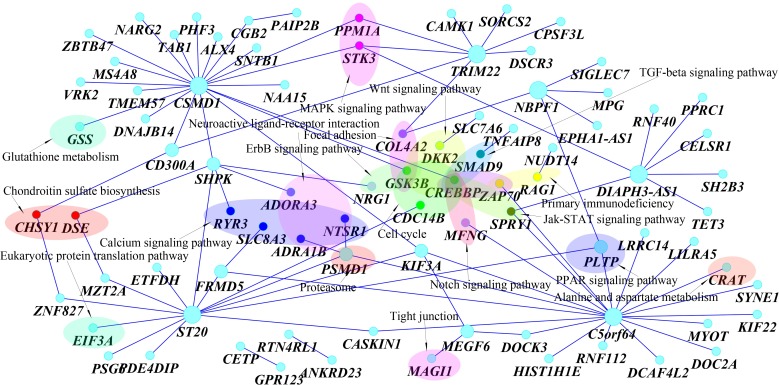
Third, pairs of SNPs between two genes jointly have significant interaction effects, but individually each pair of SNPs make mild contributions to the interaction effects as shown in Table 6. There were a total of 60 pairs of SNPs between genes CETP on chromosome 16 and GPR123 on chromosome 10 with P-values < 0.0488. None of the 60 pairs of SNPs showed strong evidence of interaction. However, a number of pairs of SNPs between genes CETP and GPR123 collectively demonstrated significant interaction influencing the traits HDL and LDL. Fourth, 91 pairs of interacting genes formed a network (Fig 8). The genes C5orf64 that had interactions with 19 genes, CSMD1 that had interactions with 20 genes, were hub genes in the network. 26 genes out of total 85 genes in the network were mainly located in 18 pathways. Each of 12 pathways included at least two interacting genes. However, the majority of interacting genes are located in different pathways. Among 18 pathways, calcium signaling pathway mediates the effect of LDL and plays a role in control of atherosclerosis susceptibility [36], LDL-cholesterol has multiple roles in regulating focal adhesion dynamics [37], LDL is involved in free radical induced apoptosis pathway [38], MAPK and JAK-STAT pathways are involved in dietary flavonoid protection against oxidized LDL [39], up-regulation of autophagy via AMPK/mTOR signaling pathway alleviates oxidized -LDL induced inflammation [40], PPARα holds a fundamental role in control of lipid homeostasis [41] and lectin-like ox-LDL receptor 1 mediates PKC-α/ERK/PPAR-γ/MMP pathway [42], HDL reduces the TGF-β1-induced collagen deposition [43], the Wnt pathway plays an important role in lipid storage and homeostasis [44], From the literatures, we found that both common and rare variants in CETP were associated with the HDL [45], CREBBP regulated LDL receptor transcription [46], PLTP was associated with HDL and LDL [47], TMEM57 was associated with serum lipid levels [48], SH2B3 was associated with LDL cholesterol [49]. It was also reported that CSMD1 was associated with multivariate phenotype defined as low levels of low density lipoprotein cholesterol (LDL-C < or = 100 mg/dl) and high levels of triglycerides (TG > or = 180 mg/dl) [50], associated with hypertension [51]. It was also reported that CSMD1 was associated with LDL and total cholesterol [52].
Table 6. P-values of 60 pairs of SNPs between genes CETP on chromosome 16 and GPR123 on chromosome 10 for testing interaction affecting both HDL and LDL.
| Gene 1 | Gene 2 | P-value | ||||
|---|---|---|---|---|---|---|
| CETP | GPR123 | 8.83E-11 | ||||
| SNP1 | BP | MAF | SNP2 | BP | MAF | P-Value |
| rs9930761 | 57007192 | 0.0672123 | rs367825198 | 134940686 | 0.00024802 | 4.53E-05 |
| rs5883 | 57007353 | 0.0577877 | rs367825198 | 134940686 | 0.00024802 | 4.90E-05 |
| rs148628525 | 56995963 | 0.00024802 | rs11101914 | 134910629 | 0.25198413 | 1.14E-03 |
| rs1800777 | 57017319 | 0.03497024 | rs2806452 | 134942166 | 0.36383929 | 2.58E-03 |
| rs1800774 | 57015545 | 0.34945437 | rs115735367 | 134940724 | 0.0014881 | 3.03E-03 |
| rs140547417 | 57009022 | 0.00124008 | rs2806452 | 134942166 | 0.36383929 | 3.46E-03 |
| rs5883 | 57007353 | 0.0577877 | rs12219529 | 134916366 | 0.14409722 | 3.69E-03 |
| rs140547417 | 57009022 | 0.00124008 | rs11101914 | 134910629 | 0.25198413 | 3.94E-03 |
| rs5883 | 57007353 | 0.0577877 | rs2806453 | 134942319 | 0.04464286 | 5.22E-03 |
| rs9930761 | 57007192 | 0.0672123 | rs115735367 | 134940724 | 0.0014881 | 5.98E-03 |
| rs5883 | 57007353 | 0.0577877 | rs2806452 | 134942166 | 0.36383929 | 6.07E-03 |
| rs1800777 | 57017319 | 0.03497024 | rs10776696 | 134942340 | 0.11433532 | 6.12E-03 |
| rs1532625 | 57005301 | 0.41914683 | rs115735367 | 134940724 | 0.0014881 | 7.16E-03 |
| rs9930761 | 57007192 | 0.0672123 | rs118125186 | 134912135 | 0.00421627 | 7.23E-03 |
| rs5883 | 57007353 | 0.0577877 | rs118125186 | 134912135 | 0.00421627 | 7.50E-03 |
