Abstract
All multicellular hosts form associations with groups of microorganisms. These microbial communities can be taxonomically diverse and dynamic, and their persistence is due to robust, and sometimes co-evolved, host microbe and microbe microbe interactions. Chemical and physical sources of stress are prominently situated in this molecular exchange, as cues for cellular responses in symbiotic microbes. Stress in the symbiotic environment may arise from three sources: host tissues, microbe-induced immune responses, or other microbes in the host environment. The responses of microbes to these stresses can be general or highly specialized, and collectively may contribute to the stability of the symbiotic system. In this review, we highlight recent work that emphasizes the role of stress as a cue in the symbiotic environment of plants and animals.
Keywords: stress response, symbiosis, host microbe, microbiota
Stress Structures Beneficial Host-Microbe and Microbe-Microbe Interactions
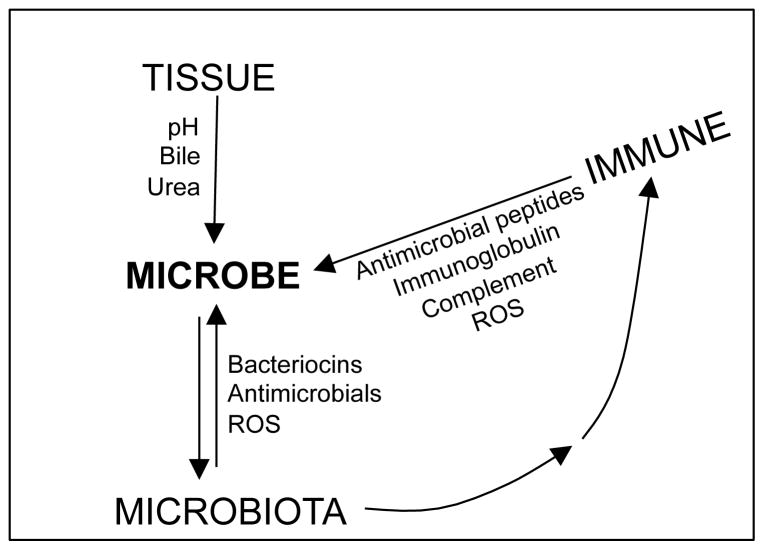
All plants and animals form beneficial associations with microbes. These host associated communities, or microbiota (see Glossary), induce processes such as tissue development [1, 2], that positively influence the physiology of the whole organism [3, 4]. To sustain mutualism, an ecological structure must be maintained such that host and microbiota derive benefit [5, 6]. Host signaling molecules [7], nutrients [8], and sources of chemical or physical stress [9], contribute to this structure (Figure 1, Key Figure). The survival of a microbe in the symbiotic milieu is a function of its resistance to host-associated and microbiota-associated stress (Figure 1). The host-tissue environment imposes chemical and physical stresses that constrain community composition [10]. In this context, the host immune system adds further chemical stresses in response to microbe-associated molecular patterns (MAMPs) and metabolic activity (Figure 1). Thus, to understand the function of symbiotic microbial communities, it will be necessary to first recognize the how microbes shape, and are shaped by, the host tissue environment.
Figure 1. Three Sources of Stress Define the Host Environment.
Host tissues may be a source of chemical and/or physical stress. The presence of tissue-associated stress is not dependent on microbes. Microbes that can grow within the constraints of the host tissues may, themselves, produce stress through competitive or antagonistic interactions. Microbial activity in host tissues, or interactions with specific microbial taxa within the community, may induce immune-associated stress. Abbreviation: ROS; reactive oxygen species.
Defining the functions of the beneficial microbiota of plants and animals is a frontier field in microbiology [11, 12] and the ecological principles that promote and maintain these communities are still poorly understood. Thus, it is of great interest to identify the mechanisms by which the beneficial microbiota interact both with the host, and with other members of the community. In this review, we focus on recent work that illustrates the role of stress as a cue (Box 1) for bacteria in a diversity of model plant and animal symbioses (Figure 2). Although we do not touch on the response of yeast and archaea to host-associated stress, a comprehensive understanding of the role of stress as a cue for microbial symbionts will require investigation of a diversity of microorganisms. We will consider three sources of stress in the context of symbiosis: (i) host tissues, (ii) microbe-stimulated immune responses, and (iii) antagonism or chemical manipulation by other microbes. The microbial response to these three sources of stress may be integrated, and we discuss the importance of understanding stress in the symbiotic environment as a cue that may originate from multiple sources.
Box 1. Is Stress a Signal or Cue?
In this review, we emphasize the function of stress in the symbiotic environment. To understand the role of stress in this environment, it is first necessary to appreciate that stress is generally a cue, but not a signal. Whereas signals evolve to elicit a response in a target organism, cues are not subject to such selective pressure [96]. It is also worth noting that to perceive a signal, there must be selective pressure for the target organism to evolve a receptor. In contrast, a receptive organism may exploit a chemical or physical cue in its environment to surveil neighboring microbes, or the host environment. In this framework, it is clear that the cue need not have evolved in the producing organism for the purpose of eliciting a response in the responding organism. Cues can be sufficient to coordinate a microbe’s response to its environment. Host-derived reactive oxygen species, cationic antimicrobial peptides, and microbially produced antibiotics, siderophores, or bacteriocins may act as cues to coordinate the response of the responding organism to the complex stresses of its environment.
