Abstract
Double-stranded RNA viruses sequester their genomes within a protein shell, called the polymerase complex. Translocation of ssRNA into (packaging) and out (transcription) of the polymerase complex are essential steps in the life cycle of the dsRNA bacteriophages of the Cystoviridae family (φ6–φ14). Both processes require a viral molecular motor P4, an NTPase, which bears structural and functional similarities to hexameric helicases. In effect, switching between the packaging and the transcription mode requires the translocation direction of the P4 motor to reverse. However, the mechanism of the reversal remains elusive. Here we characterize the P4 protein from bacteriophage φ12 and exploit its purine nucleotide specificity to delineate P4 role in transcription. The results indicate that while P4 actively translocates RNA during packaging it acts as a passive conduit for RNA export. The directionality switching is accomplished via the regulation of P4 NTPase activity within the polymerase core.
INTRODUCTION
Energy-driven translocation of nucleic acids (NAs) plays an important role in many biological processes, from chromosomal replication and segregation to virus genome encapsidation (packaging). A unifying theme in all these processes is the involvement of molecular motors, which convert chemical energy into mechanical work. Prominent examples of such motors are hexameric helicases (1). The basic activity of a helicase is a unidirectional unwinding of nucleic acid duplexes, a process that is coupled to the hydrolysis of nucleoside triphosphates (NTP) (2). Helicases are found in all living organisms and are essential for nucleic acid metabolism [e.g. DnaB, T7 gp4, T4 gp41 (3), Rho (4) RuvB, (2)].
Viral packaging motors (portal complexes) constitute another class of nucleic acid transporters. Portal complexes perform viral genome packaging into preformed capsids (procapsids) (5). Typical examples are portal complexes from dsDNA viruses (herpesviruses, adenoviruses and tailed bacteriophages φ29, λ, P22 and T4) (5–7). In these cases, an oligomeric portal protein [usually a dodecamer (8)] is an integral part of the capsid and acts as a bidirectional conduit for both DNA packaging (translocation into the capsid) and ejection (exit from the capsid). Bidirectionality is accomplished via temporal association of the packaging NTPase [terminase (9)] with the portal complex during DNA encapsidation (10). After packaging the terminase dissociates and enables dsDNA exit during infection. The exit is initially driven by osmotic pressure of the highly condensed dsDNA within the capsid (6) and is sometimes facilitated by the host restriction enzyme machinery (11).
Bidirectionality of RNA transport is also essential for dsRNA viruses, of which bacteriophage φ6 from the Cystoviridae family serves as a model for packaging and replication (12,13). Cystoviruses have a ∼13 kb genome comprising three segments, which are enclosed in an icosahedral polymerase complex which is further coated by a nucleocapsid (NC) protein shell and a lipid envelope (14). The polymerase complex (core) is composed of 120 copies of major structural protein P1, 12 monomers of RNA-dependent RNA polymerase P2, 12 hexamers of packaging motor P4 and 30 dimers of assembly factor P7 (15). Proteins P2 and P4 constitute the enzymatic machinery for genome transcription, replication and packaging (Figure 1) (16,17).
Figure 1.
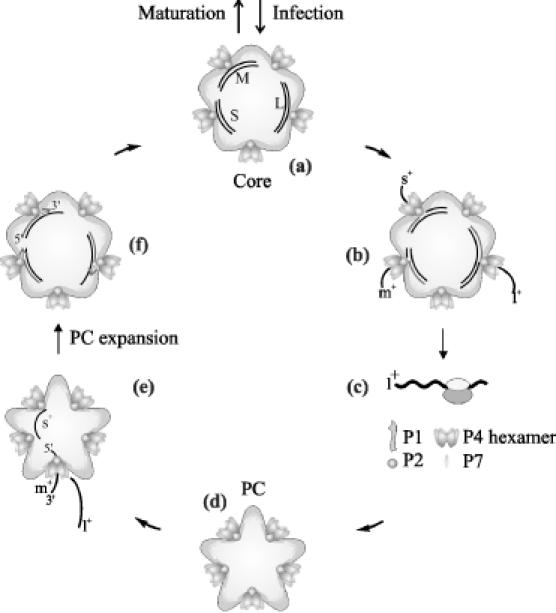
Simplified scheme of the cystoviral life cycle. The three dsRNA genomic segments of a cystovirus are brought into the host cell inside the viral core (a). Upon cell entry, the core catalyzes semi-conservative dsRNA transcription and (+) sense ssRNA transcripts (l+, m+ and s+) are extruded into the cytoplasm (b). The cellular protein synthesis machinery translates l+ RNA (c) giving rise to proteins P1, P2, P4 and P7. The newly produced proteins assemble into empty polymerase complexes (PC) (d), which are capable of packaging specifically one copy of each l+, m+ and s+ segments (e). Upon packaging PC expands and replication is initiated (f). The dsRNA-filled PC (core) can enter additional rounds of transcription or mature into infectious virions. The latter pathway uses proteins produced by the translation of m+ and s+ ssRNA segments, which is followed by the acquisition of the rest of the viral structural proteins together with the lipid membrane (not shown). The mature virus particles are released by lysis of the host cell (32).