| rs1532625 | 57005301 | 0.41914683 | rs145543174 | 134941843 | 0.00049603 | 7.90E-03 |
| rs1532625 | 57005301 | 0.41914683 | rs118125186 | 134912135 | 0.00421627 | 8.49E-03 |
| rs140547417 | 57009022 | 0.00124008 | rs45586231 | 134942832 | 0.06547619 | 8.93E-03 |
| rs9930761 | 57007192 | 0.0672123 | rs12219529 | 134916366 | 0.14409722 | 9.17E-03 |
| rs1532625 | 57005301 | 0.41914683 | rs45586231 | 134942832 | 0.06547619 | 9.23E-03 |
| rs34065661 | 56995935 | 0.00124008 | rs115735367 | 134940724 | 0.0014881 | 9.56E-03 |
| rs140547417 | 57009022 | 0.00124008 | rs11101942 | 134940862 | 0.11929563 | 9.62E-03 |
| rs9930761 | 57007192 | 0.0672123 | rs2806452 | 134942166 | 0.36383929 | 9.69E-03 |
| rs140547417 | 57009022 | 0.00124008 | rs10776696 | 134942340 | 0.11433532 | 9.76E-03 |
| rs1532625 | 57005301 | 0.41914683 | rs4838796 | 134912098 | 0.03298611 | 1.12E-02 |
| rs1800774 | 57015545 | 0.34945437 | rs118125186 | 134912135 | 0.00421627 | 1.19E-02 |
| rs1800774 | 57015545 | 0.34945437 | rs2806452 | 134942166 | 0.36383929 | 1.33E-02 |
| rs5883 | 57007353 | 0.0577877 | rs11101916 | 134912314 | 0.1703869 | 1.37E-02 |
| rs5883 | 57007353 | 0.0577877 | rs11101942 | 134940862 | 0.11929563 | 1.46E-02 |
| rs5883 | 57007353 | 0.0577877 | rs45586231 | 134942832 | 0.06547619 | 1.59E-02 |
| rs371233223 | 57005272 | 0.00024802 | rs11101916 | 134912314 | 0.1703869 | 1.68E-02 |
| rs371233223 | 57005272 | 0.00024802 | rs2806452 | 134942166 | 0.36383929 | 1.70E-02 |
| rs1532625 | 57005301 | 0.41914683 | rs2806452 | 134942166 | 0.36383929 | 1.75E-02 |
| rs13306230 | 57003250 | 0.00124008 | rs2806452 | 134942166 | 0.36383929 | 2.21E-02 |
| rs34611098 | 57004951 | 0.00124008 | rs2806452 | 134942166 | 0.36383929 | 2.21E-02 |
| rs5880 | 57015091 | 0.04861111 | rs2806452 | 134942166 | 0.36383929 | 2.23E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs11101941 | 134940779 | 0.01116071 | 2.28E-02 |
| rs1800777 | 57017319 | 0.03497024 | rs118125186 | 134912135 | 0.00421627 | 2.33E-02 |
| rs5880 | 57015091 | 0.04861111 | rs118125186 | 134912135 | 0.00421627 | 2.35E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs2806453 | 134942319 | 0.04464286 | 2.47E-02 |
| rs182237338 | 57012174 | 0.00198413 | rs12219529 | 134916366 | 0.14409722 | 2.57E-02 |
| rs5883 | 57007353 | 0.0577877 | rs11101941 | 134940779 | 0.01116071 | 2.68E-02 |
| rs5880 | 57015091 | 0.04861111 | rs11101916 | 134912314 | 0.1703869 | 2.94E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs45586231 | 134942832 | 0.06547619 | 2.95E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs11101914 | 134910629 | 0.25198413 | 3.19E-02 |
| rs376545293 | 57016085 | 0.00024802 | rs11101916 | 134912314 | 0.1703869 | 3.23E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs11101942 | 134940862 | 0.11929563 | 3.24E-02 |
| rs376545293 | 57016085 | 0.00024802 | rs11101914 | 134910629 | 0.25198413 | 3.28E-02 |
| rs139594305 | 57007286 | 0.00024802 | rs4838796 | 134912098 | 0.03298611 | 3.46E-02 |
| rs376545293 | 57016085 | 0.00024802 | rs2806452 | 134942166 | 0.36383929 | 3.48E-02 |
| rs139594305 | 57007286 | 0.00024802 | rs12219529 | 134916366 | 0.14409722 | 3.55E-02 |
| rs201267603 | 57005220 | 0.00099206 | rs12219529 | 134916366 | 0.14409722 | 3.85E-02 |
| rs13306230 | 57003250 | 0.00124008 | rs11101916 | 134912314 | 0.1703869 | 4.00E-02 |
| rs34611098 | 57004951 | 0.00124008 | rs11101916 | 134912314 | 0.1703869 | 4.00E-02 |
| rs9930761 | 57007192 | 0.0672123 | rs11101916 | 134912314 | 0.1703869 | 4.11E-02 |
| rs1800774 | 57015545 | 0.34945437 | rs189113844 | 134941821 | 0.00173611 | 4.35E-02 |
| rs1800774 | 57015545 | 0.34945437 | rs11101941 | 134940779 | 0.01116071 | 4.38E-02 |
| rs28381708 | 57007413 | 0.00124008 | rs11101914 | 134910629 | 0.25198413 | 4.57E-02 |
| rs34855278 | 57015076 | 0.0014881 | rs11101914 | 134910629 | 0.25198413 | 4.69E-02 |
| rs1532625 | 57005301 | 0.41914683 | rs10776696 | 134942340 | 0.11433532 | 4.88E-02 |
Fig 8. Networks of 91 pairs of genes showing significant evidence of interactions as identified by MFRG.