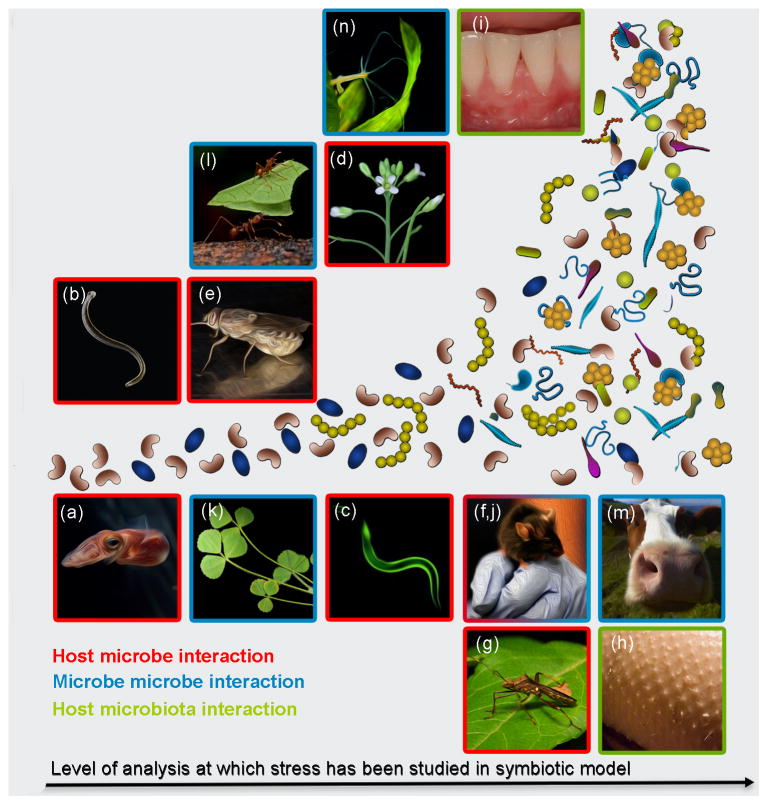
Figure 2. Host-Microbe Interactions May be Studied at the Level of an Individual Microbial Taxon, or at the Community Level.
Examples of model systems for investigating how a single species of microbe colonizes a host: (a) squid-Vibrio fischeri, (b) leach-Aeromonas veronii, (c) nematode-Xenorhabdus spp., (d) legume-Sinorhizobium meliloti, (e) tsetse fly-Sodalis glossinalus, (f) gnotobiotic mouse stomach-Helicobacter pylori, and mouse gut-diverse microbes, (g) stink bug-Burkholderia spp. Tissue sites that have been used to study microbe microbe interactions: (h) skin and (i) teeth. Examples of host that have been used to study microbial communities: (j) the mouse gut, (k) Arabidopsis thaliana, (l) leaf-cutter ants, (m) the bovine rumen, and (n) Hydra vulgaris.
Host Tissues are a Source of Physical and Chemical Stress for Symbiotic Microbes
The chemical and physical heterogeneity of host tissues defines symbiotic habitats within the host. Secreted products such as urea, bile [13], mucus, and gastric acid [14] in animals, or wax and lignocellulose in plants [15] create chemical and physical barriers that restrict colonization. Abiotic stresses, such as desiccation on skin and leaves [16, 17], ultraviolet radiation, or variations in body temperature among host-tissue sites may also restrict colonization to the subset of microbes that can grow within these parameters. In this section, we highlight recent studies that reveal both variable and conserved attributes of microbial responses to tissue-derived stress in different plants and animals.
Induction of microbial responses to tissue-associated stress may be either specific or general, and the mechanisms of resistance in different types of tissues may be equally varied. For instance, specific responses may help to promote the colonization of the gastric mucosa, or of root nodules, where the kind of stress is either relatively stable, or predictable. In the mammalian stomach, the presence of gastric acid creates a chemical barrier that restricts colonization by most microbes. To overcome this specific tissue chemistry, Helicobacter pylori both buffers its proximal environment by expressing urease, and migrates by chemotaxis towards the less acidic crypts of the gastric mucosa [18]. In contrast, responses that can be induced in a variety of environments, and that confer protection against multiple stresses, might be of benefit to symbiotic microbes that colonize tissues that experience variable sources and types of stress, such as the integument, the intestinal lumen, or root or leaf surfaces (Figure 2). For instance, symbiotic species of Burkholderia expresses polyhydroxyalkanoate (PHA) to persist in the gut of the stinkbug Riptortus pedestris (Figure 2) [19]. PHA is a storage polymer that may help R. pedestris persist in an environment where starvation, or diverse stresses that limit nutrient uptake, may arise unpredictably. Thus, the formation of PHA granules may be considered a general protective response. Further characterization of the mechanisms by which symbiotic microbes respond to tissue-derived stress may reveal core strategies by which these responses are cued to promote survival in both stochastic and predictable environments.