Packaging is performed by the hexameric NTPase P4. Purified P4 hexamers from bacteriophages φ8 and φ13 possess helicase activity which is mediated by unidirectional movement (5′ to 3′) along ssRNA (18). Furthermore, low-resolution structures and activities of φ6, φ8 and φ13 P4 are similar to those of hexameric helicases (18–20). Taken together, these findings indicate that the RNA packaging mechanism might be remarkably similar to that of nucleic acid translocation by hexameric helicases, such as transcription-termination factor Rho, cellular DNA helicase DnaB, or bacteriophage T7 helicase gp4 (3). P4 protein is also required for the export of mRNA from the core particles during semi-conservative transcription, i.e. transport in the direction opposite to that of packaging (21). The bidirectionality may be accomplished in three principal ways: (i) P4 is involved in the strand separation at the replication fork as observed for helicases in Reoviridae (22); (ii) P4 acts as a passive but necessary conduit for the newly transcribed ssRNA; (iii) P4 actively translocates RNA out of the core, i.e. its directionality is switched after packaging (e.g. the transcription hexamers may be in opposite orientation).
To test these hypotheses, we have developed a direct method based on unique enzymatic properties of P4 from bacteriophage φ12. In this work, we show that φ12 P4 is a hexameric NTPase with purine nucleotide specificity. The specificity allowed for comparison of transcriptional activity of isolated φ12 core particles under conditions favorable for P4 NTPase and in the presence of non-hydrolyzable analogs. The comparison shows that P4 does not utilize the energy of NTP hydrolysis for translocation of newly synthesized mRNA and acts instead as a passive conduit for nascent transcript export. The results also show that tight regulation of the ATPase activity within the core controls the translocation directionality of the P4 hexamer.
MATERIALS AND METHODS
φ12 virus purification and isolation of the viral core
Bacteriophage φ12 was grown on strain LM2333, a mutant of Pseudomonas syringae pv. phaseolicola HB10Y, and the core was purified as described previously (23). Briefly, 20 plate lysates of φ12 were prepared by plating phage dilutions into soft agar using an overnight bacterial culture. Plates were incubated overnight at room temperature and the top agar was collected. The cell debris and remaining agar were removed by centrifugation (T647.5 rotor, 15 000 r.p.m., 15 min, 4°C) and the virus particles in the supernatant were concentrated by centrifugation (SW41 rotor, 30 000 r.p.m., 2 h, 4°C). The phage pellet was resuspended in 1 ml ACN buffer (10 mM KH2PO4, pH 7.2, 1 mM MgSO4, 200 mM NaCl and 0.5 mM CaCl2) and further purified by equilibrium density gradient centrifugation in CsCl (average density 1.28 g/ml, Beckman SW41 rotor, 30 000 r.p.m., 8 h, 4°C). The phage band was collected and dialyzed overnight against ACN buffer. The virus was collected from the dialyzed sample by centrifugation (SW41 rotor, 33 000 r.p.m., 2 h, 4°C) and the phage pellet was resuspended in 300 μl ACN buffer.
In order to isolate cores free of the lipid and P8 shell, the purified phage was extracted with 1% Triton X-114 (24) in 1 mM EDTA, 10 mM Tris–HCl, pH 8.0, and collected by centrifugation (SW41 rotor, 30 000 r.p.m., for 2 h, 4°C). The core pellet was resuspended in buffer containing 20 mM Tris–HCl, pH 8.0, 100 mM NaCl and 2 mM MgCl2, and stored at 4°C. SDS–PAGE analysis of the core protein composition exhibited the lack of envelope components and protein P8 but had the expected ratio of proteins P1, P2, P4 and P7 (23).