Next we analyzed five traits: HDL, LDL, SBP, DBP and TOTCHOL. Again, for each trait, inverse normal rank transformation was conducted to ensure that the normality assumption of the transformed trait variable was valid. To examine the behavior of the MFRG, we plotted QQ plot of the test (S8 Fig). The QQ plots showed that the false positive rate of the MFRG for detection of interaction is controlled.
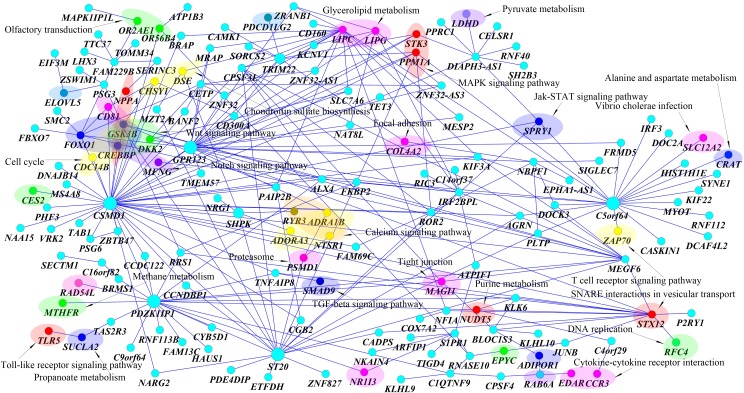
A total of 267 pairs of genes which were derived from 160 genes showed significant evidence of epistasis influencing five traits with P-values < 1.96×10−10 which were calculated using the MFRG model (S11 Table). Of them formed a largest connected subnetwork (Fig 9). The top 25 pairs of significantly interacted genes with five traits were listed in Table 7. We observed the same pattern as was observed for the two traits: HDL and LDL. 46 genes out of 160 genes in the networks were mainly located in 42 pathways including 15 signaling pathways. Among them, 14 pathways were in Fig 8. The interacting genes may be involved in the same biological pathway or in the different biological pathways. We observed 12 pathways, each of which contained at least two genes connected via interaction. However, the majority of interacting genes were not located in the same pathways.
Fig 9. Networks of 267 pairs of genes showing significant evidence of interactions as identified by MFRG.
Table 7. P-values of top 25 pairs of significantly interacted genes with five traits.
| Gene 1 | Gene 2 | P-values | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Five Traits | LDL | HDL | SBP | DBP | TOTCHOL | ||||
| MFRG | Pair-wise(min) | PCA | FRG | FRG | FRG | FRG | FRG | ||
| PDZK1IP1 | CSMD1 | 4.29E-35 | 5.61E-16 | 1.36E-10 | 4.52E-03 | 6.44E-02 | 8.62E-03 | 2.43E-03 | 1.19E-03 |
| STK3 | CSMD1 | 5.84E-34 | 3.39E-05 | 5.81E-03 | 6.06E-05 | 2.82E-07 | 1.37E-05 | 1.79E-05 | 3.22E-04 |
| MEGF6 | IRF2BPL | 1.51E-31 | 2.55E-23 | 3.33E-18 | 7.65E-01 | 8.99E-02 | 2.75E-03 | 4.07E-02 | 1.25E-01 |
| PLTP | C5orf64 | 1.52E-31 | 2.96E-03 | 9.32E-01 | 1.77E-04 | 3.39E-07 | 3.37E-08 | 1.61E-06 | 3.26E-04 |
| CSMD1 | CCNDBP1 | 3.57E-31 | 1.66E-05 | 9.71E-04 | 1.14E-04 | 3.57E-03 | 8.80E-05 | 4.84E-04 | 8.23E-04 |
| KIF3A | C5orf64 | 1.06E-30 | 4.94E-05 | 5.63E-02 | 7.44E-05 | 1.21E-06 | 2.03E-03 | 1.50E-03 | 1.05E-03 |
| CSMD1 | KIF3A | 1.10E-30 | 3.97E-05 | 1.13E-05 | 1.16E-02 | 7.04E-03 | 3.60E-01 | 3.54E-01 | 1.42E-02 |
| ST20 | PDE4DIP | 1.93E-30 | 1.14E-06 | 6.03E-02 | 1.95E-08 | 1.88E-03 | 2.06E-03 | 3.22E-03 | 2.11E-06 |
| CSMD1 | NARG2 | 2.86E-30 | 3.97E-05 | 1.90E-06 | 1.11E-04 | 2.24E-03 | 1.78E-02 | 1.90E-03 | 3.56E-04 |
| PDZK1IP1 | ST20 | 3.17E-30 | 9.82E-16 | 2.09E-03 | 6.15E-03 | 1.34E-02 | 2.30E-03 | 7.63E-03 | 3.30E-03 |
| CSMD1 | FOXO1 | 3.32E-30 | 3.01E-19 | 2.59E-08 | 1.49E-06 | 2.61E-06 | 1.49E-05 | 9.21E-07 | 8.65E-07 |
| SHPK | ST20 | 1.11E-29 | 3.27E-03 | 9.68E-02 | 6.48E-11 | 2.36E-08 | 4.26E-04 | 8.34E-05 | 1.63E-09 |