The Immune Response of Plants and Animals is a Source of Stress for Microbial Symbionts
The immune system of plants and animals has evolved to sense and respond to environmental perturbations such as wounding or colonization by microbes. Immune signaling coordinates the response to these perturbations. Receptors for damage-associated molecular patterns (DAMPs) and for MAMPs are broadly conserved within the plant and animal kingdoms. A detailed consideration of microbe-immune signaling is outside the scope of this review, and recent reviews have comprehensively addressed this topic in invertebrate [20], and vertebrate [21] animals, and in plants [22]. Instead, we highlight here several recent studies that illustrate how stresses originating from MAMP-induced immune responses act as cues for the symbiotic microbes of plants and animals.
To colonize the rhizosphere of plants, where microbes associate with a host at the root surface or within the root tissue as a nodule, symbiotic microbes induce responses that protect against immune-associated stresses. One well studied example is the legume Medicago truncatula (clover), which recruits its nodule-forming symbiont, S. meliloti, in a multi-step signaling process that culminates with the terminal differentiation of S. meliloti within the symbiosis-induced root nodules (Figure 2) [23]. Host-associated stresses such as reactive-oxygen species and antimicrobial peptides (AMPs; Box 2) promote the association of beneficial microbes in both the rhizosphere and nodule environment. Whereas S. meliloti responds to reactive-oxygen species by inducing a general stress response, more specialized responses may be induced in response to AMPs.
Box 2. Antimicrobial Peptides: Conserved Stresses in the Symbiotic Environment.
Peptides produced by the immune function of plants and animals represent an emerging class of specific, and selective, innate-immune effectors that function across an evolutionarily vast spectrum of plant- and animal-microbe mutualisms. Immune peptides contribute to the specificity of animal and plant hosts towards microbial symbionts. In animals, immune peptides are generally antimicrobial. Antimicrobial peptides (AMPs), such as the lectin RegIII-gamma, kill microbes by targeting the bacterial membrane [97, 98]]. Modifications made to membrane biomolecules such as lipopolysaccharide (LPS) and peptidoglycan (PGN) change the charge-distribution on the membrane’s surface, and lead to electrostatic repulsion of cationic AMPs: a strategy used by pathogenic Salmonella to evade innate-immune killing [99]. Both pathogenic and non-pathogenic species of Enterococcus [100], Vibrio [101, 102] and Staphylococcus [103], incorporate host-derived lipids into the bacterial cell membrane. The gut microbe Bacteriodes thetaiotamicron [9] modifies its LPS to resist AMPs, while to persist in the gut of the stinkbug, R. pedestris, the β-proteobacterium Burkholderia sp. modifies its LPS so that the structure is more sensitive to AMPs, but no longer contains the immune-reactive O-antigen [104]. Future characterization of other classes of lectins, and secreted peptides are likely to reveal additional mechanisms by which these immune proteins contribute to stress in the symbiotic environment. In legumes, nodule-specific cysteine-rich peptides (NCR) perform functions that affect nodulation by nitrogen-fixing Sinorhizobium meliloti. Multiple NCR peptides are expressed in the root nodule, yet they perform non-redundant functions: the deletion of the NCR169 abolishes root nodule formation in M. truncatula [105], while the NCR211 peptide is required to promote the survival of rhizobial bacterioids in a terminally differentiated, nitrogen-fixing state [27]. Elucidating the mechanisms by which the immune peptides of plants and animals act as specific agents of stress, or perhaps cues in the host environment, is an ongoing area of active research.
Upon first contact of S. meliloti with the roots of M. truncatula, LPS produced by these bacteria signals plant immune receptors, inducing an oxidative burst. In response to this stressful cue, S. meliloti produces two extracellular polymeric substances (EPS), succinoglycan and galactoglucan [24] (Table 1). The resulting EPS coating on the bacteria is sufficient to confer resistance to hydrogen peroxide in cultured S. meliloti [24], and is required for nodule formation [25], suggesting that EPS may promote symbiont survival in response to prolonged exposure to the plant oxidative burst. In other microbes, EPS confers resistance to diverse stresses in addition to reactive oxygen species, such as antibiotics [26]. Once inside the M. truncatula root nodule, the differentiation and senescence of S. meliloti are cued by nodule-specific cysteine-rich (NCR) peptides [27] (Box 2). Members of this class of peptide are broadly antimicrobial [28], and can both inhibit S. meliloti cell division, and induce transcriptional responses consistent with membrane and cytoplasmic stress [29]. The different responses elicited by NCR peptides suggest that resistance mechanisms may be tailored towards specific classes of peptide. Further characterization of the targets of NCR peptides are needed to reveal the mechanisms by which these peptides act as antimicrobials, and to determine how combinations of these peptides contribute to establishing specific associations with host tissues.
Table 1.