In vitro transcription of the φ12 dsRNA
Viral cores were utilized for in vitro transcription of the three viral dsRNA segments (25). The transcription reaction mixture contained 50 mM NH4Ac, 100 mM KCl, 50 mM Tris–HCl, pH 9.0, 1 mM MgCl2, 2.5 mM MnCl2, 4% (w/v) PEG 6000, 5 mM DTT, 0.2 mg/ml BSA, 0.4 U/μl RNAsin (Promega), 2.5 mM each of ATP, GTP, CTP and UTP, or AMPPNP, GMPPNP, CTP and UTP (non-hydrolyzable by P4 but good substrates for RNA synthesis). Reactions (100 μl total volume) were started by addition of ∼1.5 μg of core, incubated at 30°C for 0, 15, 30, 45, 60, 90 and 120 min and transferred to ice. The synthesized transcripts were phenol:chloroform (1:1 v/v) extracted and ethanol-precipitated. The samples were dissolved in TE buffer (Tris–EDTA) and analyzed on 1% agarose gel (in standard Tris–Borate–EDTA buffer, TBE) at room temperature.
To confirm that RNA synthesis occurred within the core, the reaction mixture containing 2.5 mM each of AMPPNP, GMPPNP, CTP and 0.1 mM UTP was supplemented with 0.1 mCi of [α-32P]UTP (3000 Ci/mmol; Amersham) and incubated at 30°C for 120 min. ‘Plus’ strand synthesis was detected by incorporation of [α-32P]UTP into the nascent RNA. The sample was sedimented through a 5–20% sucrose gradient in 20 mM Tris–HCl, pH 8.0, 50 mM NaCl and 2 mM MgCl2 (SW41 rotor, 33 000 r.p.m., 2 h, 4°C), fractionated, and the fractions were analyzed by 1% agarose gel (in TBE). The gel was dried and imaged by autoradiography.
NTPase activity assay of φ12 P4
φ12 P4 was purified as described (26). P4 concentration was determined by A280 using an extinction coefficient of 26 930 M−1 cm−1 that was calculated based on the amino acid composition (27).
The steady-state rate of NTP hydrolysis of isolated P4 and viral cores was measured using EnzChek Phosphate Assay Kit E-6646 (Molecular Probes, Inc.) as described previously (20). Because the kit has limited pH range (6.5–8.5) and strict requirements for divalent cations, NTPase activity under different pH and metal concentration was assayed by thin layer chromatography (TLC) followed by autoradiography (28). Typically, reaction mixtures contained 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5 mM MgCl2, 2 mM unlabeled ATP or GTP, and 0.5 μCi of the [α-32P]ATP or GTP (3000 Ci/mmol; Amersham Biosciences). The final P4 concentration was 0.25 mg/ml. Reactions were incubated for 1 h at 37°C, and 1 μl of the reactions was analyzed on polyethyleneimine (PEI) cellulose F TLC plates (Merck) and the amount of hydrolyzed nucleotide was determined using a phosphorimager (Fuji BAS 1500).
Translocation/helicase activity and agarose gel-shift assay
Complementary oligonucleotide displacement (COD) assay has been used to detect RNA helicase activity of isolated P4 proteins (18). An RNA substrate was prepared by annealing an unlabeled 700 nt 5′ terminal fragment of the s+ segment from phage φ6 (sR5 RNA) with the 32P-labeled 66mer of RNA oligonucleotide (RNA1). RNA1 was designed to target the 3′ proximal region of sR5. Only the middle 18 nt of RNA1 can form a duplex with the complementary sequence (nucleotides 590–607 in sR5 sequence), thus both the 5′ and 3′ termini of RNA1 form single-stranded overhangs. RNA1 displacement from sR5 indicates translocation and helicase activity (18). The liberated RNA1 probe can be separated from the duplex on 7% PAGE under native conditions (TBE).
Nucleic acid binding to P4 protein was assessed by an electrophoretic mobility shift assay (gel-shift), as described (18). Briefly, φ12 P4 (at 0.1 and 0.01 mg/ml, respectively) was incubated for 15 min on ice with RNA (0.1 mg/ml) in 7.5 μl mixtures containing 20 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 50 mM NaCl. Thereafter, 2.5 μl of 15% glycerol, 20 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 50 mM NaCl, 0.25% bromophenol blue was added and the samples were electrophoresed on 1.2% agarose gel (TBE, 5 V/cm, 90 min).
