Abstract
More than 400 genes have been noted to modulate C. elegans longevity. Recent studies testing the role of proposed secreted insulin binding proteins unexpectedly revealed a potent role for the EGF signaling pathway in promoting healthy aging and longevity in C. elegans. Activation of EGF receptor LET-23 is associated with increased mean and maximum lifespan, maintained pharyngeal pumping, extended locomotory function, and low lipofuscin and advanced glycation end product accumulation. Conversely, reducing the activity of the EGF pathway is associated with system-wide evidence of progeria. The EGF pathway appears to work in a manner largely independent of the insulin/IGF-like pathway, in that effects are additive with reduction of DAF-2/InsR activity and are not affected by DAF-16/FOXO transcription factor deficiency. Two novel regulators of EGF signaling, called HPA-1 and HPA-2 (for the high performance in old age locomotory phenotypes that their disruption confers), negatively regulate EGF action, possibly by binding and sequestering EGF. Interestingly, whereas HPA-1 appears to control aging of the animal overall, HPA-2 exerts an effect primarily on locomotory aging. As such, HPA-2 is an example of a protein with an effect on healthspan but not lifespan, a gene class that may have been missed in screens focused on longevity endpoint. To date, roles for EGF signaling in adult maintenance (particularly in non-dividing tissues) have not been addressed in other organisms—should EGF signaling exert a conserved impact on healthy aging, testing this hypothesis could hold implications for anti-aging therapies.
Introduction
Aging is influenced by genetic, environmental, and stochastic processes that exert interconnected effects on lifespan. Analyses in invertebrate models have significantly advanced understanding of genetic and environmental influences that modulate lifespan, including insulin/IGF signaling, dietary restriction, mitochondrial function, sensory activity, and metabolism (Kenyon, 2010 ; Antebi, 2007). A striking theme that has emerged is that many of the pathways that influence longevity exert effects conserved from invertebrates to mammals.
A key challenge in current aging research is to increase life expectancy coupled with significantly enhanced functionality later in life. Improved healthspan—the period of functional maintenance preceding debilitating decline--is increasingly being considered as an endpoint for aging studies. Indeed, many pathways that confer longevity, such as reduced insulin signaling, are associated with robustness of function later in life, although this is not always the rule.
Our recent investigations of potential roles for candidate insulin-binding proteins in promoting C. elegans healthspan led to the discovery of an unexpected, but major, role of EGF signaling for functional maintenance in aging nematodes. The EGF pathway includes both global and tissue-specific modulators of healthy aging. Here we review how EGF signaling can influence C. elegans aging and raise the question as to whether this pathway might contribute to long-term maintenance in other organisms.
Evaluation of healthy aging in C. elegans
As a focus of aging research progresses from concentration on the longevity endpoint to increased healthspan evaluation, the issue of how to measure healthy aging becomes a front-and-center challenge. One metric that most likely indicates healthspan at the organism level is measurement of survival of an aging population at middle and middle/late life--scores such as the 50% survival time point or mean lifespan. Genetic or drug manipulations that extend mean survivorship but not necessarily maximum lifespan are likely to be of value for extending the period of healthy life prior to decline. Analysis of mortality curves is another option for analysis of changes in the “rate of aging” (Johnson, 1987).
Several C. elegans behaviors decline with age (Bolanowski et al.,1981; Chow et al., 2006; Garigan et al., 2002; Glenn et al., 2004; Herndon et al., 2002; Huang et al., 2004) and the extent of change can be used as a measure of healthspan. Pumping of the pharynx, an organ through which the animal ingests its bacterial food, declines fairly precipitously with age (Chow et al., 2006; Huang et al., 2004). A two-day-old adult pumps on the order of 300 contractions/minute, a rate that progressively slows to essentially zero at about 12 days of adult life (Collins et al., 2008). Pharyngeal contractions are easily observed through the transparent cuticle, and can be scored directly or with the aid of video analysis. Because the pharynx pumps by some autonomous mechanisms and includes specialized muscle that resembles mammalian heart muscle, the C. elegans pharynx is sometimes considered to model mammalian heart (Mango, 2007), and its age-associated decline to model mammalian cardiac aging.
The vigor of locomotion on solid support (agar plates in the lab) or in liquid (swimming) declines during C. elegans adult life. Diminished rates of movement can be measured by eye (body bends per unit time) or with computer programs that track locomotion (Huang et al., 2006; Schreiber et al., 2010; Tsechpenakis et al., 2008). Interestingly, locomotory behavior decline correlates with deterioration of body wall muscle, with fewer sarcomere units present over time and increased fat infiltration, reminiscent of mammalian sarcopenia (Garigan et al., 2002; Glenn et al., 2004). Although muscle exhibits markedly more dramatic cellular deterioration than do neurons, some neuronal influence on C. elegans sarcopenia appears operative (Glenn et al., 2004; Murakami et al., 2008).