Conserved Stress Responses Among Microbes in Diverse Host Environments
| Symbiont response | Host Environment | Symbiont | Ref. |
|---|---|---|---|
|
| |||
| EPSa production | Squid light organ | Vibrio fischeri | [35, 36] |
| Tsetse fly gut | Sodalis glossinidius | [38] | |
| Clover root | Sinorhizobium meliloti | [24] | |
| Oxidative stress resistance | Leaf surface | Various microbiota | [34] |
| Roots | Sinorhizobium meliloti | [104, 105] | |
| Mouse gut | SFBb | [45, 46] | |
| Antimicrobial peptide resistance | Hydra mucosa | Various microbiota | [106] |
| Squid mucus | Vibrio fischeri | [107] | |
| Mouse gut | Salmonella enterica serovar Typhimurium | [99] | |
| Bacteroides thetaiotamicron | [9] | ||
| Incorporation of host lipids | Squid light organ | Vibrio fischeri | [101] |
| Mouse gut | Enterococcus sp. | [100] | |
| Staphylococcus sp. | [103] | ||
EPS, extracellular polymeric substance.
SFB, segmented filamentous bacteria. The response is predicted from transcriptional profiling.
The immune response in the phyllosphere is linked to conditions in the rhizosphere; that is, MAMP signaling that takes place at the roots of plants may systemically propagate signaling molecules to aboveground tissues [30]. Apart from this induced systemic resistance, plant tissues in the phyllosphere may also directly respond to MAMP signaling by leaf surface-associated microbes. Two immune functions that contribute to structuring phyllosphere microbiota, oxidative burst and cuticle formation, have been recently studied. The oxidative burst of plants is induced by ethylene, a plant immunity hormone produced in response to MAMPs and/or DAMPs [31]. Deficiencies in ethylene signaling alter the community structure of the Arabidopsis thaliana phyllosphere microbiota [32]. Cuticle thickening is also an immune response to MAMP signaling [31], and, like the oxidative burst, the A. thaliana wax cuticle shapes microbial community composition [33]. In rice, colonization of the phyllosphere is associated with the enrichment of microbial proteins related to oxidative-stress resistance [34], suggesting that the oxidative burst may be a common immune-associated stress that structures the phyllosphere microbiota of diverse plants (Table 1). The strategies of stress resistance among phyllosphere-colonizing microbes are still not well characterized, and it remains to be seen whether the induction of general responses that are protective against multiple sources of stress dominates among microbes that colonize this habitat.
Like plant-associated microbes, the microbiota of animals is structured by stress derived from immune function. Much like the initial colonization of M. truncatula roots by S. meliloti (Table 1), the bioluminescent microbial symbiont of the bobtail squid, Vibrio fischeri, produces EPS to survive the chemical barriers created by the immune response of the squid’s light organ [35, 36]. The regulation of EPS production is specific to certain strains of V. fischeri, and a single regulatory locus is required for squid symbionts to colonize their host [37]. Similarly, the Gram-negative bacteria Sodalis glossinidius produces EPS to colonize the tsetse fly gut [38], although it is not known whether EPS production is a determinant of host specificity in this system. As in the rhizosphere of M. truncatula root nodules, the mouse immune response secretes AMP into the gut mucosa, where only microbes capable of inducing resistance, such as the gut symbiont Bacteriodes thetaiotamicron may survive [9]. The immune proteins of the complement system structure bacterial colonization in the medicinal leech, Hirudo verbana [39] (Figure 2). H. verbana ingests mammalian blood, and complement-susceptible strains of the gut symbiont Aeromonas veronii are unable to colonize the leech [39]. Thus, immune-associated stress contributes to establishing and maintaining specific and selective associations between hosts and their beneficial microbes.
The first contact between an animal and its nascent symbionts can induce immune-associated stresses that cue colonization. In the stinkbug R. pedestris, symbiotic Burkholderia cells repress the transcription of humoral immune enzymes such as lysozyme and defensin, while inducing expression of genes encoding secreted cysteine-rich antimicrobial peptides [40]. A similar transcriptional pattern has been noted in the bobtail squid Euprymna scolopes, which tailors its expression of antimicrobial factors upon contact with V. fischeri [41, 42] to promote colonization (Figure 2). The loss of symbiotic algae and other microbes from coral tissue (i.e., ‘bleaching’) impairs transcription associated with immune function, even after the symbiotic communities have been restored [43], suggesting that signaling between corals and their symbionts shapes the immune-associated stresses of host tissues. Thus, colonization, and likely other physiological processes of symbiotic microbes, are cued by host immune function, and signaling between host and microbiota determines the specificity of immune-associated stress.
The adaptive immune response of animals may produce targeted stresses. In the vertebrate gut, a subset of the microbiota are targets of the adaptive immune protein immunoglobulin A (IgA) [44]. IgA produced in the murine intestine is raised against specific members of the microbiota, including segmented filamentous bacteria (SFB). The genome of SFB encodes peroxidase, catalase and arginase: genes that mitigate oxidative stress [45, 46], suggesting that recognition of SFB by IgA may be accompanied by an oxidative burst. Immunoglobulins specific to the normal gut microbiota have also been found in murine skin lymph nodes [47], indicating that a population of host antibodies may be raised towards members of the beneficial microbiota of other body sites. The function of this population of antibodies remains largely uncharacterized, although expression of Proteobacteria-specific IgA during the development of the mouse gut restricts the abundance of this phylum among the microbiota in the adult gut [48]; in addition, Alcaligenes spp. exploit the production of gut-associated IgA to colonize restricted habitats, such as Peyer’s patches [49]. Future investigation of the population of immunoglobulins raised against non-pathogenic gut microbes is likely to provide insight into the contribution of adaptive-immune stresses to microbial community structure, and the ecological succession of symbiotic microbial communities.