Analytical methods
Gel filtration was performed at room temperature on a Superdex-200 10/30 column (Amersham Biosciences) buffered with 20 mM Tris–HCl, pH 8.0, 50 mM NaCl and 5 mM MgCl2, flow rate 0.5 ml/min. The column was calibrated using gel filtration molecular weight standards from Sigma. Molecular masses were determined using a light scattering detector (Precision Detectors) calibrated with BSA (Sigma) (29).
Hydrodynamic properties of φ12 P4 were studied using a batch dynamic light scattering instrument (Precision Detectors) equipped with deconvolution software for correlation function analysis.
RESULTS
φ12 P4 hexamer shares similarity with φ6 P4
In previous work, we compared the enzymatic properties of the φ8 P4 NTPase with those of the NTPases from φ6 and φ13 (18). φ12, the most recently characterized member of the Cystoviridae family, offers an additional opportunity to examine functional properties of the NTPase subunit.
Biochemical and biophysical properties of φ12 P4 were found to be similar to those of φ6 P4 (Table 1 and following sections). The molecular mass of φ12 P4 deduced by light scattering was ∼205 kDa, consistent with a hexameric arrangement (210 kDa). The P4 protein forms a hexamer without NTP/NDP but requires Mg2+ ions for stability. A hydrodynamic radius (Rh = 5.5 ± 0.5 nm) indicated that φ12 P4 is a ring-shaped hexamer.
Table 1. Properties of purified P4 proteins from φ12 and φ6.
| P4 | φ12 | φ6 |
|---|---|---|
| Subunit mass (kDa)a | 35.1 | 35.1 |
| Multimeric statusb | Hexamer | Hexamere |
| Hydrodynamic radius (nm) and shapec | Rh = 5.5; ring | Rh = 5.9; ringe |
| Nucleic acid binding | Undetectable | Undetectablef |
| COD activity | Undetectable | Undetectablef |
| kcat [s−1], no RNAd | 0.17 ± 0.06 | 0.19 ± 0.06f |
| kcat [s−1], poly(C)d | 0.50 ± 0.06 | 1.12 ± 0.06f |
| kcatpoly(C)/kcatnoRNA | 2.96 | 5.89f |
φ12 P4 NTPase activity is stimulated by ssRNA but does not bind ssRNA or dsRNA
NTPase activity of purified P4 was compared with that of φ6 P4 using conditions optimal for the latter protein (18). Isolated φ12 P4 possesses a readily detectable NTPase activity hydrolyzing ATP into ADP and inorganic phosphate (Figure 2F). Basal activities of both enzymes are similar (Table 1) and are weakly stimulated by poly(C) ssRNA (kcatpoly(C)/kcatnoRNA; Table 1). We also studied the possible effect of dsRNA, ssDNA and dsDNA on the NTPase activity of φ12 P4 (not shown). No stimulation was detected.
Figure 2.
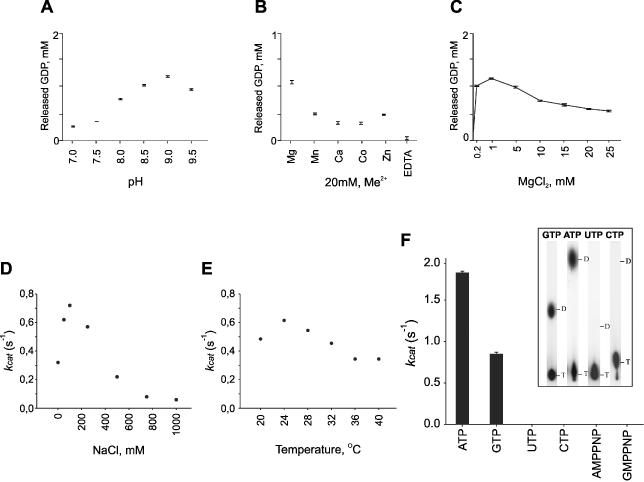
Effects of the reaction conditions on the activity of the φ12 P4 NTPase. (A–C) Quantitative analysis by TLC showing the effects of pH [(A) 20 mM Tris–HCL, 50 mM NaCl, 5 mM MgCl2], divalent metals—Me2+ [(B) 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 0.2 mM MgCl2, 20 mM Me2+] and Mg2+ concentration [(C) 20 mM Tris–HCl, pH 8.0, 50 mM NaCl], on the GTPase activity of φ12 P4 (GTP concentration was 2 mM, protein concentration 0.25 mg/ml) at 37°C. (D–F) P4 NTP turnover (kcat) was measured using steady state kinetics of Pi release (NTP concentration 1 mM) under different conditions: NaCl effect [(D) 20 mM Tris–HCl, pH 9.0, 1 mM MgCl2, 24°C)], temperature dependence [(E) 20 mM Tris–HCl, pH 9.0, 1 mM MgCl2, 100 mM NaCl)], hydrolysis of different nucleotides [(F) NTPs, AMPPNP and GMPPNP concentration 1 mM, 20 mM Tris–HCl, pH 9.0, 1 mM MgCl2, 100 mM NaCl, 24°C], and TLC run showing the hydrolysis of different nucleotides under optimal conditions (insert). The analyses were repeated three times and error bars show standard deviations.