Another conserved trait that tracks with age is the accumulation of fluorescent lipofuscin and advanced glycation end products (Gerstbrein et al., 2005), collectively referred to as age pigments. In C. elegans this fluorescence is concentrated in gut granules that appear to be secondary lysosomes (Clokey et al., 1986). Since extensively cross-linked lipofuscin cannot be degraded by lysosomal machinery, lysosomes accumulate this fluorescent material and are thought to become progressively impaired as adults age. Interestingly, animals that age well (by locomotory criteria) tend to have low age pigment accumulation and those that age poorly accumulate relatively high levels (Gerstbrein et al., 2005). Likewise, animals that are long-lived (for example the daf-2(rf) insulin receptor mutant) tend to have lower age pigment scores than do progeric mutants such as daf-16/FOXO transcription factor. Thus, high age pigment scores suggest a poor healthspan and relatively low age pigments scores suggest a strong healthspan. Age pigments can be scored using an image analysis program that measures relative intensity of fluorescence in the 340nm range; a more precise approach is to use a fluorometer that can scan samples over a range of input wavelengths and quantitate emission spectra (typically peak emission is ~342 nm).
Although more detailed cellular phenotypes than those listed above can be characterized, use of the aforementioned relatively easily determined measures can indicate whether a population appears to be aging well or poorly relative to wild-type controls. Note that animals may exhibit system-wide evidence of a strong healthspan (all measures) or extended healthspan of individual tissues (only pharynx, body wall muscle, intestine, for example) for a given genetic or drug intervention.
Genetic approaches toward understanding sarcopenia
C. elegans lives only about three weeks in the lab, whereas humans live 10 decades. Thus, it is striking that the basic phenomenon of sarcopenia (progressive loss of muscle mass and strength with aging) and general cellular phenotypes appear so similar across species. Given the power of C. elegans genetic screens to identify genes required for specific aging phenotypes, it is plausible to screen for genes that influence muscle healthspan by identifying those that accelerate or delay mobility decline (Schreiber et al., 2010; Iwasa et al., 2010).
Genes encoding likely secreted molecules that resemble insulin/IGF receptor binding domains
The C. elegans genome encodes one copy of a transmembrane kinase that is related to mammalian insulin/IGF receptor (Kimura et al., 1997), called DAF-2; there are 40 insulin-like ligand genes that encode potential ligands for this receptor (Li et al., 2003; Pierce et al., 2001). To address whether additional receptor-like molecules might be encoded by the C. elegans genome, Dlakić (Dlakić, 2002) performed an in silico screen for nematode genes that exhibited primary sequence similarity to the ligand-binding domains of the DAF-2 insulin receptor (L domains). This search did not identify any additional transmembrane kinases per se, but did turn up 54 potentially secreted proteins (signal sequences, no clear transmembrane domains, no kinase motifs) that exhibited L domain homologies. These proteins could be postulated to have the capacity to bind insulin by virtue of their homologies to insulin-binding sites of insulin receptors, but are not sequence homologs of mammalian insulin-binding proteins IBP1-7.
Systematic RNAi knockdown of individual genes in this group of 54 insulin-receptor-related proteins identified 9 with a significant enhancement of old-age swimming prowess, without altered or defective swimming ability in young adulthood (Iwasa et al., 2010). Thus, these genes were of interest as they specifically impacted age-associated swimming decline. Two of these genes, encoded by predicted open reading frames H25K10.5 and T11F1.8, and for which knockdown exerted the most robust RNAi effects on old-age swimming (Fig.1A, B), were further characterized in detail. These ORFs were renamed hpa-1 and hpa-2 (for the high performance in advanced age phenotype that their knockdown confers), respectively.
Figure 1. HPA-1 and HPA-2, related to insulin receptor and EGF receptor L domains that bind ligand, affect locomotory healthspan regulation.
