ABSTRACT
In Staphylococcus aureus, peptidoglycan metabolism plays a role in the host inflammatory response and pathogenesis. Transcription of the peptidoglycan hydrolases is activated by the essential 2-component system WalKR at low cell density. During stationary growth phase, WalKR is not active and transcription of the peptidoglycan hydrolase genes is repressed. In this work, we studied regulation of expression of the glycylglycine endopeptidase LytM. We show that, in addition to the transcriptional regulation mediated by WalKR, the synthesis of LytM is negatively controlled by a unique mechanism at the stationary growth phase. We have identified 2 different mRNAs encoding lytM, which vary in the length of their 5′ untranslated (5′UTR) regions. LytM is predominantly produced from the WalKR-regulated mRNA transcript carrying a short 5′UTR. The lytM mRNA is also transcribed as part of a polycistronic operon with the upstream SA0264 gene and is constitutively expressed. Although SA0264 protein can be synthesized from the longer operon transcript, lytM cannot be translated because its ribosome-binding site is sequestered into a translationally inactive secondary structure. In addition, the effector of the agr system, RNAIII, can inhibit translation of lytM present on the operon without altering the transcript level but does not have an effect on the translation of the upstream gene. We propose that this dual regulation of lytM expression, at the transcriptional and post-transcriptional levels, contributes to prevent cell wall damage during the stationary phase of growth.
KEYWORDS: Autolysin, cell wall, LytM, peptidoglycan, peptidoglycan hydrolase, post-transcriptional regulation, RNAIII, Staphylococcus aureus
Introduction
Staphylococcus aureus has evolved multiple strategies to respond to both environmental changes and to colonize/survive within the host. Modulation of the bacterial cell surface composition is expected to play a direct role in these adaptive processes as well as in pathogenesis.1,2 The cell wall of Gram-positive bacteria contains the peptidoglycan (PG) layer, secondary polymers such as teichoic acid, and many anchored proteins, which are directly involved in adhesion, invasion and in sensing external signals. Although the PG layer is quite rigid, its synthesis and turnover are tightly regulated. The complex regulation coordinates a constant cell wall remodeling during cell growth.3,4 This dynamic property of PG is required for several essential functions such as cell growth, to maintain the cell shape, to prevent osmotic lysis, to anchor proteins (some of which have direct roles in virulence) and to facilitate the uptake of nutrients and the export of numerous exotoxins.5,6 Furthermore, PG turnover generates powerful virulence effectors in a variety of pathogenic bacteria such as Streptococcus pneumoniae, Listeria monocytogenes, Helicobacter pylori, Neisseria meningitidis1 and S. aureus.7 Remodeling of the PG in S. aureus is in part achieved by different PG hydrolases (also known as autolysins)8,9 whose functional redundancy may explain why individually, none of them are essential.10-12 The PG hydrolases have also been associated with cell division and separation, protein secretion, biofilm formation13-19 and with the bacterial programmed cell death (reviewed in20).
Within S. aureus, expression of the autolysins is repressed by the action of the 2-component systems ArlSR21 and LytSR,22 and the transcriptional regulator MgrA.23,24 The cidAB and lrgAB operons add another level of regulation since they control the transport of peptidoglycan hydrolases to the membrane (reviewed in6). Conversely, during exponential growth WalKR was identified as a positive regulator that activates the synthesis of the 2 major S. aureus autolysins AtlA (that has amidase and glucosaminidase activities) and LytM (a glycylglycine endopeptidase), as well as 9 other proteins involved in cell wall modification.8,9 Of interest, WalKR is the only essential 2-component system within S. aureus.9 WalKR is not only required for peptidoglycan biosynthesis and turnover but its activity also leads to the stimulation of the 2 component system SaeRS.25 The SaeRS system enhances expression of virulence factors that are involved in biofilm, host-pathogen interaction, and innate immune evasion.26 Hence, WalKR is considered to be a major regulatory system for cell wall metabolism, which may play an important role in switching commensal S. aureus to the pathogenic lifestyle.9,25
The regulatory RNAIII is one of the effectors of the quorum-sensing induced accessory gene regulator (agr) and a major regulator of virulence in S. aureus.27,28 Of interest, we have previously predicted that the 3′ domain of RNAIII might form basepairing interactions with the ribosome binding sites of the mRNAs encoding for the autolysins, SsaA (staphylococcal secretory antigen A), SA2353 (SsaA homolog) and LytM.29 Interestingly, SsaA and LytM are the only 2 proteins that can by-pass WalKR essentiality by modifying the peptidoglycan cross-peptide bridges.30 Other findings have indicated that the translation of lytM mRNA is regulated through RNAIII binding.31 Therefore, in addition to the regulation of several virulence factors,29 RNAIII coordinates the repression of several cell wall hydrolases at the late-exponential phase of growth. As the WalKR system is inactive in the stationary phase of growth, one could argue that an additional level of regulation would not be required to turn off the synthesis of the peptidoglycan hydrolases. However, we show here that post-transcriptional regulation of lytM is required to reinforce the transcriptional regulation mediated by WalKR.
In this work, we have identified 2 different lytM mRNAs, which vary in the length of the 5′ untranslated (5′UTR) regions. LytM is predominantly produced from the mRNA that has a short 5′UTR, which is under the control of WalKR and thus, is growth phase regulated. Additionally, we show that the lytM mRNA is also constitutively produced as part of a bicistronic operon together with the upstream gene SA0264, which encodes a putative penicillin amidase. However, LytM encoded from this operon is poorly translated as evidenced by in vivo and in vitro data. This inhibition is the result of a combination of events that involve both RNA structural changes and RNAIII binding. This unique regulatory mechanism contributes to prevent the synthesis of LytM during the stationary growth phase, which could be detrimental to the cell due to the inappropriate alteration of cell wall structure. We show here that control of one of the major autolysins in S. aureus, LytM, is mediated both at the transcriptional and post-transcriptional levels.
Results
Two lytM transcripts originate from different promoters
The transcription of lytM is activated by WalKR during the exponential phase9 and shuts down during the stationary phase of growth as soon as WalKR is inactivated.32 RNAIII, on the other hand, is predominantly expressed at high cell density, i.e. during the stationary phase of growth27 and was proposed to control the synthesis of LytM at the translational level.29,31 This dual regulation led to the following question: why is there a need for RNAIII-dependent translational repression of lytM in stationary growth phase where lytM levels drop dramatically? In the present study, we have re-evaluated lytM expression during growth in different S. aureus strain backgrounds.
First, we have monitored the steady state levels of lytM mRNA in the RN6390 wild type (wt) strain and in a mutant strain carrying a deletion of rnaIII gene (ΔRNAIII, strain LUG950) using an RNA probe specific to lytM (Fig. 1). RN6390 wt strain expresses high RNAIII levels at the stationary phase of growth due to a mutation inactivating partially σB.33 The ΔRNAIII mutant strain was subsequently transformed with the vector pLUG274 (strain LUG1662) or with the same vector expressing RNAIII under the control of its own P3 promoter (vector pLUG298, strain LUG1855) (Fig. 1). Total RNA extracts were prepared at 2 h and 6 h of growth in BHI medium. In parallel, we monitored the expression of RNAIII and of 5S rRNA using appropriate probes. The results show an abundant level of a lytM transcript around 1000 nts corresponding to the reported WalKR regulated lytM mRNA, called lytM49 as it carries 49 nts in its 5′-UTR.9 This transcript was expressed in all strains during exponential growth phase (EP) and its levels dropped dramatically at stationary phase (SP) (For quantitation of transcript amounts, see Fig. S1B). Of note, we observed a reduction in the steady-state levels of lytM49 transcript in all ΔRNAIII mutant strains as compared to the wt strain in EP (Figs. 1, S1B). This effect is not a consequence of the absence of RNAIII, as complementation with RNAIII in SP of growth does not affect lytM levels (Fig. 1, 6h, lane 4). Unexpectedly, in both phases of growth (EP and SP), we detected a much longer transcript (> 2600 nts) that could correspond to an operon containing lytM together with the upstream gene, annotated as SA0264 in S. aureus strain N315, encoding a putative penicillin amidase. The levels of this long lytM transcript, called SA0264-lytM, varied very little in the strains expressing or not RNAIII (Figs. 1, S1B). We then probed the same samples using an RNA probe specific to SA0264. The data were well correlated with the experiment performed with the lytM probe. Indeed, a transcript was detected with a size (> 2600 nts) similar to SA0264-lytM strongly suggesting that the 2 genes are co-transcribed as part of the same operon. As mentioned above, the expression of this transcript is almost constitutive during growth (Figs. 1, S1B). In another experiment, we also analyzed ΔRNAIII strain complemented with RNAIII lacking the regulatory hairpin 13 (RNAIII-ΔH13, strain LUG1663) that forms basepairings with lytM mRNA,29,34 or complemented with the 3′ domain of RNAIII (hairpins 13 and 14, strain LUG1126) (Fig. S1A). Again, we only observed marginal differences on SA0264-lytM levels between strains carrying or not different versions of RNAIII. The same experiment was performed with the NCTC8325-4 strain35,36 and the corresponding ΔRNAIII mutant strain (WA40037) transformed with the plasmid expressing or not RNAIII (Fig. S1B). The Northern blot data shows very similar expression patterns for lytM49 and SA0264-lytM mRNAs during growth phase as compared to RN6390 (Fig. S1B), and no significant effect of RNAIII was observed on the levels of lytM49 or lytM-SA0264.