Incubation of the enzyme with ssRNA, dsRNA, ssDNA and dsDNA probes in the absence of ATP did not result in the formation of stable complexes (not shown). Although φ12 P4 exhibits RNA stimulated NTP hydrolysis, it does not possess high affinity for nucleic acids, a feature similar to φ6 P4 but distinct from φ8 and φ13 P4s, which bind nucleic acids tightly (18).
P4 lacks helicase activity
The complementary oligonucleotide displacement assay has been used previously to study the RNA translocation activity of φ6, φ8 and φ13 P4s. φ12 P4 was subjected to the COD assay and no displacement was detected after 2 h of incubation (data not shown). Similar results were obtained previously for φ6 P4, which needs to be associated with the PC to efficiently translocate ssRNA (18). Overall, the results indicate that φ12 P4 is a hexameric ssRNA-stimulated NTPase with enzymatic properties similar to those of φ6 P4 (Table 1).
Purified φ12 P4 is a purine-specific NTPase
P4 φ12 NTPase activity was examined over a wide range of conditions (Figure 2A–E). The optimum activity was achieved at relatively high pH (∼9.0, Figure 2A) and moderate salt concentration (100 mM NaCl; Figure 2D). Low concentration (<2 mM MgCl2) of magnesium proved to be essential for the activity (viz. EDTA effect) while other divalent cations exhibited inhibitory effect (Figure 2B). However, higher Mg2+ concentrations were inhibitory (Figure 2C), suggesting either negative cooperativity in Mg2+ binding or a role for Mg2+ in the product release (e.g. release of Mg:ADP complex). The optimum temperature for NTPase activity was between 24 and 28°C (Figure 2E), close to the optimal cultivation temperature for φ12 (25°C).
It has been shown that the nucleotide base specificity of the P4 packaging motors from bacteriophages φ6, φ8 and φ13 is low, and both purine and pyrimidine NTPs are efficiently hydrolyzed (18,19,30). In order to establish the nucleotide specificity of φ12 P4, we determined the kcat values for the four basic NTPs under the optimal conditions (Figure 2F). Purified φ12 P4 readily hydrolyzed purine NTPs into NDP and inorganic phosphate, ATP being preferred over GTP. Notably, no hydrolysis of pyrimidine NTPs (UTP and CTP) was detected, in contrast to P4s from related phages φ6, φ8 and φ13. The specificity holds in the presence of poly(C) ssRNA substrate under optimal conditions (kcatpoly(C)/kcatnoRNA for ATP, 1.7, and for GTP, 2.3).
Transcription of viral cores does not depend on P4 NTPase activity
Recent studies have shown that φ6 P4 NTPase is not only involved in genome packaging but is also required for export of transcripts from viral cores (21). To test whether φ12 P4 acts as a passive conduit (Figure 3A, right) or actively translocates ssRNA from the core (Figure 3A, left), we compared transcriptional activity of isolated φ12 viral cores under two conditions: first, the reaction was supplemented with P4-hydrolyzable NTPs, and second, the reaction contained NTPs that are non-hydrolyzable by P4 (i.e. AMPPNP, GMPPNP, CTP and UTP). Note that the viral polymerase protein P2 could utilize the latter set of nucleotides for ‘plus’ strand synthesis (16). The time course of transcription (Figure 3C) shows that in both cases the full-length s+, m+ and l+ segments accumulated to approximately the same degree albeit with a slower rate in the case of nucleotide analogs. The slower rate could be attributed to the effect of analogs on the polymerase (16). Thus, the NTPase activity of P4 is not essential for transcription.