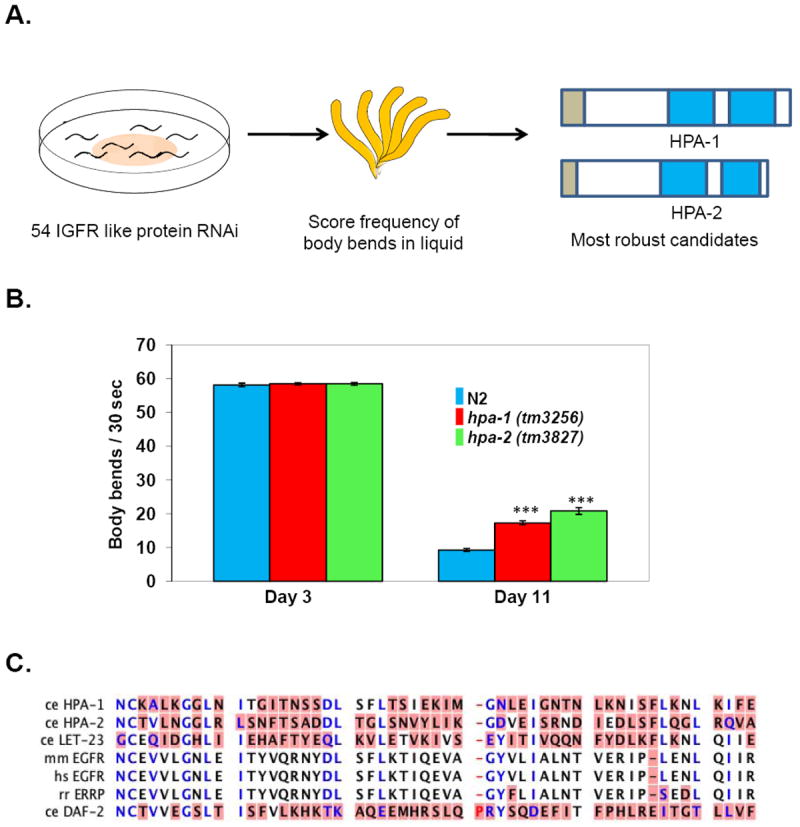
A. Summary of screen for RNAi interventions that improve late life swimming. C. elegans were fed with dsRNA to disrupt 54 IGF receptor-like genes (Dlakić, 2002) and scored for frequency of body bends in liquid on day11 (mid/late age) (Iwasa et al., 2010). Knockdown of 9 out of 54 genes resulted in better swimming in the population of old animals. Knockdown of hpa-1 and hpa-2 resulted in the most robust effects on late-life swimming. Signal sequences indicated in brown, L domains in blue.
B. Both hpa-1(tm3256) and hpa-2(tm3827) deletion mutants exhibit enhanced swimming performance in old age, confirming RNAi studies. Day 3 is the first day of adult life; day11 is mid/late life. ***p< 0.0001, compared to N2 wild type in each assay. Error bars represent standard error of the mean (SEM).
C. Domains of HPA-1 and HPA-2 proteins are similar in sequence to the L domain of C. elegans EGF receptor LET-23, mammalian ERRP and EGFR proteins, and C. elegans insulin receptor ligand binding domains (DAF-2). Blue amino acids indicate highly conserved residues; amino acids with red background represent different residues in the alignment. Alignment result is based on CLC sequence viewer. C. elegans (Ce), Homo sapiens (Hs), Mus musculus (Mm) and Rattus norvegicus (Rn).
Figures are remade from Iwasa et al., 2010.
Deciphering the mechanism by which hpa gene knockdown enhances healthy aging
Given the homologies of HPA-1 and HPA-2 to insulin-binding domains, the logical hypothesis was that these secreted proteins might influence swimming healthspan through the insulin/IGF pathway. However, testing of this hypothesis revealed that hpa-1 and hpa-2 RNAi-knockdown can still extend swimming healthspan in a daf-16/FOXO null mutant background, and that hpa-1 and hpa-2 knockdown benefits are additive with those of daf-2/InsR or age-1/PI3K reduction-of-function. These data argue against a central role for insulin/IGF signaling in HPA-1 and HPA-2 locomotory healthspan; instead insulin pathway benefits may run in parallel to HPA action. Unexpectedly, then, it appeared that hpa-1 and hpa-2 deficiencies exert major effects on locomotory aging via a pathway distinct from the conserved insulin/IGF pathway.
Insight into the mechanism of action came with the revisiting of the homologies of HPA-1 and HPA-2 to known signaling receptors. Close inspection suggested a greater sequence similarity of HPA-1 and HPA-2 to the EGF receptor EGF-binding domain than to the insulin receptor binding domain (Fig. 1C). Moreover, HPA-1 is most similar to mammalian EGF receptor-related protein (ERRP, homologous to mammalian EGFR), which is a secreted negative regulator of mammalian EGF family signaling (Yu et al., 2001). Sequence relationships thus suggested that, rather than insulin signaling, EGF signaling might be involved in C. elegans locomotory healthspan extension.
Activation of the EGF signaling pathway extends C. elegans healthspan
Given the extensive effort devoted to the identification of longevity genes in the C. elegans field, it was surprising that no work had previously examined the role of EGF signaling in lifespan extension. However, elegant work by Sternberg and colleagues had characterized the C. elegans EGF pathway in detail, with a focus on its role in developmental fate specification (Emmons, 2005; Sternberg, 2005; Sundaram, 2006) and more recently for its role in regulating the period of quiescence that precedes developmental molts (Van Buskirk et al., 2007). Thus, a wealth of genetic reagents was available for testing healthspan and lifespan phenotypes.