Figure 1.
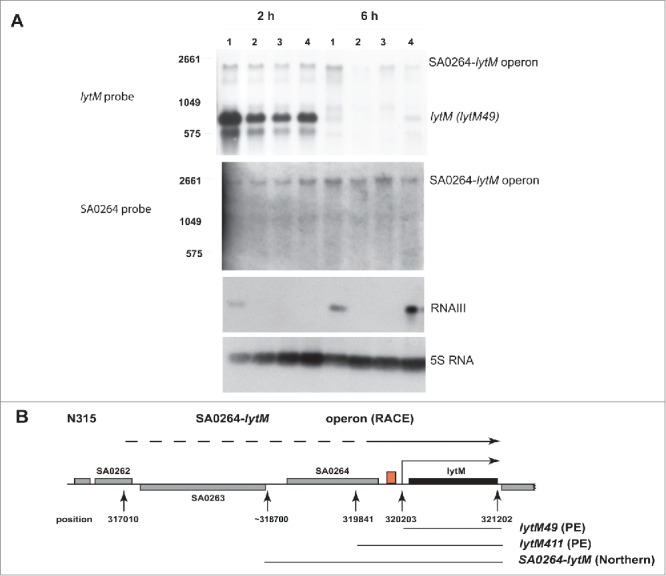
Analysis of lytM expression. (A) Northern blot experiments showing the steady-state levels of lytM mRNA in various strains. Total RNAs were prepared after 2 and 6 h of culture in BHI at 37°C: lane 1, RN6390 WT strain; lane 2, ΔrnaIII mutant strain (LUG950); lane 3, ΔrnaIII mutant strain transformed with the vector with no insert (LUG1662); lane 4, ΔrnaIII mutant strain transformed with the vector expressing wild type RNAIII (LUG1855). RNA probes were used to detect either lytM or SA0264. The transcription of RNAIII and 5S rRNA was also probed. (B) Schematic diagram of the genomic organization at the lytM locus of the S. aureus strain N315. The arrows below the figure indicate the WalKR dependent start site (320203 nts position on the genome, corresponding to lytM49), the primer extension (319841, LytM411), the Northern experiment (∼318700, bicistronic operon) and the 5′RACE (317010, bicistronic operon) identified +1 start sites, respectively. A red box between SA0264 and lytM indicates the position of the previously annotated WalR binding motif and PlytM promoter. 9 The end of the 3′UTR of lytM (321202) is also indicated. The position of the nucleotides is based on the annotation given for S. aureus N315 strain.
We independently evaluated the results by performing qRT-PCR analysis to monitor the expression of lytM in the RN6390 wt and mutant strains (Table 1). For this experiment, we used primers that hybridized either to the coding sequence of lytM (OSA218/OSA219) or between SA0263 and SA0264 (OSA547/OSA548). The qRT-PCR data reproducibly showed that the expression of SA0264-lytM is slightly enhanced at the SP of growth compared to the EP in both the wt and mutant strains whereas lytM49 was 3 times more abundant at the EP than at the SP in the wt strain (Table 1). The overall yield of the lytM49 mRNA in the mutant strains in EP was found decreased compared to the wt strain in agreement with the Northern blot experiment (Fig. 1 and S1B).
Table 1.
Expression of the 2 different lytM transcripts assessed by qRT-PCR in different strains and different phases of growth.
| lytM49 | SA0264-lytM | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean expression ratio (a.u.)1 ± SD |
Mean expression ratio (a.u.)1 ± SD |
|||||||
| WT | ΔRNAIII | ΔRNAIII/p | ΔRNAIII/pRNAIII | WT | ΔRNAIII | ΔRNAIII/p | ΔRNAIII/pRNAIII | |
| EP2 | 3.534±0.25 | 0.982±0.07 | 1.969±0.13 | 0.960±0.08 | 0.630±0.05 | 0.275±0.01 | 0.302±0.03 | 0.169±0.03 |
| SP2 | 1.000±0.7 | 0.500±0.10 | 0.715±0.06 | 1.898±0.36 | 1.000±0.09 | 0.505±0.08 | 0.841±0.08 | 0.765±0.16 |
Mean expression ratios (arbitrary units, a.u.) were calculated using the gene expression of the wild type strain in stationary phase as the reference condition.
EP: exponential phase; SP: stationary phase.
To characterize the 5′ start of the larger SA0264-lytM transcript, we used primer extension assays performed in strains RN6390, WA400 and HG001 (the latter has σB levels restored36 and expresses lower levels of RNAIII compared to RN6390) (Fig. S2). The experiment was also conducted on Δrnc strain carrying a deletion of the double-stranded RNA specific endoribonuclease III (RNase III). RNase III often acts conjointly with RNAIII to regulate expression of virulence factors,29,34 therefore we analyzed the possible impact of RNase III on lytM expression. At the EP of growth, all strains expressed the previously reported +1 transcriptional start of lytM499 (Fig. S2A). As expected, this transcript was not detected by primer extension at the late EP (Fig. S2A). In strains WA400 and HG001, a signal corresponding to a longer transcript was in addition visualized (lytM411, Fig. S2A). The 5′ end of this transcript was mapped within SA0264, at C-411 with regard to the AUG start codon of lytM (Fig. S2B). We additionally performed RACE analysis on total RNA extracts prepared from WA400 strain in SP of growth. Unexpectedly, we identified a long transcript originating upstream SA0264 at position −3242 with regard to the lytM starting codon. This latter transcript begins within SA0262 (encoding a hypothetical protein) and is antisense to SA0263 (encoding a hypothetical protein, similar to proton antiporter efflux pump). A σA promoter consensus was found centered 10 nts upstream the +1 start of the transcript originating at nt −3242. This longer transcript correlates with recent transcriptomic data,38 corroborating our findings that LytM is expressed as part of a large operon including SA0264 (SA0264-lytM). Additionally, we identified a transcript starting at C-356 with regard to the lytM AUG. We suggest that the transcripts with 5′ ends at C-411 and C-356, that were identified by primer extension and RACE, respectively, correspond to processed transcripts from the longer SA0264-lytM transcript (Fig. 1B).
All in all, our data support the existence of 2 mRNAs containing lytM. As expected the WalKR regulated promoter9 leads to a mRNA with a short 5′UTR (lytM49 mRNA) that is primarily expressed during exponential growth. In addition, we show that lytM is also transcribed as part of a polycistronic operon together with SA0264 at both the EP and SP of growth. The steady-state levels of the 2 mRNAs are not significantly dependent on RNAIII.
LytM is mainly synthesized from the short lytM49 transcript
We monitored the levels of LytM protein in the different growth phases using a FLAG-tagged version of LytM expressed from its native locus on the chromosome of S. aureus. We engineered strain HG001 to accept directly DNA from Escherichia coli. The two restriction modification systems reported to inhibit uptake of foreign DNA have been deleted, resulting in strain BCJ100 (ΔhsdR ΔTψπϵιιι).39 Subsequently, a FLAG-tag was introduced in the C-terminus of LytM (strain EL100) to monitor the synthesis of LytM by Western blotting. The same experiment was performed on RN4220 (strain EL101), which is capable of accepting foreign DNA,39,40 and which produces low levels of RNAIII.41
First, we have verified by Northern blot analysis the levels of lytM mRNA in backgrounds HG001 and RN4220 (Fig. 2A). RNAIII and 5S RNA levels were analyzed in parallel using specific probes. As described above, the WalKR driven lytM49 transcript was also the most abundant mRNA observed during exponential growth (Fig. 2A, lanes 1–2) in strain BCJ100, and its yield decreased progressively (Fig. 2A). In the RN4220 background, lytM49 levels were significantly lower compared to the HG001 background (Fig. 2A, lanes 3–4), in agreement with previously published data.30 A much longer transcript with a size corresponding to SA0264-lytM was detected at all growth phases with slight variation in the 2 genetic (HG001 and RN4220) backgrounds (Fig. 2A, lanes 1–12). We noticed that strain EL100 expressed lower levels of RNAIII at the beginning of the growth than its parent background BCJ100 (Fig. 2A, lanes 2 and 6), while the levels were identical after 6 h of growth in both strains (Fig. 2A, lanes 9–10).
Figure 2.