| DIAPH3-AS1 | SPRY1 | 2.65E-29 | 5.40E-08 | 2.07E-06 | 1.34E-06 | 1.35E-04 | 2.47E-02 | 8.94E-02 | 7.67E-05 |
| PLTP | NBPF1 | 4.60E-29 | 2.92E-03 | 4.62E-01 | 6.34E-05 | 6.77E-06 | 2.85E-04 | 8.16E-03 | 3.93E-05 |
| CREBBP | CSMD1 | 2.18E-28 | 2.34E-05 | 5.30E-26 | 3.16E-03 | 2.78E-03 | 1.33E-01 | 6.30E-02 | 2.21E-03 |
| TAB1 | CSMD1 | 2.21E-28 | 1.33E-03 | 1.34E-04 | 5.12E-02 | 2.51E-02 | 1.48E-02 | 6.86E-02 | 4.17E-02 |
| PAIP2B | CSMD1 | 2.79E-28 | 2.54E-07 | 3.24E-12 | 1.02E-06 | 3.85E-06 | 1.24E-08 | 8.12E-08 | 1.38E-05 |
| CSMD1 | PHF3 | 2.95E-28 | 1.31E-04 | 7.20E-09 | 3.61E-03 | 6.28E-02 | 9.38E-02 | 6.87E-02 | 1.82E-03 |
| CCNDBP1 | ST20 | 3.19E-28 | 3.57E-03 | 1.32E-03 | 2.08E-05 | 1.32E-07 | 2.77E-06 | 1.17E-06 | 2.30E-05 |
| SHPK | CSMD1 | 8.06E-28 | 1.90E-03 | 1.77E-05 | 2.93E-04 | 1.26E-03 | 2.88E-02 | 2.68E-02 | 4.14E-04 |
| ROR2 | IRF2BPL | 9.44E-28 | 2.02E-23 | 2.71E-17 | 5.04E-02 | 1.17E-01 | 2.30E-01 | 2.20E-01 | 9.30E-03 |
| NARG2 | ST20 | 1.25E-27 | 4.69E-05 | 1.07E-02 | 1.00E-04 | 2.46E-04 | 6.28E-05 | 1.28E-05 | 3.14E-05 |
| TRIM22 | CPSF3L | 1.96E-27 | 1.03E-18 | 4.10E-05 | 2.39E-05 | 9.32E-05 | 5.34E-05 | 2.50E-05 | 3.09E-05 |
| ST20 | PSMD1 | 5.40E-27 | 6.32E-08 | 2.73E-02 | 4.93E-06 | 4.41E-06 | 2.31E-03 | 1.66E-05 | 1.24E-05 |
| KIF3A | MEGF6 | 7.65E-27 | 4.46E-06 | 6.84E-03 | 3.22E-04 | 1.07E-04 | 3.68E-02 | 7.89E-02 | 6.58E-04 |
Again, we observed that pairs of SNPs between two genes jointly have significant interaction effects, but individually each pair of SNPs might make mild contributions to the interaction effects as shown in S12 Table. There were a total of 6,766 pairs of SNPs between genes CSMD1 and FOXO1. S12 Table listed 101 pairs of SNPs with P-values < 0.049. The majority of the 101 pairs of SNPs showed no strong evidence of interaction. However, they collectively demonstrated significant interaction influencing five traits.
Among 42 pathways, in the previous sections we reported that 14 pathways were associated with HDL and LDL. From the literatures, we also know that unsaturated fatty acids stimulated the uptake of the LDL particles [53], PPAR signaling pathway was correlated with blood pressure [54], purine metabolism was associated with SBP [55], Wnt signaling pathway mediated cholesterol transportation [56], glycerolipid metabolism pathway was correlated with total cholesterol [57], focal adhesion pathway was involved in lipid modulation [58], Cell adhesion molecules was correlated with blood pressure [59].
We also observed from the literatures that a number of genes that appeared in the list of interacted genes with five traits had major genetic effects with single trait. Many reports showed that CETP, LIPC and LIPG were associated with HDL and LDL [60–62] and that MTHRR had known main effects for LDL [63] and blood pressure [64], NR1I3 for lipid metabolism [65], PLTP for LDL [66],[67], FOXO1 for LDL [68] and hypertension [69], SMAD9 for hypertension [70], and CSMD1 for SBP [51].