To address potential roles of the EGF pathway in healthy aging, consequences of genetic manipulation of the EGF ligand (encoded by lin-3 (Hill et al., 1992)) and EGF receptor (encoded by let-23 (Aroian et al., 1990)) were tested (Iwasa et al., 2010). An EGFR/let-23(gf) gain-of-function mutant retained late-life swimming vigor and muscle integrity, maintained pharyngeal pumping capacity late into life, exhibited low rates of age pigment accumulation, and survived more robustly in midlife and late life, revealing a role for EGFR activation in organism-wide healthy aging (Fig. 2). Conversely, a reduction-of-function mutation in EGFR/let-23 confers accelerated swimming decline and diminished survival at mid life; as does mutation of EGF/lin-3(rf). Thus, activation of EGF signaling positively influences healthspan as well as lifespan in C. elegans.
Figure 2. An activated EGF receptor gain-of-function mutant exhibits enhanced swimming prowess late in life, increased longevity, and lower age pigment accumulation in aging animals.
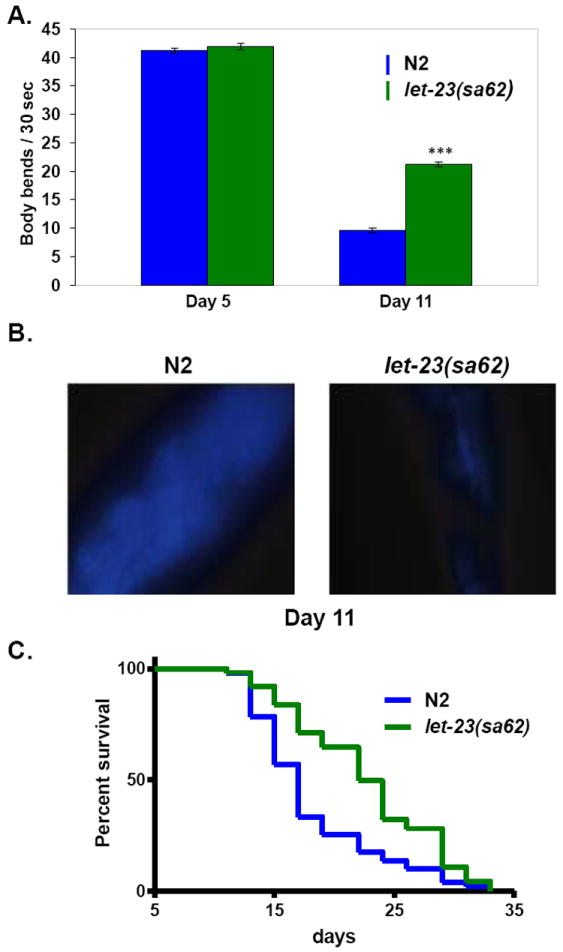
A. An activated EGFR / let-23 gain-of-function mutant exhibits a slower decline in swimming prowess in old age as compared to wild type N2.
B. Accumulation of lipofuscin in the intestine is visible here as fluorescent granules. An activated EGFR / let-23 gain-of-function mutant has reduced accumulation of lipofuscin and advanced glycation endproducts compared to wild type N2.
C. An activated EGFR / let-23 gain-of-function mutant shows an increase in mean and maximum lifespan as compared to wild type N2.
Together, these data support that activation of EGF pathway promotes system-wide healthy aging. ***p< 0.0001. Error bars represents standard error of mean (SEM).
Figures are remade from Iwasa et al., 2010.
A specific downstream signaling pathway involving phopholipase Cγ and the ER IP3 receptor calcium-release channel mediates EGF healthspan benefits
There are three distinct downstream signal transduction pathways for C. elegans EGF signaling that act through LIN-3/EGF and LET-23/EGFR. A RAS-MAPK (let-60-mek-1) pathway affects several cell fate decisions, including the well-studied vulval development (Moghal et al., 2003). LET-23/EGFR (LET-23) signaling also induces behavioral quiescence at the molt through plc-3 (phospholipase Cγ), unc-13, and tpa-1(both encoding diacylglycerol-binding proteins) (Van Buskirk et al., 2007). In addition, EGFR acts through PLC-γ (plc-3) and IP3-inositol (1,4,5) triphosphate receptor (IP3R, encoded by itr-1) signaling (endoplasmic reticulum (ER) calcium release) to affect ovulation and defecation cycles (Clandinin et al., 1998; Yin et al., 2004). Testing of representative genes in each downstream signaling branch established that it is the PLC- γ/plc-3 and IP3 receptor/itr-1 signaling that mediates healthy aging (Iwasa et al., 2010). Supporting data include that an IP3R gain-of-function shows a markedly enhanced general healthspan and significantly extended lifespan. Conversely, IP3R reduction-of-function confers a short life and accelerated swimming decline and RNAi-knockdown can block the beneficial effects of EGFR activation. Because the IP3R modulates intracellular calcium by regulating ER calcium release, data implicate appropriate/maintained calcium homeostasis in healthy aging.