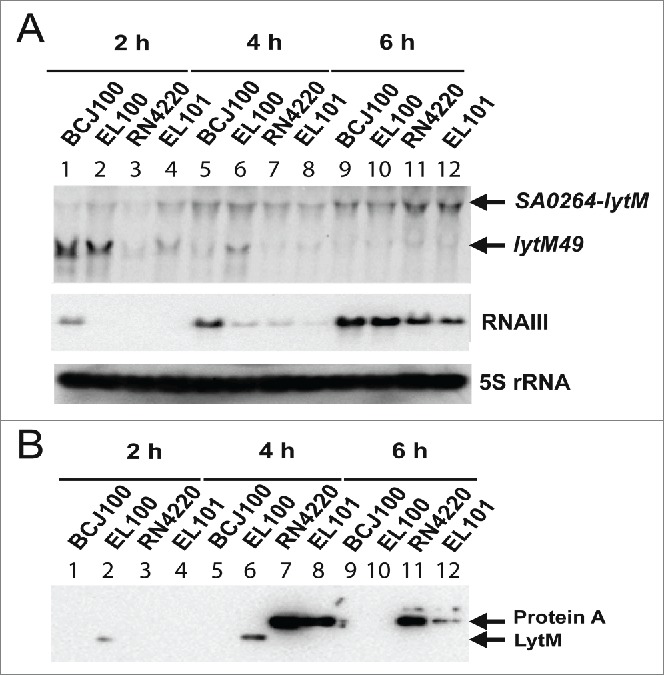
Analysis of LytM synthesis in various strain backgrounds. (A) Northern blot experiments showing the steady state levels of lytM mRNAs (top), of RNAIII (middle) and of 5S RNA (loading control, bottom). Total RNAs were prepared after 2 h (lanes 1 to 4), 4 h (lanes 5 to 8) and 6 h (lanes 9 to 12) of culture in BHI at 37°C from 2 strain backgrounds, BCJ100 (HG001, ΔhsdR Δtype III) and RN4220. A FLAG tag was introduced at the 3′ end of lytM gene in BCJ100 (EL100) and in RN4220 (EL101) to analyze the synthesis of LytM in the 2 strain backgrounds. Experiments were performed on samples prepared from BCJ100 (lanes 1, 5, 9), EL100 (lanes 2, 6, 10), RN4220 (lanes 3, 7, 11) and EL101 (lanes 4, 8, 12). lytM49, corresponds to the WalKR-induced lytM mRNA. (B) Western blot performed using monoclonal antibodies against FLAG tag on protein extracts prepared from the same strains as described for the Northern experiment (see also Material and Methods). For the 2 h samples, the equivalent of 0.8 OD units was loaded, while for the 4 and 6 h samples, the equivalent of 1 OD unit was loaded in order to visualize LytM. As a control, a gel run in parallel with the same samples was stained with Coomassie blue to verify that each lane contained comparable amounts of protein (not shown). The legend is the same as for Fig. 2A. Lanes 7–8 and 11–12: protein A reacted with the anti-Flag antibody in RN4220 and EL101 because RNAIII is weakly expressed. RNAIII is known to repress the translation of spa mRNA encoding protein A through basepairing interactions.34
In vivo expression of LytM was evaluated by Western blotting using an anti-FLAG antibody. LytM was clearly detected during exponential and post-exponential growth phases in the HG001 background (Fig. 2B, lanes 2 and 6, EL100 strain), and its synthesis strongly decreased at the entry into SP (Fig. 2B, lane 10). LytM could also be detected in the RN4220 background after 4 and 6 h of growth but only after prolonged exposure of the blot (data not shown). This observation correlates with the very low levels of lytM49 mRNA in RN4220 background while the level of the longer SA0264-lytM transcript remained almost unchanged in the various strains (Fig. 2A). A non-specific signal was detected on the western blot, which may correspond to protein A (lanes 7–8 and 11–12).
All in all, we suggest that the detected LytM protein in HG001 background (EL100 strain) is mainly produced from the lytM49 transcript while the longer SA0264-lytM mRNA is most likely poorly translatable in vivo. We propose that the very low levels of LytM observed at the late exponential phase of growth (6 h) may correspond to the early expressed protein that remained stable.
Translation of the long lytM mRNA is not efficient and is repressed by RNAIII in vitro
We evaluated the in vitro translational efficiency of lytM mRNAs carrying 5′ UTRs with various sizes, because the in vivo data suggested that LytM is poorly synthesized from the long SA0264-lytM transcript,. In addition, we studied the formation of a possible RNAIII-lytM interaction and its consequence on translation (Fig. 3). Four lytM transcripts of various lengths were produced by in vitro T7 transcription. The first transcript (lytM49) begins at the WalKR-dependent transcription start (-49 with respect to the AUG). The second transcript (lytM211) carries a longer 5′-UTR which adds 160 nts to the lytM transcript (-211). The third, corresponding to the processed transcript (lytM411), starts at position C-411 mapped by primer extension, increasing the 5′UTR of lytM by 360 nts. The fourth transcript comprises the SA0264-lytM operon starting 80 nts upstream the start codon of SA0264.
Figure 3.

RNAIII-dependant repression of lytM mRNA translation. (A) In vitro translation assays using the PURESYSTEM performed with lytM mRNAs (100 nM) carrying various lengths of the 5′ untranslated regions (lytM49, lytM211) or with the SA0264-lytM operon (200 nM). A Flag tag was introduced at the C-terminal of LytM or of SA0264 to analyze the synthesis of the corresponding proteins. (Left) For LytM translation, the experiments were done on the mRNAs alone (lanes 1, 6) or in the presence of increasing concentrations of the 3′ domain of RNAIII: 100 nM (lanes 2, 7), 250 nM (lanes 3, 8), 500 nM (lanes 4, 9) and 1 µM (lanes 5, 10). (Right) For SA0264 translation, the following amounts of the 3′ regulatory domain of RNAIII were used: 200 nM (lane 2), 500 nM (lane 3) and 1 mM (lane 4). A control assay, performed without SA0264-lytM mRNA, shows an unspecific signal (lane 5, (*)). (B) In vitro translation assays with a PCR fragment corresponding to lytM211 (1 nM). Translation on lytM211 alone (lane 1) or in the presence of increasing quantities of wild type RNAIII or of RNAIII deleted of the hairpin 13 (RNAIII-ΔH13): 30 nM (lane 2), 60 nM (lane 3), 120 nM (lane 4), 240 nM (lane 5), 360 nM (lane 6) and 480 nM (lane 7). The same experiment was performed with YFP protein as the negative control. The proteins were separated on a SDS-PAGE 10% and were revealed using FLAG-specific antibodies. Northern blot experiments verified the quality of lytM mRNA and RNAIII using DNA-DIG labeled probes prepared with the primers LytM-EcoRI-T7/LytM-FlagR and RNAIII sense/RNAIII antisense, respectively. (C) Formation of the initiation ribosomal complex monitored by toeprinting using lytM49 and lytM411 as mRNAs. Lane 1: incubation control of lytM mRNAs; lane 2: formation of the initiation ribosomal complex containing lytM mRNA, the initiator tRNA, and S. aureus 70S; lane 3: formation of the initiation ribosomal complex in the presence of the 3′ domain of RNAIII (50 nM); lane 4: formation of the initiation ribosomal complex in the presence of RNAIII deleted of the hairpin H13 (RNAIII-ΔH13; 50 nM); lanes U, C, G, A: sequencing ladders. For the reverse transcription reaction, the oligonucleotide LytM-REV3 was used.
The translation efficiency of the different mRNAs was evaluated in vitro using translation assays (Figs. 3A, 3B). The Puresystem is a coupled transcription-translation system that allows monitoring of protein synthesis by Western blot analysis. We performed the assays using RNA templates that were transcribed and purified before the addition of the Puresystem mix (Fig. 3A) or PCR fragments that were transcribed in vitro by the Puresystem T7 polymerase (Fig. 3B). For all of the constructs, we added a FLAG epitope to the C-terminus of LytM to facilitate detection by an anti-FLAG antibody. We additionally introduced a FLAG-tag at the C-terminus of SA0264 in the long transcript including LytM (Fig. 3A). The Western blot experiments revealed that the WalKR-dependent lytM49 mRNA is significantly more efficiently translated than lytM211 (Fig. 3A) while LytM was not translated from the SA0264-lytM transcript (result not shown). Therefore, increasing the length of lytM 5′UTR had a strong negative impact on translation efficiency. In contrast, the synthesis of SA0264 was efficient. This result suggests that LytM synthesis is not coupled to that of SA0264 (Fig. 3A).
To analyze the effect of RNAIII on lytM mRNA translation, we added increasing amounts of RNAIII to the in vitro translation assays. The assays were also performed in the presence of the 3′ regulatory domain of RNAIII or RNAIII deleted of hairpin 13 (RNAIII-ΔH13) (Figs. 3A, 3B). The data show that the synthesis of LytM was dramatically decreased by the addition of RNAIII or its 3′-domain (Figs. 3A, 3B). Conversely, LytM levels were only mildly affected at the highest concentration of RNAIII-Δ13 (Fig. 3B). The inhibitory effect of RNAIII and the 3′ domain on lytM translation is specific because the synthesis of Yfp protein was not altered (Fig. 3B). Northern blot analysis was also performed on lytM211 mRNA and RNAIII (Fig. 3B) to demonstrate the absence of RNase contamination in the assays. These data also showed that full RNAIII-dependent repression is obtained when the concentration of lytM is approximately equal to the highest amount of RNAIII (480 nM). Contrary to the effect on LytM, the translation of SA0264 within the SA0264-lytM operon was only slightly altered by the addition of increasing amounts of the 3′ domain (Fig. 3A).