The host may also modify MAMPs produced by the symbiont to modulate its immune responses, and thereby promote colonization. This effect is achieved through enzymes that modify the chemical structures of MAMPs, such as alkaline phosphatase and peptidoglycan-recognition protein. Specifically, the expression of E. scolopes alkaline phosphatase (AP) is required to maintain a stable colonization by V. fischeri. The MAMP lipopolysaccharide (LPS) is a substrate of AP, suggesting that dephosphorylation of symbiont LPS may help maintain a stable colonization [50]. Similarly, tsetse fly peptidoglycan-recognition protein protects the obligate endosymbiont Wiggelsworthia from damaging antimicrobial peptides during the transmission of the microbe to progeny in adult fly milk, presumably by altering the immune-activating structures of the MAMP peptidoglycan (PGN) [51]. Thus, context-dependent modifications made to conserved MAMP signals may contribute to the establishment of specific symbiotic associations. To the best of our knowledge, it remains to be seen whether similar modifications occur in plants.
Microbe-Derived Stress Shapes Symbiotic Communities
Nutrient secretions from the epidermis of plants or the epithelium of animals are sufficient to scaffold a beneficial microbial community [52, 53]. Within the scaffolding provided by host-derived nutrients, competition among members of the microbiota may make the community more resilient to external perturbation by pathogens, or chemical agents [54]. Bacteriocins [55], antimicrobials [56], reactive oxygen species [57], and bacteriophage [58, 59] are stresses derived from microbes in the host environment that can structure the competition for nutrients among communties. The production of the bacteriocin coproporphyrin III by a human skin-specific Propionibacterium sp. induces EPS production by Staphylococcus aureus, leading to co-aggregation of the two species [60] (Figure 2). Similarly, antimicrobial production by the symbionts of leaf-cutter ants constrains the composition of the ant’s fungal garden [56] (Figure 2). Hydrogen peroxide produced by Streptococcus oligofermentans counteracts lactic acid production by Streptococcus mutans in the oral microbiota [57], suggesting that the two species may interact within the diffusive limits of their environment. Bacteriophage-mucin binding interactions have been shown to structure colonization of mucosal layers in the mammalian gut [58, 59], and the accumulation of bacteriophage in sputum or EPS may enhance the protective function of this barrier towards antimicrobials or other chemical stresses [61]. In addition, a functional CRISPR foreign DNA defense system is required for Xenorhabdus nematophila to colonize the gut of its nematode host [62] (Figure 2), suggesting that bacteriophage may be a stressful attribute of the nematode gut microbiota. It is likely that these and other microbe-associated stresses, combined with tissue and immune-derived stress, constrain the growth and partitioning of nutrients among microbes in the host environment.
Recent work has documented how stable gut communities typically express robustness and resiliency to invasion by pathogens. Specifically, mutualists in the mouse gut prevent colonization by the pathogen Citrobacter rodentum by competing for nutrients [63], while the microbial community that inhabits the epithelial surface of the freshwater cnidarian Hydra vulgaris is required for resistance to fungal pathogens [64] (Figure 2). In this context, stress is a cue that contributes to the barrier function of the beneficial microbiota. Several other studies highlight the function of microbe-associated stress in colonization resistance. The beneficial mouse-gut microbe Clostridium scindens hydrolyzes bile acids, resulting in the inhibition of C. difficile [65]. The production of acetate and EPS by Bifidobacteria spp. in the gut attenuates the virulence of pathogenic Escherichia coli [66] and C. rodentum [67], respectively. Antibiotic treatment-induced perturbation of the mouse gut leads to the accumulation of sialic acid, which is liberated from the mucosa by the beneficial microbiota. This sugar promotes the invasion of intestinal pathogens such as Salmonella enterica serovar Typhimurium and Clostridium difficile [68]. Fermentation by the beneficial microbe B. thetaiotaomicron produces succinate, whose accumulation within the antibiotic-perturbed mouse gut creates an additional source of nutrition for enteropathogens like C. difficile [69], E. coli, and C. rodentum [70]. A similar phenomenon has been observed in the antibiotic-treated guts of broiler hens [71, 72], and is a well-known aspect of rumen microbial ecology [14]. In addition, pumpkins have a native microbiota (including members of both the γ-proteobacteria and Gram-positive bacilli) that is antagonistic towards bacterial pathogens of this plant [73], although the nature of the antagonism is not yet known. Thus, any perturbations of the natural microbial ecology can disturb the flow of substrates in symbiotic communities, thereby either providing nutrition for opportunistic pathogens, or creating a dysbiotic community unable to maintain a stable colonization of the host. Future work that characterizes both the mechanisms underlying these interspecies interactions, and the extent to which microbial community interactions are mediated by the induction of stress responses, will surely provide insight into the role of stress as a cue for the assembly and resilience of symbiotic communities.