Discussion
Most genetic analyses of phenotypes have focused on analyzing single traits or, analyzing each phenotype independently. However, multiple phenotypes are highly correlated. Genetic variants can be associated with more than one trait. Genetic pleiotropic effects likely play a crucial role in the molecular basis of correlated phenotypes. To address these central themes and critical barriers in interaction analysis of multiple phenotypes, we shift the paradigm of interaction analysis from individual interaction analysis to pleiotropic interaction analysis and uncover the global organization of biological systems. MFRG was used to develop a novel statistical framework for joint interaction analysis of multiple correlated phenotypes. By large simulations and real data analysis the merits and limitations of the proposed new paradigm of joint interaction analysis of multiple phenotypes were demonstrated.
The new approach fully uses all phenotype correlation information to jointly analyze interaction of multiple phenotypes. By large simulations and real data analysis, we showed that the proposed MFRG for joint interaction analysis of correlated multiple phenotypes substantially increased the power to detect interaction while keeping the Type 1 error rates of the test statistics under control. In real data analysis, we observed that although pairs of genes showed no strong evidence of interactions influencing individual trait, they indeed demonstrated significant interactions if interactions were simultaneously analyzed for correlated multiple traits.
Due to lack of power of the widely used statistics for testing interaction between loci and its computational intensity, exploration of genome-wide gene-gene interaction has been limited. Few significant interaction results have been observed. Many geneticists question the universe presence of significant gene-gene interaction. Our analysis showed that although the number of significantly interacted genes for single phenotype was small, the number of significantly interacted genes for multiple phenotypes substantially increased. Our results suggested that joint interaction analysis of multiple phenotypes should be advocated in future genetic studies of complex traits.
The interaction analysis for multiple phenotypes has been limited to common variants in carefully controlled experimental crosses and has mainly focused on the pair-wise interaction analysis. Although pair-wise interaction analysis is suitable for common variants, it is difficult to use to test interaction between rare and rare variants, and rare and common variants. There is an increasing need to develop statistics that can be used to test interactions among the entire allelic spectrum of variants for joint interaction analysis of multiple phenotypes. The MFRG utilizes the merits of taking genotype as functions and decomposes position varying genotype function into orthogonal eigenfunctions of genomic position. Only a few eigenfunctions that capture major information on genetic variation across the gene, are used to model the genetic variation. This substantially reduces the dimension in genetic variation of the data. The MFRG can efficiently test the interaction between rare and rare, rare and common, and common and common variants.
In both real data analysis of two phenotypes and five phenotypes, the interacted genes formed interaction networks. Hub genes in the interaction networks were also observed. These hub genes usually play an important biological role in causing phenotype variation.
An essential issue for interaction analysis of a large number of phenotypes is how to reduce dimension while fully exploiting complementary information in multiple phenotypes. The standard multivariate regression models for joint interaction analysis of multiple phenotypes do not explore the correlation structures of multiple phenotypes and reduce the dimensions of the phenotypes, and hence have limited power to detect pleotropic interaction effects due to large degrees of freedom. Data reduction techniques such as principal component analysis should be explored in the future interaction analysis of multiple phenotypes.
The results in this paper are preliminary. The current marginal approaches for interaction analysis cannot distinguish between direct and indirect interactions, which will decrease our power to unravel mechanisms underlying complex traits. To overcome these limitations, causal inference tools should be explored for the joint interaction analysis of multiple phenotypes. The purpose of this paper is to stimulate further discussions regarding great challenges we are facing in the interaction analysis of high dimensional phenotypic and genomic data produced by modern sensors and next-generation sequencing.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
Authors thank two anonymous reviewers for the thorough reading of the manuscript and thoughtful suggestions that improved the manuscript.
Data Availability
This study uses data from the NHLBI’s Exome Sequencing Project (ESP). These confidential data are available to interested researchers through application to NIH data Access Committees for authorization. Data access is provided via dbGaP Authorized Access. Contact information for the NIH data access commitees can be found at https://gds.nih.gov/04po2_1DAC.html
Funding Statement
This work was supported by the National Institute of Health, Grants 1R01AR057120–01 and 1R01HL106034-01 to MX (http://www.nih.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wei WH, Hemani G, Haley CS (2014) Detecting epistasis in human complex traits. Nat Rev Genet 15: 722–733. 10.1038/nrg3747 [DOI] [PubMed] [Google Scholar]