To measure the efficiency of the formation of translation initiation complexes, toeprinting assays were used.42 Briefly, primer extension is performed on a complex formed by 30S ribosomal subunits, lytM mRNA and the initiator tRNAMet. Reverse transcription stops when it encounters the 30S initiation complex. The data shows a toeprint at +16 relatively to the AUG of the various forms of lytM mRNAs (Fig. 3C). In agreement with the in vitro translation assays, the toeprint formed with lytM411 was strongly decreased as compared to lytM49 mRNA (Fig. 3C). Thus, increasing the length of the 5′UTR has most likely altered the accessibility of the ribosome-binding site. The addition of increasing concentrations of the 3′ domain of RNAIII strongly decreased the formation of the toeprint and concomitantly reverse transcriptase pauses occurred at U-10 and G-11, which are the signatures of RNAIII-lytM mRNA duplex formation (Fig. 3C). Conversely, the addition of RNAIII-ΔH13, which fails to bind lytM mRNA, did not alter the yield of the toeprint (Fig. 3C).
All in all, we confirmed that the lytM49 transcript originating from the WalKR regulated promoter has the potential to be efficiently translated in vivo, but the extension of the 5′UTR (lytM211, lytM411, SA0264-lytM) decreased considerably the translation efficiency. The 3′ domain of RNAIII is sufficient to bind lytM mRNA and to occlude ribosome binding.
Increasing the 5′ UTR region of lytM changes the accessibility of ribosome binding site
To analyze whether the extension of the 5′UTR of lytM changes the accessibility of the ribosome binding site, we probed the secondary structure of the 5′ untranslated regions of lytM49, lytM211 and lytM411 using enzymatic (Fig. S3) and chemical probes (Fig. S4). The conformation of the mRNAs was analyzed using RNase T1 (specific for single-stranded guanines), RNase T2 (specific for unpaired nucleotides with a preference for adenines) and RNase V1 (specific for double-stranded regions). As chemicals, we used DMS to map the reactivity of adenines at N1 and cytosines at N3, as well as the selective 2′-hydroxyl acylation of ribose by 1M7 to gain knowledge on the flexibility of ribose for each nucleotide of lytM49 and lytM411 RNAs.43 The secondary structure models for the 5′UTR of lytM49 and lytM411 mRNAs were built taking into account experimental data (Fig. 4).
Figure 4.
Secondary structure models of lytM mRNA carrying a short (lytM49) and long (lytM411) 5′ untranslated region. (A and B) Summary of the enzymatic cleavages for lytM49 and lytM411, respectively: strong and moderate RNase V1 cuts are represented by black and gray arrowheads, respectively. RNase T1 cuts are shown by black arrows and RNase T2 by open arrows. For RNases T1 and T2 cleavages, plain and dashed lines represent strong and moderate cuts, respectively. Changes induced by the binding of RNAIII or its 3′ domain are indicated as follows: black circles denote strong protection, enhancements and new RNase V1 cleavages are represented by asterisks. Summary of the DMS modifications performed on lytM49: circled nucleotides are reactive toward DMS; plain and dashed lines represent moderate and weak reactivities, respectively. Protections induced by RNAIII or its 3′ domain are shown by black circles. Binding of the 3′ domain of RNAIII to lytM411 induces protection of riboses against SHAPE (gray squares). (C) The secondary structure of the 3′ domain of RNAIII is given. RNAIII binding protected 3 regions of lytM411 mRNA, they are given as motifs 1 to 3. The Shine and Dalgarno (SD) sequence and the AUG codon are in green. Nucleotides in red can potentially form basepairing interactions between lytM mRNA and the 3′ domain of RNAIII.
In the short 5′UTR of lytM49 mRNA transcript, the SD sequence is engaged in an unstable stem-loop structure. This hairpin motif is well supported by the enzymatic cleavage patterns and the reactivity of the N1 position of adenines located primarily in the loop regions. The AUG initiation codon is found in an unpaired region, which is susceptible to cleavages by the single-strand specific RNase T2 (Figs. 4A, S3). The same experiments were performed with lytM411 mRNA. The enzymatic and chemical probing showed that the long 5′UTR is composed of several long hairpin structures (H1 to H3). Indeed, the stems are susceptible to RNase V1 cleavages while the apical loops are sensitive to single-strand specific RNases (Figs. 4B, S3). In the proposed model, the SD sequence and part of the coding sequence are engaged in interactions with upstream nucleotides of the long 5′UTR of lytM411 (Fig. 4). We however do not exclude the possible existence of alternative structures that would allow the ribosome binding site to form either basepairing interactions with the 5′UTR (Fig. 4B) or a local hairpin structure as found in lytM49 mRNA (Fig. 4A).
Using footprinting assays, we analyzed the binding of RNAIII or of its 3′ domain to lytM49 and lytM411 mRNAs (Figs. 4, S3, S4). Increasing concentrations of RNAIII or the 3′ domain induced the same reactivity changes at distant regions of lytM mRNA transcripts. For the short lytM49 transcript, the main interaction site is around the SD sequence consistent with bioinformatic predictions that revealed complementary sequences to hairpin 13 (H13) of RNAIII.29 Strong protections against DMS and single-strand specific RNases were observed in the apical loops encompassing A-24 to A-18 and in the unpaired region containing the initiation codon (A+1 to U+9). Concomitantly, enhanced RNase V1 cuts were observed at A+6 to A+9, and the N1 position of adenines −30, −34 to −37 became more reactive toward DMS (Fig. 4). For the lytM411 mRNA, binding of RNAIII or its 3′ domain modified the cleavage and reactivity patterns toward chemicals (DMS, Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)) in 3 distinct regions of the mRNA (Fig. 4C). The first site involves a G-rich motif at U-112 to G-120, which was also strongly protected against RNase T1 hydrolysis (Fig. 4, motif 1). In this region, several RNase V1 cuts were enhanced, and riboses were also protected against SHAPE (Fig. 4B, S4). Interestingly, the GGGAAUAUU sequence is strictly complementary to the C-rich motif found in the unpaired region between H13 and H14 of RNAIII (Fig. 4C). Furthermore, RNAIII binding also caused strong protections of riboses in region +1 to +5 (motif 2) while position −14 became more reactive. These protections were specifically attributed to the formation of base-pairings with H13 because the deletion of this domain in RNAIII caused no effect on lytM411 mRNA (Fig. S4). In some experiments, the RBS was only weakly protected by RNAIII binding and a third site of interaction was observed 168 nucleotides downstream of the translation initiation site in the coding sequence (motif 3) where RNAIII or its 3′ domain protected a G-rich region against RNase T1 cleavages (Fig. S3). This third site contained the UGGGGA sequence, which is fully complementary to the apical loop of H13 (Fig. 4C). Therefore, we propose that 2 different complexes might form with the long lytM mRNA leading to the formation of either motifs 1 and 2, or motifs 1 and 3 (Fig. 4C). These data resulted most probably from the formation of alternative structures of the RBS in lytM411.
Taken together, our data showed that the extension of the 5′UTR of lytM modified the structure of the RBS to decrease the translation efficiency and provided additional binding sites for RNAIII.
Deregulation of lytM during stationary phase growth causes cell wall perturbation
We searched for cell envelope related phenotypes as a consequence of the regulatory effect of RNAIII. To evaluate membrane integrity, we utilized the Live/Dead staining assay (BacLight bacterial-viability assay, Molecular Probes). This assay allows distinguishing intact from damaged membranes by fluorescence microscopy. Only cells that have compromised membranes are stained with the red fluorescent dye propidium iodide. At early exponential phase of growth, S. aureus RN6390 and WA400 (ΔRNAIII) cells grown in BHI were almost completely stained in green indicating that the cell membranes were not altered (data not shown). However, during the late exponential growth phase, a significant number of cells from WA400 strain were stained in red (Fig. S5A), indicating damaged membranes. We analyzed whether the major autolysin LytM could be responsible for the observed phenotype. The Live/Dead staining showed that the deletion of lytM in WA400 mutant strain partially restored the wt phenotype (Fig. S5A). Indeed, statistical analysis was performed on 2 independent experiments where the cells from 3 to 4 frames were analyzed for each strain. In the late exponential phase, only 1.29 % of the cells were damaged (red) in the wt background, while in the WA400 strain 5.79 % of the cells were affected. The lytM deletion in WA400 slightly ameliorated this phenotype, as 3.67 % of the cells were stained in red. The deletion of lytM in the WT background (strain LUG1420) mirrored the WT phenotype. Reproducible data were obtained from at least 5 independent experiments.
As the agr system was found to affect autolysis of S. aureus,44 we have analyzed the potential role of RNAIII in the regulation of autolysis using Triton X-100 treatment of the different strains (Fig. S5B). The ΔRNAIII mutant exhibited a lower Triton induced autolysis rate as compared to RN6390 strain. Interestingly, complementation of cells with a plasmid expressing the 3′ domain of RNAIII restored the autolysis phenotype of the WT strain even though the 3′ domain is less stable than RNAIII (Fig. S5B). The RN6390 strain carrying a deletion of the lytM gene had a phenotype similar to the isogenic WT strain. In addition, the autolysis rate of the WA400-ΔlytM strain was identical to the parental WA400 strain showing that the absence of LytM did not cause any effect on Triton induced autolysis.