Do Microbial Communities Coordinate Their Response to Stress?
Microbial communities may also structure their response to stress through quorum signaling. The quorum-signaling pheromone AI-2 regulates cellular metabolism and stress responses: e.g., oxidative stress and urease genes are responsive to AI-2 in the gut symbiont Lactobacillus reuteri [74], while AI-2 expressed by the mammalian pathogen Streptococcus pneumoniae regulates expression of biofilm-formation [75]. AI-2 is encoded by diverse microbial taxa, and can also mediate interspecies communication; in fact, AI-2 produced by Ruminococcus obeum, a member of the normal intestinal microbiota, inhibits the transcription of colonization factors encoded by pathogenic Vibrio cholerae, thereby mitigating virulence of this intestinal pathogen in the mouse gut [76]. Following antibiotic treatment, AI-2 production by E. coli is sufficient to shift the composition of the two main phyla of microbes present in the human gut, Bacteriodes and Firmicutes, towards a ratio that more closely resembles the composition of a healthy gut [77]. Because AI-2 signaling can mediate communication among species, and targets of AI-2 signaling include elements of the microbial stress response, it will be interesting to discover whether AI-2 signaling coordinates stress responses across species, and perhaps, contributes to the resilience of polymicrobial communities.
Concluding Remarks and Future Perspectives
The contribution of stress to the physiology and ecology of beneficial microbes is an emerging frontier. Chemical and physical stresses are central to the conserved ‘language’ of symbiosis, and should be considered a normal attribute of the symbiotic milieu. Similar to MAMP signaling, the ability to produce stresses that cue beneficial host microbe interactions, such as oxidative stress and antimicrobial peptides, is broadly conserved in the plant and animal kingdoms. It is also evident that stress may target specific members of the symbiotic microbiota (e.g., through IgA), or act at the community level (e.g., oxidative burst), and that the microbial response to stress may be general (e.g., secretion of EPS), or tailored towards a specific type of stress (e.g., modifying the structure of LPS to resist host AMP). It remains to be seen whether responses to community-level stress are coordinated by interspecies or inter-kingdom signaling. The diversity of experimentally tractable animal and plant model systems (Figure 2), as well as the ability to study symbiosis in natural populations, will potentiate future work aimed at deciphering conserved and ancient molecular interactions between hosts and their beneficial microbes. Indeed, it may reveal even more central roles for stress in the context of the symbiotic environment.
What questions should direct future research (see Outstanding Questions)? Symbiotic environments are structurally heterogeneous [78], and microbial communities are characterized by their own ecological structure [79–81]. Thus, to understand how stress cues microbes in the host environment, molecular processes must be understood in the context of both host biology and microbial ecology. To make this link will require knowledge of physical and chemical rules that govern the activity of stress molecules specific to the host-tissue environment, as well as knowledge of the spatial and temporal constraints of the cue. Beneficial microbes in plants and animals form symbioses in the context of host biological rhythms and, in some cases, actually contribute to entraining these rhythms [82–87]. Techniques are being developed to visualize the spatial structure of microbiota [88–92], and to map the chemical diversity of symbiotic communities [93]. Perhaps, future initiatives to study the diverse microbial communities of plants and animals [94, 95] will catalyze efforts to fully recognize stress as a cue in the symbiotic environment.
Outstanding Questions.
What are the conserved and variable mechanisms by which stress cues bacteria, yeast and archaea in the symbiotic environment?
How do host- and microbe-associated stressors structure the assembly and maturation of beneficial microbial communities?
What are the conserved chemical and physical stresses in plants and animals, and how does the response of symbiotic microbes to a stress differ among host environments?
Do species-specific, and inter-species signaling coordinate responses of symbiotic microbes to stress?
Are stressors in the host environment subject to intrinsic biological rhythms?
Trends Box.
Stress cues the colonization of specific host niches by beneficial microbes.
Microbes cue, and respond to, immune-associated stress.
Within the beneficial microbiota, stress promotes stability and resilience.
Plant and animal symbiotic communities sense a core set of conserved stresses.
Acknowledgments
The authors would like to thank P.J. Foster, E. Haswell, and M.J. McFall-Ngai for productive discussions, José Saavedra for assistance preparing figures, and G. Teitzel for critical evaluation and suggestions to improve the article. We also thank the many authors whose work provided inspiration for this review, and apologize to those whose work we may have overlooked. JAS received funding from the Microbes in Health and Disease Training Program (UW Madison NIH-NIAID T32 AI055397). Additional funding was provided to EGR by NIH grants from NIGMS (GM099507), NIAID (AI050611) and ORIP (RR012294).