Figure 3.
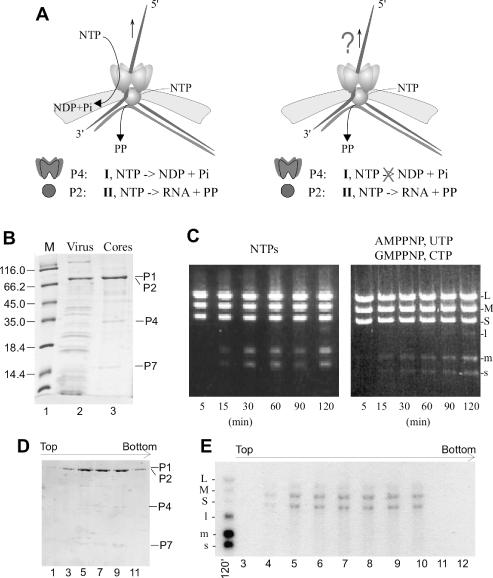
Assessment of transcriptional activity of φ12 cores in vitro under favorable conditions for P4 NTPase activity and in the presence of non-hydrolyzable analogs. (A) Two possible models of semi-conservative transcription in Cystoviruses (left—P4 actively translocates ssRNA from the core; right—P4 acts as a passive conduit). Packaging NTPase and polymerase in the context of the capsid. Polymerase is colored red and packaging NTPase is green. (B) SDS–PAGE analyses of purified φ12 virus and core components. (C) Time course of transcription reaction. Reaction products in the presence of P4-hydrolyzable and non-hydrolyzable nucleotides were analyzed on agarose gel and auotoradiographed. (D and E) Sedimentation analysis of the cores after in vitro transcription in the presence of P4-non-hydrolyzable nucleotides: protein composition analyzed on SDS–PAGE (D) and autoradiograph of RNA products separated on agarose gel (E). Lane 120′ corresponds to labeled products of core transcription which where loaded onto gradient.
Since P2 polymerase can carry out semi-conservative transcription in the absence of other core proteins (16), the following control was performed to show that the transcription occurred within the cores. The transcription reaction was repeated with P4 non-hydrolysable nucleotides in the presence of 0.1 mCi of [α-32P]. After 2 h the sample was sedimented through a sucrose gradient and the protein content of the collected fractions was assayed (Figure 3D). The peak appeared to be relatively sharp, indicating that cores remained intact. Semi-conservative transcription also implies that the newly synthesized RNA displaces the non-template strand from the dsRNA substrate and should remain associated with the core. The RNA analysis of the gradient fractions shows that the labeled dsRNA segments L, M and S co-sedimented with the core-containing fractions (Figure 3E) indicating that transcription took place in association with the particles.
φ12 P4 does not exhibit NTPase activity within the polymerase core
φ6 nucleocapsids and NC core particles (particles lacking surface protein P8) have been shown to hydrolyze all four NTPs to NDPs (28,30). The following experiment was carried out to test the NTPase activity of isolated φ12 cores. For this purpose, ATP, GTP, CTP or UTP hydrolysis by core were assayed in the P4-optimal buffer (20 mM Tris–HCl, pH 9.0, 100 mM NaCl, 1 mM MgCl2, at 28°C) or under the conditions used for transcription (see Materials and Methods). No phosphate release was detected (data not shown). Because the NTPase assay was designed to detect less than one active P4 hexamer per core particle, we can conclude that all P4 hexamers are inactivated within the core.
DISCUSSION
The strategy of transcription differs between different families of dsRNA viruses: cystoviruses (three genome segments) and birnaviruses (two genome segments) act semi-conservatively, whereas the Reoviridae (with 10 to 12 genome segments and a larger polymerase subunit) carry out conservative transcription. The conservative strategy requires that the dsRNA emerging from the polymerase must be separated and the newly made ‘plus’ strand must be extruded from the core and the parent ‘minus’ strand reannealed with the parental one. Consequently, RNA helicase activity is required to assist conservative RNA synthesis in Reoviridae.