HPA-1 and HPA-2 act via the EGF pathway
Although the aforementioned experiments reveal a role for an EGF pathway in healthy aging, they do not address whether the newly identified HPA-1 and HPA-2 secreted proteins actually exert healthspan effects via this pathway. A key experiment was to test whether hpa-1 or hpa-2 knockdown could confer healthspan benefits when the EGF signaling pathway was disrupted. Indeed, if hpa-1 or hpa-2 are knocked down in the let-23/EGFR(rf) background, lifespan and swimming prowess benefits later in life are no longer observed. Healthspan benefits of hpa-1 and hpa-2 knockdown are also eliminated when IP3R signaling is down-regulated. Together, data support that hpa-1 and hpa-2 disruption must act via the EGFR/IP3R pathway to confer healthy aging. Note that since genetic deletion or RNAi targeting of hpa-1 and hpa-2 improves healthy aging, HPA-1 and HPA-2 normally function as negative regulators of the EGF pathway and negative regulators of healthy aging. The HPA-1 and HPA-2 homologies to EGF binding sites suggest they may bind circulating EGF to limit availability for signaling. HPA-1-related mammalian ERRP is thought to bind EGF in mammals to limit EGFR signaling (Marciniak et al., 2004), supporting such a hypothesis, and suggesting a conserved regulatory mechanism.
A model for EGF signaling in maintaining aging C. elegans
Analysis of EGF signaling in C. elegans leads to a working model for this pathway in modulation of healthy aging (Fig. 3). In the wild-type aging adult, candidate EGF-binding proteins HPA-1 and HPA-2 bind circulating EGF to limit maintenance signaling. Disruption of HPA proteins releases EGF for increased EGFR binding and activation of downstream PLCγ/IP3R signaling, which significantly improves maintenance and function during adult life. Note that many details of this mechanism remain to be addressed, including the questions of whether HPA-1 or HPA-2 bind EGF, which of the known C. elegans EGF isoforms modulates healthy aging, in what tissues EGF acts to promote healthspan, and what happens after IP3R modulation of intracellular calcium to improve maintenance and extend healthy aging.
Figure 3. Model of C. elegans EGF pathway modulation in aging.
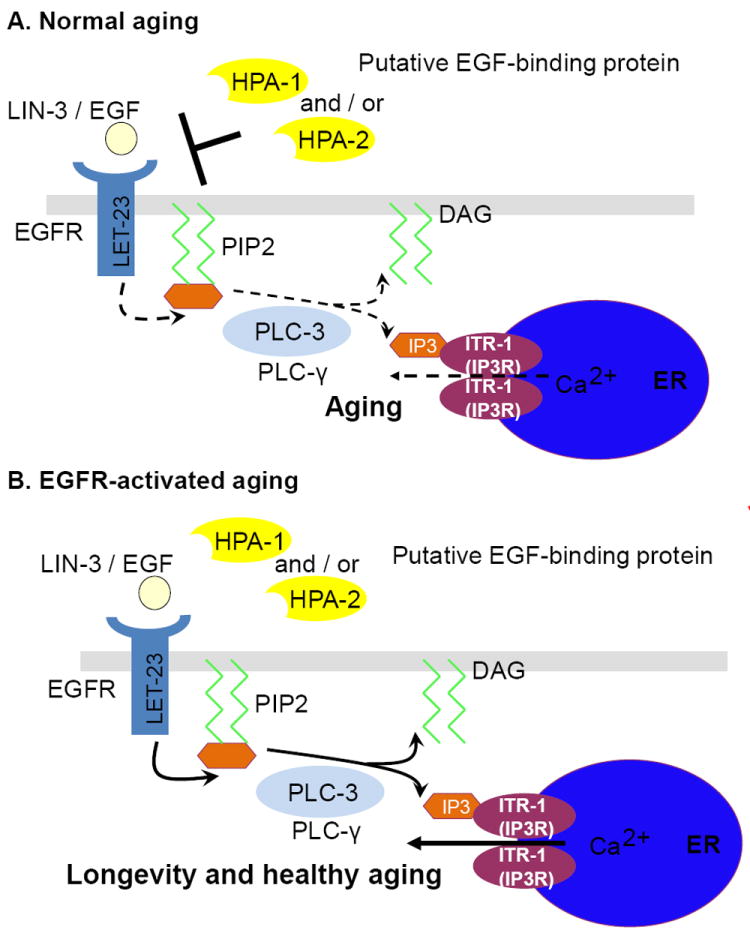
A. In wild type, putative EGF-binding proteins HPA-1 and HPA-2 function as negative regulators (possibly sequestering EGF ligand) to inhibit LIN-3/EGF LET-23/EGFR signal transduction, conferring the wild-type aging rate.
B. When HPA-1 or HPA-2 are knocked down or absent, EGF signaling through EGFR is increased, activating downstream phospholipase Cγ and IP3 receptor (more ER calcium release) to promote extended healthspan and lifespan. Note that HPA-1 knockdown impacts all assays of animal healthspan, whereas HPA-2 exerts its effect primarily on locomotory healthspan.