Glossary
- Abiotic
pertaining to a physical or chemical attribute of an environment that is not derived from living entities within that environment
- Antagonism
an interaction in which the behavior of one organism damages or inhibits another
- Bacteriocin
a peptide toxin produced by one microbe that generally targets other microbes
- Complement system
small proteins of the vertebrate immune system that circulate in the blood. Complement proteins that are activated by immune signaling bind to microbial targets to enhance the function of antibodies or phagocytic cells
- Chemical manipulation
a compound produced by one organism that alters the gene expression or physiology of another target organism, thereby decreasing the fitness of the target
- Chemotaxis
the migration of an organism in response to a gradient of a chemical stimulus. In bacteria, chemical stimuli are sensed by surface receptors, triggering a signaling cascade that regulates the direction of rotation of the flagellar motor
- Clustered
regularly interspaced short palindromic repeats (CRISPR), a CRISPR element is a chromosomal locus encoded in many microbes that confers immunity towards mobile genetic elements of foreign origin. In conjunction with CRISPR-associated enzymes, short sequences of DNA complementary to foreign DNA are incorporated as spacers into the CRISPR locus. When expressed, the short sequences bind to target foreign DNA, targeting that DNA for cleavage by CRISPR-associated enzymes. These short segments act as a form of adaptive immunity
- Cue
a chemical or physical stimulus sensed by an organism, and that conveys information about the organism’s environment, resulting in a coordinate physiological response. In contrast to a signal, cues do not appear or evolve primarily to produce a response in the organism
- Damage-associated molecular pattern (DAMP)
host molecules that, when detected outside of the cell, trigger unfavorable inflammatory responses. For example, extracellular host DNA may trigger inflammation
- Extracellular polymeric substances (EPS)
molecules secreted by microbes that coat the cell surface. EPS can be a variety of biomolecules, including DNA, protein, and carbohydrate. Some forms of EPS contribute to stress resistance
- Habitat
an environment generally inhabited by a particular species
- Humoral immunity
the branch of adaptive immunity mediated by macromolecules (as opposed to cells) found in extracellular fluids such as secreted antibodies, complement proteins, and certain antimicrobial peptides
- Induced systemic resistance
a type of signaling occurring between select beneficial microbes and plants. The microbes induce an immune response that protects both local and systemic plant tissues from damage by either pathogenic microbes or insect predators
- Lectin
a type of protein that can bind to cell membranes. They are sugar binding and become the “glyco” portion of glycoconjugates on the membranes. Lectins perform recognition on the cellular and molecular level, and play numerous roles in biological recognition phenomena involving cells, carbohydrates, and proteins. Lectins also mediate attachment and binding of bacteria and viruses to their intended targets
- Microbiota
the community of microorganisms (beneficial, pathogenic, or commensal) that colonize a particular environment, such as the tissues of a plant or animal
- Microbe-associated molecular pattern (MAMP)
conserved signal molecules produced by both beneficial and pathogenic microbes that elicit host innate-immune signaling
- Mutualism
an association of one or more organisms, wherein each partner provides, and derives a benefit, in relation to the other partners
- Oxidative burst
the rapid release of reactive oxygen species from cells, often as part of an immune response
- Phyllosphere
all parts of a plant that are aboveground (stems, leaves, fruit), and that can be colonized by microbes
- Quorum signaling
a type of signaling in which microbes coordinate group responses by sensing the accumulation of specific secreted molecules (e.g., pheromones)
- Rhizosphere
the plant root surface and zone of the surrounding soil into which roots secrete nutrients and signal molecules
- Signal
a chemical or physical stimulus produced by one organism, with the primary purpose of eliciting a response in a target organism
- Specificity
the characteristic of a symbiotic association that results in the interaction being restricted to the members of particular species
- Stress
a chemical or physical agent that, unless mitigated by the induction of a physiological response, will damage the fitness of an organism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kabat AM, et al. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014;35:507–517. doi: 10.1016/j.it.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer F, Bäckhed F. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 4.Limpens E, et al. Lipo-chitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu Rev Phytopathol. 2015:53. doi: 10.1146/annurev-phyto-080614-120149. [DOI] [PubMed] [Google Scholar]
- 5.Axelrod R. The evolution of cooperation. Basic Books; 1984. [Google Scholar]
- 6.Hussa EA, Goodrich-Blair H. It takes a village: ecological and fitness impacts of multipartite mutualism. Annu Review Microbiol. 2013;67:161–178. doi: 10.1146/annurev-micro-092412-155723. [DOI] [PubMed] [Google Scholar]
- 7.Mullard A. Microbiology: Tinker, bacteria, eukaryote, spy. Nature News. 2009;459:159–161. doi: 10.1038/459159a. [DOI] [PubMed] [Google Scholar]
- 8.Pickard JM, Chervonsky AV. Intestinal fucose as a mediator of host–microbe symbiosis. J Immunol. 2015;194:5588–5593. doi: 10.4049/jimmunol.1500395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen T, et al. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 11.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulgarelli D, et al. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 13.Begley M, et al. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Russell JB, Rychlik JL. Factors that alter rumen microbial ecology. Science. 2001;292:1119–1122. doi: 10.1126/science.1058830. [DOI] [PubMed] [Google Scholar]