In semi-conservative transcription (cystoviruses) the polymerase P2 can act as a helicase during the first of these steps. The rate of transcription elongation by isolated P2 [30 nt/s; (16)] is similar to that reported for the core system [19–24 nt/s; (31)], suggesting that P2 does not require the assistance of other proteins to unwind RNA duplex during elongation. Intriguingly, it was shown that hexameric protein P4 from φ6 is needed for the exit of the newly synthesized RNA (21). However, it was not clear whether the RNA exit is coupled to NTP hydrolysis by P4, e.g. whether this step constitutes an active translocation in the direction opposite to that of packaging. The purine specificity of φ12 P4 enabled us to test this hypothesis. The results clearly show that P4 NTPase activity is turned off in the φ12 core and consequently is not needed for RNA export during transcription (Figure 4). In other words, P4 hexamer might act as a passive conduit for RNA export. The RNA polymerase P2 that also acts as a helicase powers the export. In effect, the directionality of the RNA transport via P4 is determined by the NTPase activity, which in turn is regulated by the core. Most likely, P4 adopts an inactive conformation as a result of polymerase complex expansion, which is associated with packaging and replication and activates the polymerase subunit (14) (Figure 4B).
Figure 4.
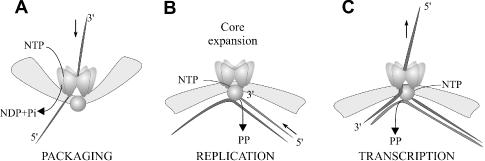
A model for switching between RNA packaging and semi-conservative transcription.
The absence of P4 NTPase activity within φ12 cores is in contrast with the bacteriophage φ6, for which activities comparable to the isolated protein were reported (28,30). Two classes of P4 hexamers were identified for the φ6 polymerase complex, a tightly bound and a weakly bound class, respectively (21). The former class was shown to be essential for packaging while the latter is important for transcription but does not support packaging. In the light of the present results it remains to be seen whether such distinction between packaging and transcription vertices applies also to the bacteriophage φ12. Conversely, NTP-independent transcription may be limited to the φ12 bacteriophage.
Similar regulatory effects by other proteins in the multiprotein replicative complexes have been reported for the related hexameric helicases (1). For example, the helicase (translocation) activity regulation plays an important role in replication and transcription termination. Perhaps, packaging motors and hexameric helicases are not only structurally related but regulation of their activities may also exhibit similar features.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Paul Gottlieb for providing the strain LM2333 and the bacteriophage φ12 stock. D.E.K. is a fellow of the National Graduate School in Informational and Structural Biology. J.L. is a fellow of Viikki Graduate School in Biosciences. This work was supported by the Academy of Finland [‘Finnish Center of Excellence Program 2000–2005’, grants 206926 (RT), 1202855 and 1202108 (DHB)].
REFERENCES
- 1.Delagoutte E. and von Hippel,P.H. (2003) Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Quart. Rev. Biophys., 36, 1–69. [DOI] [PubMed] [Google Scholar]
- 2.Caruthers J.M. and McKay,D.B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol., 12, 123–133. [DOI] [PubMed] [Google Scholar]
- 3.Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 4.Skordalakes E. and Berger,J.M. (2003) Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell, 114, 135–146. [DOI] [PubMed] [Google Scholar]
- 5.Catalano C.E., Cue,D. and Feiss,M. (1995) Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol., 16, 1075–1086. [DOI] [PubMed] [Google Scholar]
- 6.Smith D.E., Tans,S.J., Smith,S.B., Grimes,S., Anderson,D.L. and Bustamante,C. (2001) The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature, 413, 748–752. [DOI] [PubMed] [Google Scholar]
- 7.Sheaffer A.K., Newcomb,W.W., Gao,M., Yu,D., Weller,S.K., Brown,J.C. and Tenney,D.J. (2001) Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol., 75, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson A.A., Tao,Y., Leiman,P.G., Badasso,M.O., He,Y., Jardine,P.J., Olson,N.H., Morais,M.C., Grimes,S., Anderson,D.L. et al. (2000) Structure of the bacteriophage φ29 DNA packaging motor. Nature, 408, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catalano C.E. (2000) The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci., 57, 128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimes S., Jardine,P.J. and Anderson,D. (2002) Bacteriophage φ29 DNA packaging. Adv. Virus Res., 58, 255–294. [DOI] [PubMed] [Google Scholar]