Taken together these data collectively suggest that deletion of RNAIII induces perturbation in the cell envelope that can be partially restored by the deletion of lytM gene. This perturbation however, does not result into higher autolysis rates.
Discussion
Genome-wide mapping of bacterial transcriptional start sites facilitated by advanced next-generation sequencing technologies revealed in many cases multiple promoters derived from the same genetic locus. This leads to complex RNA profiles in genomes including S. aureus.45-47 Such information is helpful to further analyze the regulation of individual genes. Here, we have defined how LytM expression is regulated during growth. We showed that lytM mRNA is produced from at least 2 different transcriptional start sites. In addition to the transcriptional control, we demonstrated that LytM synthesis is regulated at the post-transcriptional level through different mechanisms involving both the quorum sensing-induced RNAIII and mRNA structural changes mediated by the length of its 5′UTR (Fig. 5).
Figure 5.
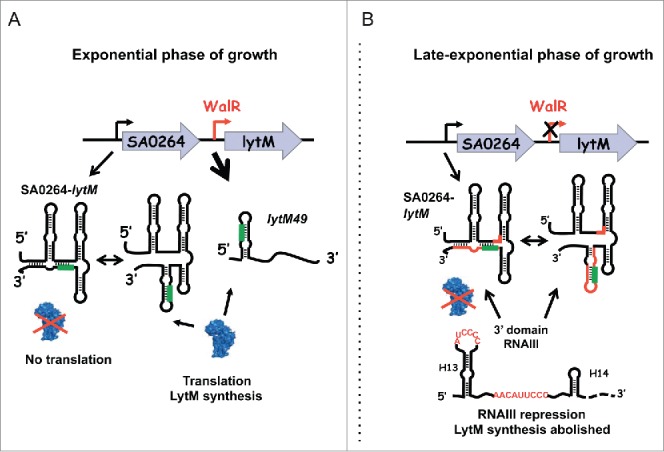
Model of the regulatory mechanism of LytM synthesis. (A) During exponential growth, the response regulatory protein WalR activates the expression of lytM49 mRNA to generate a mRNA with a short 5′ untranslated region (5′UTR). The Shine and Dalgarno (SD) sequence colored in green is involved in an unstable hairpin structure allowing translation by the ribosome. A long bicistronic operon SA0264-lytM is also weakly produced. This long RNA transcript adopts a secondary structure that sequesters the SD sequence and part of the coding sequence into a more stable structure. In vitro probing revealed the presence of an alternative structure where the SD sequence could be engaged in a hairpin structure similar to lytM49. This structure is not efficiently recognized by the ribosome. Our data confirm that the pool of LytM present during exponential growth is predominantly produced from the highly expressed lytM49. (B) During late-exponential phase, WalR is inactive and the expression of lytM49 is abolished while the bicistronic operon is constitutively expressed. Although LytM is weakly translated from the bicistronic operon, RNAIII is able to bind at 2 distinct sites of the 5′UTR of lytM411 to inhibit translation. Hence, LytM synthesis is regulated at both the transcriptional and post-transcriptional levels. The 3′ domain of RNAIII contains 2 sequences (in red) which are complementary to mRNA regions in red: H13 forms basepairing interactions with the SD while the C-rich motif located in the interhelical region of the 3′ domain is complementary to a G-rich motif of the 5′UTR. The 30S subunit is shown in blue.
In many microorganisms, including S. aureus, the processes of synthesis and hydrolysis of the covalent peptidoglycan bonds during cell growth are intimately balanced. This suggests that expression of both cell wall biosynthetic- and hydrolytic- enzymes has to be tightly and coordinately regulated during growth.9,48 LytM and other peptidoglycan hydrolases are strongly repressed during the stationary phase of growth.4 At the transcriptional level, lytM expression is activated by the 2-component system WalKR during early exponential growth.9 WalKR-dependent activation of lytM expression gives rise to a mRNA carrying a short 5′-UTR of 49 nucleotides that is efficiently translated (Figs. 1 and 2). During the stationary growth phase, the WalR regulatory protein becomes inactive and lytM transcription is additionally repressed by several other regulators like the 2 component systems ArlSR and LytSR, and the transcriptional regulator MgrA (Fig. S6).21-24 However, very recent data suggest that in B. subtilis prolonged activation of WalR can occur due to alternate phosphorylation by a eukaryotic-like Ser/Thr kinase. This can lead to enhanced activation or repression of WalR targets at stationary phase.49 As this Ser/Thr kinase is conserved in S. aureus (PknB), alternate expression profiles of WalR targets could also occur. Intriguingly, we have observed that lytM is also produced from a bicistronic operon, which includes the upstream SA0264 gene encoding for a protein similar to penicillin amidase. The same transcriptional start site was also observed in S. aureus 15981 strain38 indicating that the presence of a weakly expressed bicistronic operon encompassing SA0264 and lytM is conserved in S. aureus strains. We observed this longer lytM transcript at both the exponential and stationary growth phases and found that the steady state yield does not depend on RNAIII levels (Fig. 1, S2 and Table 1). In agreement with this observation, the level of lytM mRNA did not significantly vary in a transcriptomic analysis of an agr mutant.50 Using different experimental approaches such as primer extension and RACE, we detected various RNA fragments emanating from the >3kB long transcript. Notably, the total RNA samples that were used for the RACE experiment were treated with Tobacco Acid Pyrophosphatase (TAP) to remove processed RNA. However, we have previously found that TAP treatment does not completely remove processed RNA.51 The 2 shorter transcripts carrying a 411 nts and 356 nts long 5′UTR, respectively, that were identified in ΔRNAIII and in HG001 strains are most likely products of a processing event of the longer bicistronic operon and not primary transcripts for the following reasons. First, there is no obvious promoter preceding the 5′ ends of these transcripts. Second, a recent RNA sequencing study evaluating the biological functions of RNases J1/J2 in S. aureus revealed several 5′-monophosphorylated RNAs within SA0264. These cleaved products were only observed in the J1 and J2 deletion strains but not in the WT strain.52 Taken together, these transcripts are most likely generated by cleavages and are further removed by RNase J. The various RNA fragments we observed reflect a complex degradation pattern of the long SA0264-lytM transcript.
We show here that the bicistronic mRNA and the processed transcript are translated less efficiently than the WalKR-dependent lytM mRNA (Figs. 2 and 3). This finding correlates well with the observation that the levels of LytM drop considerably during stationary growth phase.53 Probing the structure of the large mRNA revealed that the RBS could form alternative basepairing interactions with upstream nucleotides located in the 5′UTR to occlude ribosome binding. In vitro translation assays suggested that the synthesis of LytM is not coupled to SA0264 and that the bicistronic operon generates a translationally inactive lytM mRNA (Fig. 3). In addition, we have observed a second level of regulation during the late exponential phase of growth when the cell division slows down (Fig. 5B). The regulatory RNAIII, and more precisely its 3′ domain, behaves as an antisense RNA that binds to several sites of lytM mRNA and prevents translation (Figs. 4, 5B). Therefore both RNA structural changes and the quorum sensing-induced RNAIII contribute to prevent the synthesis of LytM during stationary phase. Regulation at both the transcriptional and the translational levels has the advantage to generate a tight regulation of synthesis particularly when leaky gene expression could be detrimental to the cell.34
We have observed that the cell membrane integrity was compromised in many cells of the ΔRNAIII strain during the late exponential growth phase. This phenotype was partially alleviated in the double mutant ΔRNAIII ΔlytM strain (Fig. S5A). These data suggest that the deregulation of LytM synthesis caused by the deletion of RNAIII contributes to cell envelope perturbation during stationary phase. Alterations in the membrane could affect cell wall synthesis and morphology by affecting access to precursors, as well as binding and transport of cell wall modifying enzymes. We searched to identify whether possible alterations in cell wall could lead to altered resistance in lysis. We used Triton-X as the lysis reagent. However, somehow non-intuitively, we observed that deregulation of cell wall homeostasis due to the absence of RNAIII results in cells that are less susceptible to lysis (Fig. S5B), in agreement to previous results.44 LytM does not seem to contribute to the observed lysis phenotype consistently with previous work.54 The effect of RNAIII on autolysis appears to be restricted to its 3′ domain, which is the main repressor and conserved domain of RNAIII.29,34,55
The observed phenotypic outcomes related to the deletion of RNAIII may be due to the deregulation of several other cell-wall modifying enzymes. Recent data has shown that the V8 exo-protease (SspA, indirectly activated by RNAIII through the Rot regulator56) cleaves and deactivates AtlA (Fig. S6) resulting in decreased biofilm formation.17 Therefore, it is possible that the quorum sensing RNAIII-mediated synthesis of proteases may impact the production of biofilm to favor cell dissemination.27 Besides LytM, it was previously shown that RNAIII repressed the synthesis of SA2353, encoding an amidase.29 In addition, conserved base-pairing interactions among S. aureus strains have been predicted between the 3′ domain of RNAIII and several mRNAs encoding enzymes involved in cell wall metabolism, namely the protein SA2093 (an homolog of the staphylococcal secretory antigen SsaA) and the 2 transglycosylases SceD and IsaA (Fig. S7). In S. epidermidis RNAIII represses the expression of one of the major hydrolases, AtlE, though the mechanism was not studied.57 A recently published work defines an important function of LytM for the release of protein A from the cell wall into the extracellular milieu.58 Previously we have shown that RNAIII is repressing the production of protein A34 and here we show that RNAIII additionally represses LytM. As a result, RNAIII controls not only expression of protein A but also its release from the cell surface (Fig. S6). Therefore, in addition to its known role in the regulation of virulence factors, RNAIII emerges as a major player in cell wall metabolism either through direct base-pairing interactions with mRNAs (as for lytM) or indirectly through the production of proteases (Fig. S6).