- 2.Cordell HJ (2002) Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet 11: 2463–2468. [DOI] [PubMed] [Google Scholar]
- 3.Cordell HJ (2009) Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet 10: 392–404. 10.1038/nrg2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemani G, Theocharidis A, Wei W, Haley C (2011) EpiGPU: exhaustive pairwise epistasis scans parallelized on consumer level graphics cards. Bioinformatics 27: 1462–1465. 10.1093/bioinformatics/btr172 [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Clark AG, Keinan A (2013) Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet 9: e1003321 10.1371/journal.pgen.1003321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schupbach T, Xenarios I, Bergmann S, Kapur K (2010) FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics 26: 1468–1469. 10.1093/bioinformatics/btq147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yung LS, Yang C, Wan X, Yu W (2011) GBOOST: a GPU-based tool for detecting gene-gene interactions in genome-wide case control studies. Bioinformatics 27: 1309–1310. 10.1093/bioinformatics/btr114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Boerwinkle E, Xiong M (2014) Epistasis analysis for quantitative traits by functional regression model. Genome Res 24: 989–998. 10.1101/gr.161760.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu JK, Wang X, Wang P (2014) Testing gene-gene interactions in genome wide association studies. Genet Epidemiol 38: 123–134. 10.1002/gepi.21786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kam-Thong T, Czamara D, Tsuda K, Borgwardt K, Lewis CM, et al. (2011) EPIBLASTER-fast exhaustive two-locus epistasis detection strategy using graphical processing units. Eur J Hum Genet 19: 465–471. 10.1038/ejhg.2010.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Zhang K, Yi N (2011) A Bayesian hierarchical model for detecting haplotype-haplotype and haplotype-environment interactions in genetic association studies. Hum Hered 71: 148–160. 10.1159/000324841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueki M, Cordell HJ (2012) Improved statistics for genome-wide interaction analysis. PLoS Genet 8: e1002625 10.1371/journal.pgen.1002625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Dong H, Luo L, Zhu Y, Peng G, et al. (2010) A novel statistic for genome-wide interaction analysis. PLoS Genet 6: e1001131 10.1371/journal.pgen.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y (2012) A novel bayesian graphical model for genome-wide multi-SNP association mapping. Genet Epidemiol 36: 36–47. 10.1002/gepi.20661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Jin L, Xiong M (2006) Test for interaction between two unlinked loci. Am J Hum Genet 79: 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu T, Chen Y, Kiralis JW, Collins RL, Wejse C, et al. (2013) An information-gain approach to detecting three-way epistatic interactions in genetic association studies. J Am Med Inform Assoc 20: 630–636. 10.1136/amiajnl-2012-001525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knights J, Yang J, Chanda P, Zhang A, Ramanathan M (2013) SYMPHONY, an information-theoretic method for gene-gene and gene-environment interaction analysis of disease syndromes. Heredity (Edinb) 110: 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahachie John JM, Van Lishout F, Van Steen K (2011) Model-Based Multifactor Dimensionality Reduction to detect epistasis for quantitative traits in the presence of error-free and noisy data. Eur J Hum Genet 19: 696–703. 10.1038/ejhg.2011.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lishout F, Mahachie John JM, Gusareva ES, Urrea V, Cleynen I, et al. (2013) An efficient algorithm to perform multiple testing in epistasis screening. BMC Bioinformatics 14: 138 10.1186/1471-2105-14-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Tong X, Zhu Z, Liang M, Cui W, et al. (2013) Development of GMDR-GPU for gene-gene interaction analysis and its application to WTCCC GWAS data for type 2 diabetes. PLoS One 8: e61943 10.1371/journal.pone.0061943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens M (2013) A unified framework for association analysis with multiple related phenotypes. PLoS One 8: e65245 10.1371/journal.pone.0065245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW (2013) Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 14: 483–495. 10.1038/nrg3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W, Chen D, Zhao M, Zou Y, Zeng Y, et al. (2015) Genepleio software for effective estimation of gene pleiotropy from protein sequences. Biomed Res Int 2015: 269150 10.1155/2015/269150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill WG, Zhang XS (2012) On the pleiotropic structure of the genotype-phenotype map and the evolvability of complex organisms. Genetics 190: 1131–1137. 10.1534/genetics.111.135681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ (1992) Major depression and generalized anxiety disorder. Same genes, (partly) different environments? Arch Gen Psychiatry 49: 716–722. [DOI] [PubMed] [Google Scholar]
- 26.Wagner GP, Zhang J (2011) The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet 12: 204–213. 10.1038/nrg2949 [DOI] [PubMed] [Google Scholar]