A tissue-specific component of EGF signaling—HPA-2 impacts locomotory healthspan without great effect on age-pigment accumulation or lifespan
There is an important distinction between hpa-1 and hpa-2 activities in healthspan promotion (Iwasa et al., 2010). HPA-1 and HPA-2 were both identified by virtue of their capacities to influence locomotory aging. Interestingly, these two sequence-related proteins are co-expressed during adulthood by posterior intestine, glial amphid and plasmid socket cells, and a few neurons, which most likely secrete HPA-1 and HPA-2. Like the EGF pathway mutations, the hpa-1 deletion mutation impacts aging broadly—enhancing overall quality in all healthspan assays. Notably, this is not the case for hpa-2, deletion of which exerts a strong impact on locomotory aging, but not on age pigment accumulation or lifespan (Iwasa et al., 2010). In other words, hpa-2 effects are fairly specific to the aging of the locomotory system and do not confer systemic benefits. Models by which HPA-2 might act differentially include a distinct binding to a specific EGF isoform or an affinity to muscle to regulate expression in that tissue.
It should be emphasized that hpa-2 represents one of the first documented examples of a gene with tissue-specific effects on aging quality. The identification of a healthspan-specific gene suggests that there may exist a class of genes that when manipulated might change the aging of individual tissues without a major impact on the longevity endpoint. As such, healthspan-only genes may have been missed by genetic screens for longevity per se. New screens for tissue–specific betterment of aging phenotypes might reveal a class of genes that might be manipulated to promote healthy aging of individual tissues.
Perspectives
Insulin and EGF: signaling pathways that can influence C. elegans aging quality
Experiments with C. elegans temperature-sensitive signaling mutations support that the EGF pathway can be activated only in the adult in order to positively influence aging (Iwasa et al., 2010). Up-regulation confers benefits system-wide in the locomotory systems, the heart muscle system, and intestinal maintenance, as well as survival. Thus, in addition to the insulin/IGF pathway (which can also act in the adult to modulate lifespan (Dillin et al., 2002)), EGF signaling appears to be another signaling pathway with major impact on the quality of aging.
It should be emphasized that seven additional hpa genes identified as related to the extracellular domain of InsR can be knocked down by RNAi to extend healthy locomotory aging (Iwasa et al., 2010). As proposed for HPA-1 and HPA-2, these might also bind EGF to regulate that signaling pathway; alternatively, these other proteins might bind insulins as originally suggested (Dlakić, 2002). Further investigation of these aging modulators is needed to clarify roles in regulating EGF, insulin, or both pathways to promote healthy aging.
Might enhanced EGF family signaling promote healthy aging and maintained function across species?
To date, there is no evidence for roles of EGF signaling in healthy aging in other experimental models, but the specific question of age-associated maintenance has not yet been directly addressed. In Drosophila, EGF family signaling is mediated by TGFα-related ligands Spitz (ventral ectoderm patterning (Schweitzer et al., 1995)), Keren (Reich et al., 2002), Gurken (oogenesis (Nilson et al., 1999)), and Vein (oogenesis role, neuregulin homolog (Schnepp et al., 1996)) and EGFR homolog DER (Shilo, 2003) (Fig. 4). One secreted EGF-binding protein, Argos, is known to sequester Spitz ligand to limit signaling (Klein et al., 2004), as is proposed for HPA interaction with LIN-3/EGF. It should be noted that Argos is not homologous to HPA proteins and actually binds EGF via an interaction domain that is distinct from other known EGF-binding domains (Klein et al., 2008). Roles for enhanced EGF signaling in adult flies have not yet been probed. If Argos serves a function analogous to HPA-1, its genetic disruption in adults might enhance EGF signaling and confer healthy aging in flies.
Figure 4. Summary of ligands and receptors in the EGF signaling pathways in C. elegans, Drosophia melanogaster, and humans.
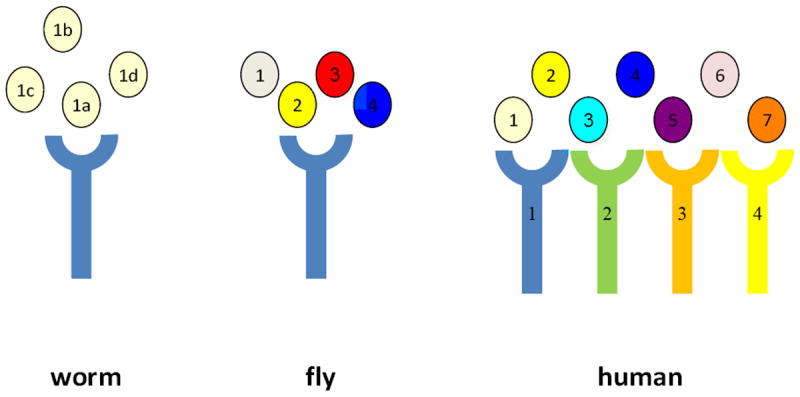
There is only one EGF ligand gene, lin-3, encoded by the C. elegans genome, although 4 splice-generated isoforms can be produced (noted a-d) (Van Buskirk et al., 2007). Four ligand genes are found in Drosophila: 1. Keren, 2. Spitz (TGF-α related), 3. Gurken, 4. Vein (neuregulin homolog). There are seven ligands in human: 1. EGF, 2. TGF-α, 3. Amphregulin, 4. Neuregulin, 5. Epigerulin, 6. Epigen, 7. Betacellulin. Both C. elegans and Drosophila genomes encode single EGFR receptors, LET-23 and DER respectively. Four ErbB subfamily receptors in human are: 1. EGFR, 2. HER-2, 3. HER-3, 4. HER-4. Human ligands cross-bind to multiple receptors.