This work highlights the functional consequences that the varying lengths of the 5′UTRs of lytM mRNA have on both the RNA folding and the translation efficiency of LytM. Such RNA structural changes mediated by the length of the 5′UTR have previously been shown to favor the choice of an alternative initiation codon59 or to affect the mRNA stability.51,60 Hence, diverse 5′UTR-dependent regulatory mechanisms are expected to be more widespread than previously anticipated due to the complexity of the RNA profiles observed in bacterial genomes.
Materials and methods
Strains and plasmids
Staphylococcus aureus strains and plasmids used in this study are listed in Table S1. Escherichia coli strains DH5α or XL1-Blue were used as host strains for plasmid constructions. S. aureus strain RN4220 was used as the recipient strain of constructed plasmids to avoid the endogenous modification systems. The deletion/replacement ΔlytM/aphA-3 mutant of S. aureus RN6390 (LUG1420) and of S. aureus WA400 (LUG1432) were obtained by using pMAD, a thermosensitive plasmid, which contains a constitutively expressed ß-galactosidase gene. This allows positive selection for the double crossover by following the ß-galactosidase activity on X-gal agar plates.61 The kanamycin resistance gene (amplified from the pCRTM2.1 plasmid, INVITROGEN) was cloned in pMAD between 2 DNA fragments corresponding respectively to the chromosomal regions upstream and downstream the lytM coding sequence. Primer pairs LytM1186/LytM1626 and LytM2679/LytM3241 were used to amplify the 2 regions. The resulting plasmid, pLUG759, was transformed by electroporation successively into RN4220, and RN6390 or WA400. Transformants were grown at the non-permissive temperature (37°C) to select cells that have the plasmid integrated into the chromosome by homologous recombination. To favor the second recombination event, a single colony was grown at 30°C for 10 generations and plated at 37°C overnight. Cells that lost the plasmid through a double cross over event were selected on Xgal agar plates. PCR was used to confirm the loss of lytM gene.
We constructed a strain of S. aureus HG001 that could be transformed with plasmids isolated from E. coli by deleting the endogenous restriction barriers using the targetron approach.39,62 The targetron plasmids pNKL62-hsdR and pNKL62-type III were a kind gift from P. Linder and A.R. Corvaglia. We verified that the genome of HG001 encodes both the putative hsdR and type III genes by sequencing PCR fragments amplified from the genome using appropriate oligonucleotides (o15–16 and o18–19) (Table S2). Targetron plasmids were passed successively through RN4220 and HG001 strains. The final strain that could be transformed with plasmids isolated from E. coli (BCJ100) was constructed by combining the 2 mutations (hsdR and type III) into a single strain by performing 2 sequential rounds of the mutagenesis protocol using the 2 different targetron plasmids.
To express a Flag-tagged version of LytM, the vector pMutin-FLAG was used. First, lytM was amplified from RN6390 genomic DNA using oligonucleotides LytM-fw-HindIII/LytM-rev2-KpnI and cloned into the pJet 1.2 vector resulting in plasmid pEL90. The lytM gene was then eluted from pEL90 as a HindIII/KpnI fragment and cloned into the corresponding sites of pMutin-FLAG resulting in plasmid pEL93 (Table S1). Direct transformation of pEL93 into BCJ100 and RN4220 gave strains EL100 and EL101, respectively (Table S1). A strand overlap PCR approach was used to produce a template for an in vitro RNA transcript that would include SA0264-flag and lytM. Briefly, primers SA0264-T7/SA0264-flag and LytM-overhang/LytM-rev2-KpnI were used to amplify SA0264 and lytM, respectively. The purified PCR fragments were used as template for PCR using external primers SA0264-T7/LytM-rev2-KpnI. This assembled PCR fragment was then cloned into pJet 1.2 giving plasmid pEL103. For the second subclone, the PCR fragment containing SA0264 and lytM-flag was amplified using primers SA0264-T7/LytM-rev-EcoRI and cloned into pJet 1.2 resulting in plasmid pEL104. The operon cassettes SA0264-flag-lytM and SA0264-lytM-flag were eluted from subclones as either BamHI-KpnI or BamHI-EcoRI fragments and cloned into the corresponding sites of vector pUT7,63 which is a pUC119 derivative carrying the T7 promoter to facilitate in vitro transcription. This resulted in plasmids pEL105 and pEL106, respectively.
S. aureus was grown either on blood-agar plates or in brain-heart infusion medium (BHI) supplemented with erythromycin (10 μg/ml) or chloramphenicol (10 µg/ml) when appropriate.
Preparation of RNAs
The various lytM RNAs (lytM49, lytM211 and lytM411, the number corresponding to the length of the respective 5′-UTR, see Results section) were transcribed in vitro using T7 RNA polymerase from PCR fragments, which included the T7 promoter sequence on the forward primer. To amplify lytM49, lytM211 and lytM411, the forward oligonucleotides LytM-T7c, LytM-EcoRI-T7 and 497-T7 were used, respectively, while in all cases the same reverse oligonucleotide LytM-FlagR was used (Table S2). RNA fragments SA0264-flag-lytM and SA0264-lytM-flag were transcribed using the TranscriptAid T7 High Yield Transcription kit (ThermoScientific) from pEL105 and pEL106 after linearization with EcoRI. RNAIII and derivatives were in vitro transcribed as described previously.29,64. The RNAs were purified by 8% polyacrylamide-8 M urea gel electrophoresis. After elution in 0.5 M ammonium acetate/1 mM EDTA buffer, the RNAs were precipitated twice with ethanol. Before use, RNAs were denatured by incubation at 90°C for 2 min in the absence of magnesium and salt, chilled 1 min on ice, then renatured at 37°C for 15 min in TMN (20 mM Tris-acetate pH 7.5, 10 mM magnesium-acetate, 150 mM Na-acetate) or TMK (10 mM Tris-HCl pH 8, 10 mM MgCl2, 100 mM KCl) buffer.
Northern, primer extension and RACE experiments
Total RNAs from various phases of growth were isolated for Northern, primer extension and RACE experiments as described previously.51 Total RNA samples were separated on 5% polyacrylamide gels containing 8 M urea or 1.2 % agarose gels containing 20 mM guanidine thiocyanate. Hybridizations with specific digoxigenin-labeled RNA or DNA probes and detection were carried out as described previously.51 For all experiments, the quantity of 5S rRNA was verified using a digoxigenin-labeled RNA probe (prepared from the respective PCR template that was amplified with oligonucleotides 5S-T7trans/5S-T7trans rev).
Primer extension was carried out on 10–20 µg of total RNA prepared from S. aureus strains as described.51 The 5′ start and the size of the RNA was evaluated by running sequencing ladders in parallel. Sequencing was performed either on purified RNAs or on DNA fragments.
The 5′ end of the lytM mRNAs was determined by rapid amplification of cDNA ends (RACE) using the First Choice RLM-RACE kit (Ambion). Briefly, total RNA was first treated with Tobacco Acid Pyrophosphatase (TAP) to remove processed RNA. Then a 5′-RACE Acceptor oligo provided by the manufacturer was ligated to the RNA. Subsequently, reverse transcription was performed with random primers to obtain cDNA. Finally, a nested PCR was performed: the first PCR reaction was done with primers 5′-RACE_outer and LytM PE and the second with primers 5′-RACE_inner and LytM RACE2, respectively. The PCR products were purified and cloned. Sequencing was performed on 9 colonies.
Quantitative RT-PCR analysis
The RN6390 wild type and the mutant strains LUG950 (ΔRNAIII), LUG1662 (ΔRNAIII/p) and LUG1855 (ΔRNAIII/pRNAIII) were grown in BHI rich medium until OD600nm ≈1.2 for the exponential phase (EP) study and 5 for the stationary phase (SP) study. RNA extractions were performed as previously described,65 followed by a DNase I treatment with the TURBO DNA-free reagent (Ambion, Austin, TX) to eliminate residual genomic DNA. cDNAs (cDNAs) were synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), in a 20 µl reaction volume containing 1 µg total RNA. Oligonucleotides were designed for 100–200 bp amplicons using BEACON Designer 4.02 software (Premier Biosoft International, Palo Alto, CA). qRT-PCRs, critical threshold cycles (CT) and n-fold changes in transcript levels were performed and determined as previously described9 using the SsoFast™ EvaGreen Supermix (Bio-Rad, Hercules, CA) and normalized with respect to 16S rRNA whose levels did not vary under our experimental conditions. The expression of 16S RNA was probed using oligos OSA161, OSA162. All assays were performed using quadruplicate technical replicates and repeated with 2 independent biological samples. The data are presented as the mean values and standard deviation (SD).