- 27.Aschard H, Vilhjalmsson BJ, Greliche N, Morange PE, Tregouet DA, et al. (2014) Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies. Am J Hum Genet 94: 662–676. 10.1016/j.ajhg.2014.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schifano ED, Li L, Christiani DC, Lin X (2013) Genome-wide association analysis for multiple continuous secondary phenotypes. Am J Hum Genet 92: 744–759. 10.1016/j.ajhg.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter GW, Hays M, Sherman A, Galitski T (2012) Use of pleiotropy to model genetic interactions in a population. PLoS Genet 8: e1003010 10.1371/journal.pgen.1003010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snitkin ES, Segre D (2011) Epistatic interaction maps relative to multiple metabolic phenotypes. PLoS Genet 7: e1001294 10.1371/journal.pgen.1001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo L, Zhu Y, Xiong M (2012) Quantitative trait locus analysis for next-generation sequencing with the functional linear models. J Med Genet 49: 513–524. 10.1136/jmedgenet-2012-100798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferraty F, Romain Y (2011) The Oxford Handbook of Functional Data Analysis: Oxford University Press. [Google Scholar]
- 33.Vintem AP, Price NT, Silverman RB, Ramsay RR (2005) Mutation of surface cysteine 374 to alanine in monoamine oxidase A alters substrate turnover and inactivation by cyclopropylamines. Bioorg Med Chem 13: 3487–3495. [DOI] [PubMed] [Google Scholar]
- 34.Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, et al. (2012) Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69. 10.1126/science.1219240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beasley TM, Erickson S, Allison DB (2009) Rank-based inverse normal transformations are increasingly used, but are they merited? Behav Genet 39: 580–595. 10.1007/s10519-009-9281-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song W, Wang H, Wu Q (2015) Atrial natriuretic peptide in cardiovascular biology and disease (NPPA). Gene 569: 1–6. 10.1016/j.gene.2015.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Z, Miyoshi T, Bao Y, Sheehan JP, Matsumoto AH, et al. (2009) Microarray analysis of gene expression in mouse aorta reveals role of the calcium signaling pathway in control of atherosclerosis susceptibility. Am J Physiol Heart Circ Physiol 296: H1336–1343. 10.1152/ajpheart.01095.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoque M, Rentero C, Conway JR, Murray RZ, Timpson P, et al. (2015) The cross-talk of LDL-cholesterol with cell motility: insights from the Niemann Pick Type C1 mutation and altered integrin trafficking. Cell Adh Migr 9: 384–391. 10.1080/19336918.2015.1019996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gajaria TK, Patel DK, Devkar RV, Ramachandran AV (2015) Flavonoid rich extract of Murraya Koenigii alleviates in-vitro LDL oxidation and oxidized LDL induced apoptosis in raw 264.7 Murine macrophage cells. J Food Sci Technol 52: 3367–3375. 10.1007/s13197-014-1399-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan X, Wang J, Hou J, Lin C, Bensoussan A, et al. (2015) Berberine alleviates ox-LDL induced inflammatory factors by up-regulation of autophagy via AMPK/mTOR signaling pathway. J Transl Med 13: 92 10.1186/s12967-015-0450-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konstandi M, Shah YM, Matsubara T, Gonzalez FJ (2013) Role of PPARalpha and HNF4alpha in stress-mediated alterations in lipid homeostasis. PLoS One 8: e70675 10.1371/journal.pone.0070675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai KL, Chang YL, Huang PH, Cheng YH, Liu DH, et al. (2016) Ginkgo biloba extract inhibits oxidized low-density lipoprotein (oxLDL)-induced matrix metalloproteinase activation by the modulation of the lectin-like oxLDL receptor 1-regulated signaling pathway in human umbilical vein endothelial cells. J Vasc Surg 63: 204–215 e201. 10.1016/j.jvs.2014.05.098 [DOI] [PubMed] [Google Scholar]
- 43.Spillmann F, Miteva K, Pieske B, Tschope C, Van Linthout S (2015) High-density lipoproteins reduce endothelial-to-mesenchymal transition. Arterioscler Thromb Vasc Biol 35: 1774–1777. 10.1161/ATVBAHA.115.305887 [DOI] [PubMed] [Google Scholar]
- 44.Scott CC, Vossio S, Vacca F, Snijder B, Larios J, et al. (2015) Wnt directs the endosomal flux of LDL-derived cholesterol and lipid droplet homeostasis. EMBO Rep 16: 741–752. 10.15252/embr.201540081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirim D, Wang X, Niemsiri V, Radwan ZH, Bunker CH, et al. (2016) Resequencing of the CETP gene in American whites and African blacks: Association of rare and common variants with HDL-cholesterol levels. Metabolism 65: 36–47. 10.1016/j.metabol.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapoor GS, Golden C, Atkins B, Mehta KD (2003) pp90RSK- and protein kinase C-dependent pathway regulates p42/44MAPK-induced LDL receptor transcription in HepG2 cells. J Lipid Res 44: 584–593. [DOI] [PubMed] [Google Scholar]
- 47.Yan D, Navab M, Bruce C, Fogelman AM, Jiang XC (2004) PLTP deficiency improves the anti-inflammatory properties of HDL and reduces the ability of LDL to induce monocyte chemotactic activity. J Lipid Res 45: 1852–1858. [DOI] [PubMed] [Google Scholar]
- 48.Guo T, Yin RX, Lin QZ, Wu J, Shen SW, et al. (2014) Polymorphism of rs873308 near the transmembrane protein 57 gene is associated with serum lipid levels. Biosci Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, et al. (2009) Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet 85: 628–642. 10.1016/j.ajhg.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SH, Kim S (2012) Pattern discovery of multivariate phenotypes by association rule mining and its scheme for genome-wide association studies. Int J Data Min Bioinform 6: 505–520. [PubMed] [Google Scholar]