An HPA-1 sequence-related mammalian protein is the rat EGF receptor-related protein (ERRP) (Iwasa et al., 2010). Interestingly, in mammalian cell culture, ERRP negatively regulates EGFR cell proliferation activity (Yu et al., 2001) and can be an effective therapeutic agent for prostate (Marciniak et al., 2004) and colon cancer models, the latter of which is suggested to occur via sequestration of EGFR ligand TGF-α (Schmelz et al., 2007). There is no clear human ERRP homolog (Reiter et al., 2006), but, by analogy to C. elegans studies, a protein functionally related to mouse ERRP or fly Argos might influence the maintenance of aging tissues in other organisms. The C. elegans findings raise interest in developing an ERRP knockout mouse (possibly adult-specific conditional) to address potential impact on aging biology.
Mouse genetics have not yet clearly delineated specific roles of EGFs and EGFRs in biology. The epidermal growth factor receptor (EGFR) transmembrane receptor tyrosine kinase belongs to the mammalian ErbB subfamily (EGFR, Erb-2/HER-2; Erb-3/HER-3; Erb-4/HER-4). When mammalian EGF family ligands bind (EGF, TGF-α, HB-EGF, amphiregulin, epiregulin, epigen, betacellulin, neuregulin) (Fig.4), EGFRs regulate cellular signal transduction pathways that regulate cell proliferation, survival, and migration (Citri et al., 2006). Receptor knockouts have revealed developmental lethality but some ligand knockouts are viable. Knockout mice for TGF-α ligand exhibit excessive hair loss and weight loss with aging (Luetteke et al., 1999). Triple knockout of ligands EGF, TGF- α, and amphiregulin exhibit a higher penetrance of hair and weight loss compared to the TGF- α single knockout mutant (Luetteke et al., 1999). Whether these phenotypes might suggest accelerated segmental aging awaits a more focused look at phenotypes and an experimental separation of adult phenotypes from developmental defects by adult-specific and possibly tissue-specific disruption.
Enhanced EGFR signaling has been associated with epithelial tumors in mammals (Herbst, 2004), which raises significant questions regarding value of enhanced EGF signaling as an anti-aging therapy. In C. elegans, cell divisions are completed by the time the animal reaches adulthood, which means that all aging studies are focused on post-mitotic cells. Thus, distinguishing EGF impact on non-dividing cell types such as neurons might be critical for evaluating potential for age-associated maintenance. The fact that some EGF signaling regulators appear to be tissue-restricted (e.g. HPA-2 on locomotory aging), is a proof-of-principle that beneficial tissue-restricted manipulations might be capable of promoting healthy aging of specific declining systems. It is also possible that a specific mammalian EGF family member might prove to be potent for maintenance signaling rather than proliferation. Serum levels of EGF decrease in healthy human aging (Shurin et al., 2007)—if EGF impact on human aging is conserved, novel mechanisms of increasing EGF action, such as HPA disruption might extend healthy aging.