In vitro translation assays and western analysis
The in vitro translation assays were carried out using lytM49, lytM211, lytM411, SA0264-flag-lytM and SA0264-lytM-flag PCR fragments or RNA transcripts, and the PURESYSTEMII classic kit (Cosmo Bio Co, Japan). All constructs carry an additional sequence corresponding to the Flag peptide, which was inserted at the 3′ end of lytM, SA0264 or Yfp. The reaction was done at 37°C for 1 h in the presence of 1 pmol of mRNA. Experiments were also carried out in the presence of increasing concentrations of RNAIII or its derivatives (1 to 10 pmoles). Briefly, the 2 RNAs were denatured separately, chilled on ice, diluted in 1X TMK containing 1 mM DTT and then renatured separately. The two RNAs were then mixed and incubated at 37°C for 10 min. Then 7 µl of the Puresystem reaction mix was added (5 µl of the solution A and 2 µl of the solution B) and the samples were incubated at 37°C for 1 h. The final volume of the reaction was adjusted to 10 µl if necessary by addition of 1X TMK buffer. The tubes were placed on ice to stop the reaction, and the proteins were detected by western blot using antibodies against the FLAG tag. Each experiment was repeated at least 3 times with different samples.
Toeprinting
The preparation of the S. aureus 30S subunits, the formation of a simplified translational initiation complex with mRNA, and the extension inhibition conditions were performed as previously described.42 Experimental details are given in the Supplementary Information.
Structure probing
Formation of lytM mRNA-RNAIII complex was carried out at 37°C for 15 min in the TMN buffer. Enzymatic hydrolysis was performed on cold lytM (1 pmole) in 10 µl of TMN, in the presence of 1 µg carrier tRNA at 37°C for 5 min, RNase T1 (0.0025 units), RNase V1 (0.5 units) and RNase T2 (0.05 U). DMS modifications of positions N1 of adenines and N3 of cytosines were done in the presence of 1 µl DMS diluted in ethanol (1:8) for 5 min at 37°C. Ribose modifications were done with benzoyl cyanide for 1 s as described.66 The enzymatic reactions were stopped by phenol extraction followed by RNA precipitation. The enzymatic cleavages and ribose modifications were detected by primer extension with reverse transcriptase as described.67
Autolysis assays
To eliminate possible variations due to growth in nutrient media, Triton X-100 was used to lyse the S. aureus strain sets according to Fujimoto and Bayles.44 In brief, cells were grown to mid-exponential phase, pelleted by centrifugation, washed and resuspended in equal volume of 10 mM Tris-HCl (pH 8) containing 0.1% of Triton X-100 and incubated at 30°C with agitation. Cellular lysis was measured by determining the changes in OD570nm as a function of time and expressed as percentage of the initial OD570nm.
LIVE/DEAD staining of cells
S. aureus cells were harvested by centrifugation (13 000 g, 10 min, 4°C) after 2 and 5 h of growth, resuspended in 0.85% NaCl and incubated at 20°C for 1 h. Cells were re-pelleted and stained with a mixture of propidium iodide and SYTO 9 (nucleic acid stains) according to the manufacturers instructions (LIVE/DEAD BacLight Bacterial Viability kit, L7012, Molecular probes). Bacterial cells were visualized in an Axioplan 2 imaging microscope. Images from 2 independent experiments were analyzed with the Quantity One software (Biorad) to count the red and green cells and the percentage of damaged cells was calculated accordingly. The experiment was reproduced at least 5 times.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are thankful to Gérard Lina, and Tarek Msadek for helpful comments and critical reading of the manuscript, to Inigo Lasa, Alejandro Toledo-Arana, Alexander Smirnov and Joerg Vogel for sharing transcriptomic data on lytM operon structure and for stimulating discussions, and to Patrick Linder and Peter Redder for helpful advices concerning the construction of mutant strains.
Funding
This work was supported by the Centre National de la Recherche Scientifique (CNRS; PR), the Institut National pour la Recherche Médicale (INSERM; FV & TG), the Agence Nationale de la Recherche (ANR10-Pathogenomics-ARMSA; PR, FV & TG). EL has received a post-doctoral fellowship from FEBS. This work has been published under the framework of the LABEX: ANR-10-LABX-0036_NETRNA (PR) and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program.
References
- 1.Boneca IG. The role of peptidoglycan in pathogenesis. Curr Opin Microbiol 2005; 8:46-53; PMID:15694856; http://dx.doi.org/ 10.1016/j.mib.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 2.Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol 2014; 91:862-74; PMID:24405365; http://dx.doi.org/ 10.1111/mmi.12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology (Reading, Engl) 2000; 146 (Pt 2):249-62; PMID:10708363; http://dx.doi.org/ 10.1099/00221287-146-2-249 [DOI] [PubMed] [Google Scholar]
- 4.Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 2008; 70:1307-22; PMID:19019149; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- 5.Lee TK, Huang KC. ScienceDirectThe role of hydrolases in bacterial cell-wall growth. Curr Opin Microbiol 2013; 16:760-6; PMID:24035761; http://dx.doi.org/ 10.1016/j.mib.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice KC, Bayles KW. Molecular Control of Bacterial Death and Lysis. Microbiol Mol Biol Rev 2008; 72:85-109; PMID:18322035; http://dx.doi.org/ 10.1128/MMBR.00030-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atilano ML, Pereira PM, Vaz F, Catalão MJ, Reed P, Grilo IR, Sobral RG, Ligoxygakis P, Pinho MG, Filipe SR. Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. eLife 2014; 3:36-23; PMID:24692449; http://dx.doi.org/14762013 10.7554/eLife.02277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 2004; 186:1175-81; PMID:14762013; http://dx.doi.org/ 10.1128/JB.186.4.1175-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 2007; 189:8257-69; PMID:17827301; http://dx.doi.org/ 10.1128/JB.00645-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, et al.. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 2002; 43:1387-400; PMID:11952893; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02832.x [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, et al.. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 2009; 10:291; PMID:19570206; http://dx.doi.org/ 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. mBio 2012; 4:e00537-12-e00537-12; PMID:23404398; http://dx.doi.org/10.1128/mBio.00537-129220008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 1997; 24:1013-24; PMID:9220008; http://dx.doi.org/ 10.1046/j.1365-2958.1997.4101774.x [DOI] [PubMed] [Google Scholar]
- 14.Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infection and Immunity 2011; 79:1153-65; PMID:21189325; http://dx.doi.org/ 10.1128/IAI.00364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA 2007; 104:8113-8; PMID:17452642; http://dx.doi.org/ 10.1073/pnas.0610226104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus aureus Atl AM and GL Murein Hydrolase Activities in Cell Division, Autolysis, and Biofilm Formation. PLoS ONE 2012; 7:e42244; PMID:22860095; http://dx.doi.org/ 10.1371/journal.pone.0042244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Krishnan V, Macon K, Manne K, Narayana SVL, Schneewind O. Secreted Proteases Control Autolysin-mediated Biofilm Growth of Staphylococcus aureus. J Biol Chem 2013; 288:29440-52; PMID:23970550; http://dx.doi.org/ 10.1074/jbc.M113.502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi J-I, Komatsuzawa H, Sugai M. Identification and molecular characterization of an N-acetylmuramyl- l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol Microbiol 2005; 58:1087-101; PMID:16262792; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04881.x [DOI] [PubMed] [Google Scholar]
- 19.Sugai M. Peptidoglycan hydrolases of the staphylococci. J Infect Chemotherapy 1997; 3:113-27; http://dx.doi.org/ 10.1007/BF02491501 [DOI] [Google Scholar]
- 20.Bayles KW. PERSPECTIVES. Nat Rev Micro 2014; 12:63-9; http://dx.doi.org/ 10.1038/nrmicro3136; PMID:2433618510869073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol 2000; 182:3955-64; PMID:10869073; http://dx.doi.org/ 10.1128/JB.182.14.3955-3964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 1996; 178:611-8; PMID:8550490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingavale SS, Van Wamel W, Cheung AL. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol 2003; 48:1451-66; PMID:12791130; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03503.x [DOI] [PubMed] [Google Scholar]
- 24.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infection and Immunity 2005; 73:1423-31; PMID:15731040; http://dx.doi.org/ 10.1128/IAI.73.3.1423-1431.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaune A, Dubrac S, Blanchet C, Poupel O, Mäder U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon J-M, et al.. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infection and Immunity 2012; 80:3438-53; PMID:22825451; http://dx.doi.org/ 10.1128/IAI.00195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol 2010; 192:613-23; PMID:19933357; http://dx.doi.org/ 10.1128/JB.01242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet 2008; 42:541-64; PMID:18713030; http://dx.doi.org/ 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 28.Fechter P, Caldelari I, Lioliou E, Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett. 2014 Aug 1;588(15):2523-9; PMID:24873876; http://dx.doi.org/10.1016/j.febslet.2014.05.037 [DOI] [PubMed] [Google Scholar]