- 51.Chittani M, Zaninello R, Lanzani C, Frau F, Ortu MF, et al. (2015) TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J Hypertens 33: 1301–1309. 10.1097/HJH.0000000000000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, et al. (2012) Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 8: e1002629 10.1371/journal.pgen.1002629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo T, Velez-Carrasco W, Qi K, Hall M, Worgall TS, et al. (2002) Selective uptake from LDL is stimulated by unsaturated fatty acids and modulated by cholesterol content in the plasma membrane: role of plasma membrane composition in regulating non-SR-BI-mediated selective lipid transfer. Biochemistry 41: 7885–7894. [DOI] [PubMed] [Google Scholar]
- 54.Yousefipour Z, Newaz M (2014) PPARalpha ligand clofibrate ameliorates blood pressure and vascular reactivity in spontaneously hypertensive rats. Acta Pharmacol Sin 35: 476–482. 10.1038/aps.2013.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mallamaci F, Testa A, Leonardis D, Tripepi R, Pisano A, et al. (2014) A polymorphism in the major gene regulating serum uric acid associates with clinic SBP and the white-coat effect in a family-based study. J Hypertens 32: 1621–1628; discussion 1628. 10.1097/HJH.0000000000000224 [DOI] [PubMed] [Google Scholar]
- 56.Qin L, Hu R, Zhu N, Yao HL, Lei XY, et al. (2014) The novel role and underlying mechanism of Wnt5a in regulating cellular cholesterol accumulation. Clin Exp Pharmacol Physiol 41: 671–678. 10.1111/1440-1681.12258 [DOI] [PubMed] [Google Scholar]
- 57.Rise P, Colombo C, Galli C (1997) Effects of simvastatin on the metabolism of polyunsaturated fatty acids and on glycerolipid, cholesterol, and de novo lipid synthesis in THP-1 cells. J Lipid Res 38: 1299–1307. [PubMed] [Google Scholar]
- 58.Jeon JH, Kim SK, Kim HJ, Chang J, Ahn CM, et al. (2010) Lipid raft modulation inhibits NSCLC cell migration through delocalization of the focal adhesion complex. Lung Cancer 69: 165–171. 10.1016/j.lungcan.2009.10.014 [DOI] [PubMed] [Google Scholar]
- 59.Ballard KD, Quann EE, Kupchak BR, Volk BM, Kawiecki DM, et al. (2013) Dietary carbohydrate restriction improves insulin sensitivity, blood pressure, microvascular function, and cellular adhesion markers in individuals taking statins. Nutr Res 33: 905–912. 10.1016/j.nutres.2013.07.022 [DOI] [PubMed] [Google Scholar]
- 60.Todur SP, Ashavaid TF (2013) Association of CETP and LIPC Gene Polymorphisms with HDL and LDL Sub-fraction Levels in a Group of Indian Subjects: A Cross-Sectional Study. Indian J Clin Biochem 28: 116–123. 10.1007/s12291-012-0259-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ray KK, Vallejo-Vaz AJ (2015) The evolving role of CETP inhibition: beyond HDL cholesterol. Lancet 386: 412–414. 10.1016/S0140-6736(15)60608-0 [DOI] [PubMed] [Google Scholar]
- 62.Khetarpal SA, Edmondson AC, Raghavan A, Neeli H, Jin W, et al. (2011) Mining the LIPG allelic spectrum reveals the contribution of rare and common regulatory variants to HDL cholesterol. PLoS Genet 7: e1002393 10.1371/journal.pgen.1002393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morais CC, Alves MC, Augusto EM, Abdalla DS, Horst MA, et al. (2015) The MTHFR C677T Polymorphism Is Related to Plasma Concentration of Oxidized Low-Density Lipoprotein in Adolescents with Cardiovascular Risk Factors. J Nutrigenet Nutrigenomics 8: 105–113. 10.1159/000439218 [DOI] [PubMed] [Google Scholar]
- 64.Xi B, Shen Y, Zhao X, Chandak GR, Cheng H, et al. (2014) Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children. J Hum Hypertens 28: 32–36. 10.1038/jhh.2013.50 [DOI] [PubMed] [Google Scholar]
- 65.Lima LO, Bruxel EM, Hutz MH, Van der Sand CR, Van der Sand LC, et al. (2013) Influence of PPARA, RXRA, NR1I2 and NR1I3 gene polymorphisms on the lipid-lowering efficacy and safety of statin therapy. Arq Bras Endocrinol Metabol 57: 513–519. [DOI] [PubMed] [Google Scholar]
- 66.Yazdanyar A, Jiang XC (2012) Liver phospholipid transfer protein (PLTP) expression with a PLTP-null background promotes very low-density lipoprotein production in mice. Hepatology 56: 576–584. 10.1002/hep.25648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang XC, Jin W, Hussain MM (2012) The impact of phospholipid transfer protein (PLTP) on lipoprotein metabolism. Nutr Metab (Lond) 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka J, Qiang L, Banks AS, Welch CL, Matsumoto M, et al. (2009) Foxo1 links hyperglycemia to LDL oxidation and endothelial nitric oxide synthase dysfunction in vascular endothelial cells. Diabetes 58: 2344–2354. 10.2337/db09-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savai R, Al-Tamari HM, Sedding D, Kojonazarov B, Muecke C, et al. (2014) Pro-proliferative and inflammatory signaling converge on FoxO1 transcription factor in pulmonary hypertension. Nat Med 20: 1289–1300. 10.1038/nm.3695 [DOI] [PubMed] [Google Scholar]
- 70.Drake KM, Comhair SA, Erzurum SC, Tuder RM, Aldred MA (2015) Endothelial chromosome 13 deletion in congenital heart disease-associated pulmonary arterial hypertension dysregulates SMAD9 signaling. Am J Respir Crit Care Med 191: 850–854. 10.1164/rccm.201411-1985LE [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
This study uses data from the NHLBI’s Exome Sequencing Project (ESP). These confidential data are available to interested researchers through application to NIH data Access Committees for authorization. Data access is provided via dbGaP Authorized Access. Contact information for the NIH data access commitees can be found at https://gds.nih.gov/04po2_1DAC.html