Acknowledgments
We appreciate and acknowledge the scientific contributions of Dr. Hiroaki Iwasa in the analysis of the EGF pathway in C. elegans aging biology. We thank Dr. Matthew Buechner for critical reading of the manuscript. Work was supported by NIH R01AG024882001, the Ellison Medical Foundation, and the Glenn Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature. 1990;348:693–699. doi: 10.1038/348693a0. [DOI] [PubMed] [Google Scholar]
- Bolanowski MA, Russell RL, Jacobson LA. Quantitative measures of aging in the nematode Caenorhabditis elegans. I. Population and longitudinal studies of two behavioral parameters. Mech Ageing Dev. 1981;15:279–295. doi: 10.1016/0047-6374(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Chow DK, Glenn CF, Johnston JL, Goldberg IG, Wolkow CA. Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Exp Gerontol. 2006;41:252–260. doi: 10.1016/j.exger.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Collins JJ, Huang C, Hughes S, Kornfeld K. WormBook. The C. elegans Research Community, WormBook; 2008. The measurement and analysis of age-related changes in Caenorhabditis elegans. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dlakić M. A new family of putative insulin receptor-like proteins in C. elegans. Curr Biol. 2002;12:R155–157. doi: 10.1016/s0960-9822(02)00729-7. [DOI] [PubMed] [Google Scholar]
- Emmons SW. WormBook. The C. elegans Research Community, WormBook; 2005. Male development. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- Glenn CF, Chow DK, David L, Cooke CA, Gami MS, Iser WB, Hanselman KB, Goldberg IG, Wolkow CA. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. J Gerontol A Biol Sci Med Sci. 2004;59:1251–1256. doi: 10.1093/gerona/59.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS. Review of epidermal growth factor receptor biology. International Journal of Radiation Oncology, Biology, Physics. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Hill RJ, Sternberg PW. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KM, Cosman P, Schafer WR. Machine vision based detection of omega bends and reversals in C. elegans. J Neurosci Methods. 2006;158:323–336. doi: 10.1016/j.jneumeth.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Iwasa H, Yu S, Xue J, Driscoll M. Novel EGF pathway regulators modulate C. elegans healthspan and lifespan via EGF receptor, PLC-gamma, and IP3R activation. Aging Cell. 2010;9:490–505. doi: 10.1111/j.1474-9726.2010.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE. Aging can be genetically dissected into component processes using longlived lines of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1987;84:3777–3781. doi: 10.1073/pnas.84.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Klein DE, Nappi VM, Reeves GT, Shvartsman SY, Lemmon MA. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- Klein DE, Stayrook SE, Shi F, Narayan K, Lemmon MA. Structural basis for EGFR ligand sequestration by Argos. Nature. 2008;453:1271–1275. doi: 10.1038/nature06978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- Mango SE. WormBook. The C. elegans Research Community, WormBook; 2007. The C. elegans pharynx: a model for organogenesis. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak DJ, Rishi AK, Sarkar FH, Majumdar AP. Epidermal growth factor receptor related peptide inhibits growth of PC-3 prostate cancer cells. Mol Cancer Ther. 2004;3:1615–1621. [PubMed] [Google Scholar]
- Moghal N, Sternberg PW. The epidermal growth factor system in Caenorhabditis elegans. Exp Cell Res. 2003;284:150–159. doi: 10.1016/s0014-4827(02)00097-6. [DOI] [PubMed] [Google Scholar]
- Murakami H, Bessinger K, Hellmann J, Murakami S. Manipulation of serotonin signal suppresses early phase of behavioral aging in Caenorhabditis elegans. Neurobiol Aging. 2008;29:1093–1100. doi: 10.1016/j.neurobiolaging.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Nilson LA, Schüpbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Shilo BZ. Keren. A new ligand of the Drosophila epidermal growth factor receptor, undergoes two modes of cleavage. EMBO J. 2002;21:4287–4296. doi: 10.1093/emboj/cdf439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JL, Maihle NJ, Clinton GM. Therapeutic potential of epidermal growth factor receptor-related protein. Mol Cancer Ther. 2006;5:2954. doi: 10.1158/1535-7163.MCT-05-0404. [DOI] [PubMed] [Google Scholar]
- Schmelz EM, Xu H, Sengupta R, Du J, Banerjee S, Sarkar FH, Rishi AK, Majumdar AP. Regression of early and intermediate stages of colon cancer by targeting multiple members of the EGFR family with EGFR-related protein. Cancer Res. 2007;67:5389–5396. doi: 10.1158/0008-5472.CAN-07-0536. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, McIntire SL. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. 2010;6:e1000972. doi: 10.1371/journal.pgen.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Shilo BZ. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res. 2003;284:140–149. doi: 10.1016/s0014-4827(02)00094-0. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Yurkovetsky ZR, Chatta GS, Tourkova IL, Shurin MR, Lokshin AE. Dynamic alteration of soluble serum biomarkers in healthy aging. Cytokine. 2007;39:123–129. doi: 10.1016/j.cyto.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Sternberg PW. WormBook. The C. elegans Research Community, WormBook; 2005. Vulval development. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram MV. WormBook. The C. elegans Research Community, WormBook; 2006. RTK/Ras/MAPK signaling. http://www.wormbook.org. [DOI] [Google Scholar]
- Tsechpenakis G, Bianchi L, Metaxas D, Driscoll M. A novel computational approach for simultaneous tracking and feature extraction of C. elegans populations in fluid environments. IEEE Trans Biomed Eng. 2008;55:1539–1549. doi: 10.1109/TBME.2008.918582. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- Yin X, Gower NJ, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Rishi AK, Turner JR, Liu D, Black ED, Moshier JA, Majumdar AP. Cloning of a novel EGFR-related peptide: a putative negative regulator of EGFR. Am J Physiol Cell Physiol. 2001;280:C1083–1089. doi: 10.1152/ajpcell.2001.280.5.C1083. [DOI] [PubMed] [Google Scholar]