- 29.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al.. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes & Development 2007; 21:1353-66; PMID:17545468; http://dx.doi.org/ 10.1101/gad.423507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaune A, Poupel O, Mallet A, Coic Y-M, Msadek T, Dubrac S. Peptidoglycan Crosslinking Relaxation Plays an Important Role in Staphylococcus aureus WalKR-Dependent Cell Viability. PLoS ONE 2011; 6:e17054; PMID:21386961; http://dx.doi.org/ 10.1371/journal.pone.0017054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chunhua M, Yu L, Yaping G, Jie D, Qiang L, Xiaorong T, Guang Y. The expression of LytM is down-regulated by RNAIII in Staphylococcus aureus. J Basic Microbiol 2012; 52:636-41; PMID:22581788; http://dx.doi.org/ 10.1002/jobm.201100426 [DOI] [PubMed] [Google Scholar]
- 32.Ramadurai L, Jayaswal RK. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. Journal of Bacteriology 1997; 179:3625-31; PMID:9171409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. Journal of Bacteriology 1988; 170:4365-72; PMID:2457579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al.. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 2005; 24:824-35; PMID:15678100; http://dx.doi.org/ 10.1038/sj.emboj.7600572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick RP. ANALYSIS BY TRANSDUCTION OF MUTATIONS AFFECTING PENICILLINASE FORMATION IN STAPHYLOCOCCUS AUREUS. J Gen Microbiol 1963; 33:121-36; PMID:14072829; http://dx.doi.org/ 10.1099/00221287-33-1-121 [DOI] [PubMed] [Google Scholar]
- 36.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. Repair of Global Regulators in Staphylococcus aureus 8325 and Comparative Analysis with Other Clinical Isolates. Infection and Immunity 2010; 78:2877-89; PMID:20212089; http://dx.doi.org/ 10.1128/IAI.00088-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J 1990; 9:1391-9; PMID:2328718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, et al.. Base pairing interaction between 5″- and 3-″UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet 2013; 9:e1004001; PMID:24367275; http://dx.doi.org/ 10.1371/journal.pgen.1004001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corvaglia AR, Francois P, Hernandez D, Perron K, Linder P, Schrenzel J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc Natl Acad Sci USA 2010; 107:11954-8; PMID:20547849; http://dx.doi.org/ 10.1073/pnas.1000489107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983; 305:709-12; PMID:6226876; http://dx.doi.org/ 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 41.Traber K, Novick R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol Microbiol 2006; 59:1519-30; PMID:16468992; http://dx.doi.org/ 10.1111/j.1365-2958.2006.04986.x [DOI] [PubMed] [Google Scholar]
- 42.Fechter P, Chevalier C, Yusupova G, Yusupov M, Romby P, Marzi S. Ribosomal initiation complexes probed by toeprinting and effect of trans-acting translational regulators in bacteria. Methods Mol Biol 2009; 540:247-63; PMID:19381565; http://dx.doi.org/ 10.1007/978-1-59745-558-9_18 [DOI] [PubMed] [Google Scholar]
- 43.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005; 127:4223-31; PMID:15783204; http://dx.doi.org/ 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- 44.Fujimoto DF, Bayles KW. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol 1998; 180:3724-6; PMID:9658022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al.. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA 2011; 108:20172-7; PMID:22123973; http://dx.doi.org/ 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al.. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010; 464:250-5; PMID:20164839; http://dx.doi.org/ 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- 47.Toledo-Arana A, Solano C. Deciphering the physiological blueprint of a bacterial cell: revelations of unanticipated complexity in transcriptome and proteome. Bioessays 2010; 32:461-7; PMID:20486131; http://dx.doi.org/ 10.1002/bies.201000020 [DOI] [PubMed] [Google Scholar]
- 48.Antignac A, Sieradzki K, Tomasz A. Perturbation of cell wall synthesis suppresses autolysis in Staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. Journal of Bacteriology 2007; 189:7573-80; PMID:17827298; http://dx.doi.org/ 10.1128/JB.01048-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Libby Elizabeth A, LAGJD . The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two- Component System in Bacillus subtilis. 2015; 1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang S-J, Rice KC, Bayles KW, Smeltzer MS. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology (Reading, Engl) 2006; 152:3075-90; PMID:17005987; http://dx.doi.org/ 10.1099/mic.0.29033-0 [DOI] [PubMed] [Google Scholar]
- 51.Lioliou E, Sharma CM, Caldelari I, Helfer A-C, Fechter P, Vandenesch F, Vogel J, Romby P. Global Regulatory Functions of the Staphylococcus aureus Endoribonuclease III in Gene Expression. PLoS Genet 2012; 8:e1002782; PMID:22761586; http://dx.doi.org/ 10.1371/journal.pgen.1002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linder P, Lemeille S, Redder P. Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation. PLoS Genet 2014; 10:e1004207; PMID:24586213; http://dx.doi.org/ 10.1371/journal.pgen.1004207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramadurai L, Lockwood KJ, Nadakavukaren MJ, Jayaswal RK. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology (Reading, Engl) 1999; 145:801-8; PMID:10220159; http://dx.doi.org/ 10.1099/13500872-145-4-801 [DOI] [PubMed] [Google Scholar]
- 54.Singh VK, Carlos MR, Singh K. Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiology Letters 2010; 311:167-75; PMID:20738399; http://dx.doi.org/ 10.1111/j.1574-6968.2010.02087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 2006; 61:1038-48; PMID:16879652; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05292.x [DOI] [PubMed] [Google Scholar]
- 56.Oscarsson J, Tegmark-Wisell K, Arvidson S. Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII). Int J Med Microbiol 2006; 296:365-80; PMID:16782403; http://dx.doi.org/ 10.1016/j.ijmm.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 57.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis 2003; 188:706-18; PMID:12934187; http://dx.doi.org/ 10.1086/377239 [DOI] [PubMed] [Google Scholar]
- 58.Becker S, Frankel MB, Schneewind O, Missiakas D. Release of protein A from the cell wall of Staphylococcus aureus. 2014; 111:1574-9; PMID:24434550; http://dx.doi.org/10.1073/pnas.1317181111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omairi-Nasser A, de Gracia AG, Ajlani G. A larger transcript is required for the synthesis of the smaller isoform of ferredoxin:NADP oxidoreductase. Mol Microbiol 2011; 81:1178-89; PMID:21790803; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07739.x [DOI] [PubMed] [Google Scholar]
- 60.Obana N, Shirahama Y, Abe K, Nakamura K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol Microbiol 2010; 77:1416-28; PMID:20572941; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07258.x [DOI] [PubMed] [Google Scholar]
- 61.Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 2004; 70:6887-91; PMID:15528558; http://dx.doi.org/ 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 2006; 12:1271-81; PMID:16741231; http://dx.doi.org/10.1261/rna.687069208917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serganov A, Rak A, Garber M, Reinbolt J, Ehresmann B, Ehresmann C, Grunberg-Manago M, Portier C. Ribosomal protein S15 from Thermus thermophilus–cloning, sequencing, overexpression of the gene and RNA-binding properties of the protein. Eur J Biochem 1997; 246:291-300; PMID:9208917; http://dx.doi.org/ 10.1111/j.1432-1033.1997.00291.x [DOI] [PubMed] [Google Scholar]
- 64.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 2000; 6:668-79; PMID:10836788; http://dx.doi.org/ 10.1017/S1355838200992550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Even S, Burguière P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol 2006; 188:2184-97; PMID:16513748; http://dx.doi.org/ 10.1128/JB.188.6.2184-2197.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 2007; 129:4144-5; PMID:17367143; http://dx.doi.org/ 10.1021/ja0704028 [DOI] [PubMed] [Google Scholar]
- 67.Chevalier C, Geissmann T, Helfer A-C, Romby P. Probing mRNA structure and sRNA-mRNA interactions in bacteria using enzymes and lead(II). Methods Mol Biol 2009; 540:215-32; PMID:19381563; http://dx.doi.org/ 10.1007/978-1-59745-558-9_16 [DOI] [PubMed] [Google Scholar]
- 68.Gagnaire J, Dauwalder O, Boisset S, Khau D, Freydière A-M, Ader F, Bes M, Lina G, Tristan A, Reverdy M-E, et al.. Detection of Staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS ONE 2012; 7:e40660; PMID:22792394; http://dx.doi.org/ 10.1371/journal.pone.0040660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaltwasser M, Wiegert T, Schumann W. Construction and Application of Epitope- and Green Fluorescent Protein-Tagging Integration Vectors for Bacillus subtilis. Appl Environ Microbiol 2002; 68:2624-8; PMID:11976148; http://dx.doi.org/ 10.1128/AEM.68.5.2624-2628.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