- 11.Davies G.P., Kemp,P., Molineux,I.J. and Murray,N.E. (1999) The DNA translocation and ATPase activities of restriction-deficient mutants of Eco KI. J. Mol. Biol., 292, 787–796. [DOI] [PubMed] [Google Scholar]
- 12.Mindich L. (1999) Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage φ6. Microbiol. Mol. Biol. Rev., 63, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb P., Strassman,J., Qiao,X.Y., Frucht,A. and Mindich,L. (1990) In vitro replication, packaging, and transcription of the segmented double-stranded RNA genome of bacteriophage φ6: studies with procapsids assembled from plasmid-encoded proteins. J. Bacteriol., 172, 5774–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher S.J., Dokland,T., Ojala,P.M., Bamford,D.H. and Fuller,S.D. (1997) Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J., 16, 4477–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poranen M.M. and Tuma,R. (2004) Self-assembly of double-stranded RNA bacteriophages. Virus Res., 101, 93–100. [DOI] [PubMed] [Google Scholar]
- 16.Makeyev E.V. and Bamford,D.H. (2000) Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J., 19, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juuti J.T. and Bamford,D.H. (1995) RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage φ6. J. Mol. Biol., 249, 545–554. [DOI] [PubMed] [Google Scholar]
- 18.Kainov D.E., Pirtimaa,M., Tuma,R., Butcher,S.J., Thomas,G.J.,Jr, Bamford,D.H. and Makeyev,E.V. (2003) RNA packaging device of dsRNA bacteriophages: possibly as simple as hexamer of P4 protein. J. Biol. Chem., 278, 48084–48091. [DOI] [PubMed] [Google Scholar]
- 19.Juuti J.T., Bamford,D.H., Tuma,R. and Thomas,G.J.,Jr (1998) Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ6. J. Mol. Biol., 279, 347–359. [DOI] [PubMed] [Google Scholar]
- 20.Lisal J., Kainov,D.E., Bamford,D.H., Thomas,G.J.,Jr and Tuma,R. (2004) Enzymatic mechanism of RNA translocation in dsRNA bacteriophages. J. Biol. Chem., 279, 1343–1350. [DOI] [PubMed] [Google Scholar]
- 21.Pirttimaa M.J., Paatero,A.O., Frilander,M.J. and Bamford,D.H. (2002) Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage φ6 is required for single-stranded RNA packaging and transcription. J. Virol., 76, 10122–10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouet P., Diprose,J.M., Grimes,J.M., Malby,R., Burroughs,J.N., Zientara,S., Stuart,D.I. and Mertens,P.P. (1999) The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell, 97, 481–490. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb P., Potgieter,C., Wei,H. and Toporovsky,I. (2002) Characterization of φ12, a bacteriophage related to φ6: nucleotide sequence of the large double-stranded RNA. Virology, 295, 266–271. [DOI] [PubMed] [Google Scholar]
- 24.Ojala P.M., Juuti,J.T. and Bamford,D.H. (1993) Protein P4 of double-stranded RNA bacteriophage φ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J. Virol., 67, 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emori Y., Iba,H. and Okada,Y. (1983) Transcriptional regulation of three double-stranded RNA segments of bacteriophage φ6 in vitro. J. Virol., 46, 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancini E.J., Kainov,D.E., Wei,H., Gottlieb,P., Tuma,R., Bamford,D.H., Stuart,D.I. and Grimes,J.M. (2004) Production, crystallization and preliminary X-ray crystallographic studies of the bacteriophage φ12 packaging motor. Acta Cryst. D, 60, 588–590. [DOI] [PubMed] [Google Scholar]
- 27.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 28.Paatero A.O., Syvaoja,J.E. and Bamford,D.H. (1995) Double-stranded RNA bacteriophage φ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J. Virol., 69, 6729–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldentey J., Tuma,R. and Bamford,D.H. (2000) Assembly of bacteriophage PRD1 spike complex: role of the multidomain protein P5. Biochemistry, 39, 10566–10573. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins R.H., Tuma,R., Juuti,J.T., Bamford,D.H. and Thomas,G.Jr, (1999) A novel Raman spectrophotometric method for quantitative measurement of nucleoside triphosphate hydrolysis. Biospectroscopy, 5, 3–8. [DOI] [PubMed] [Google Scholar]
- 31.Usala S.J., Brownstein,B.H. and Haselkorn,R. (1980) Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell, 19, 855–862. [DOI] [PubMed] [Google Scholar]
- 32.Mindich L. and Bamford,D.H. (1988) Lipid-containing bacteriophages. In Calendar,R. (ed.), The Bacteriophages. Plenum Publishing Corporation, NY, Vol. 2, pp. 475–519. [Google Scholar]