- 15.Savatin DV, et al. Wounding in the plant tissue: the defense of a dangerous passage. Front Plant Sci. 2014;5:470. doi: 10.3389/fpls.2014.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 17.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang JY, et al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe. 2015;18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JK, et al. Polyester synthesis genes associated with stress resistance are involved in an insect–bacterium symbiosis. Proc Natl Acad Sci U S A. 2013;110:E2381–E2389. doi: 10.1073/pnas.1303228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 23.Oldroyd GE. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 24.Lehman AP, Long SR. Exopolysaccharides from Sinorhizobium meliloti can protect against H2O2-dependent damage. J Bacteriol. 2013;195:5362–5369. doi: 10.1128/JB.00681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leigh JA, et al. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wozniak DJ, Parsek MR. Surface-associated microbes continue to surprise us in their sophisticated strategies for assembling biofilm communities. F1000prime reports. 2014;6:26. doi: 10.12703/P6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 28.Tiricz H, et al. Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization associated changes in the transcriptome of Sinorhizobium meliloti. Appl Environ Microbiol. 2013:01791–01713. doi: 10.1128/AEM.01791-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penterman J, et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci U S A. 2014;111:3561–3566. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 31.Mersmann S, et al. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010;154:391–400. doi: 10.1104/pp.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodenhausen N, et al. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisberg EE, et al. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. 2013;8(11):e78613. doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knief C, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata S, et al. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol. 2012;194:6736–6747. doi: 10.1128/JB.00707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks JF, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A. 2014;111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandel MJ, et al. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maltz MA, et al. OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl Environ Microbiol. 2012;78:7760–7768. doi: 10.1128/AEM.01858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braschler TR, et al. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl Environ Microbiol. 2003;69:4268–4271. doi: 10.1128/AEM.69.7.4268-4271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Futahashi R, et al. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. 2013;8(5):e64557. doi: 10.1371/journal.pone.0064557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kremer N, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kremer N, et al. The dual nature of haemocyanin in the establishment and persistence of the squid-Vibrio symbiosis. Proc R Soc B. 2014:281. doi: 10.1098/rspb.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinzón JH, et al. Whole transcriptome analysis reveals changes in expression of immune-related genes during and after bleaching in a reef-building coral. Roy Soc Open Sci. 2015;2:140214. doi: 10.1098/rsos.140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pamp SJ, et al. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuwahara T, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen W, et al. Adaptive immunity to murine skin commensals. Proc Natl Acad Sci U S A. 2014;111:E2977–E2986. doi: 10.1073/pnas.1401820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obata T, et al. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci U S A. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rader BA, et al. Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-Vibrio symbiosis. mBio. 2012;3(3):e00093–12. doi: 10.1128/mBio.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Aksoy S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc Natl Acad Sci U S A. 2012;109:10552–10557. doi: 10.1073/pnas.1116431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodge A, Fitter AH. Microbial mediation of plant competition and community structure. Funct Ecol. 2013;27:865–875. [Google Scholar]
- 54.Coyte KZ, et al. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 55.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoenian I, et al. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci U S A. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, et al. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol. 2012;78:2120–2127. doi: 10.1128/AEM.07539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barr JJ, et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc Natl Acad Sci U S A. 2015;112:13675–13680. doi: 10.1073/pnas.1508355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barr JJ, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollenberg MS, et al. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio. 2014;5:e01286–01214. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Secor PR, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18:549–559. doi: 10.1016/j.chom.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veesenmeyer JL, et al. NilD CRISPR RNA contributes to Xenorhabdus nematophila colonization of symbiotic host nematodes. Mol Microbiol. 2014;93:1026–1042. doi: 10.1111/mmi.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraune S, et al. Bacteria–bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 2015;9(7):1543–1556. doi: 10.1038/ismej.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 67.Fanning S, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A. 2012;109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferreyra JA, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curtis MM, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neumann A, Suen G. Differences in major bacterial populations in the intestines of mature broilers after feeding virginiamycin or bacitracin methylene disalicylate. J Appl Microbiol. 2015;119:1515–1526. doi: 10.1111/jam.12960. [DOI] [PubMed] [Google Scholar]
- 72.LaVorgna M, et al. Performance of broilers fed a broader spectrum antibiotic (virginiamycin) or a narrower spectrum antibiotic (bacitracin methylene disalicylate) over 3 consecutive grow-out cycles. J Appl Poultry Res. 2013;22:574–582. [Google Scholar]
- 73.Fürnkranz M, et al. Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microbial Ecol. 2012;63:418–428. doi: 10.1007/s00248-011-9942-4. [DOI] [PubMed] [Google Scholar]
- 74.Wilson CM, et al. Transcriptional and metabolomic consequences of LuxS inactivation reveal a metabolic rather than quorum-sensing role for LuxS in Lactobacillus reuteri 100–23. J Bacteriol. 2012;194:1743–1746. doi: 10.1128/JB.06318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal JE, et al. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun. 2013;81:1341–1353. doi: 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thompson JA, et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 78.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 79.Yasuda K, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z, et al. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014;8:881–893. doi: 10.1038/ismej.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu HP, et al. Spatial heterogeneity of gut microbiota reveals multiple bacterial communities with distinct characteristics. Sci Rep. 2014;4:6185. doi: 10.1038/srep06185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heath-Heckman EA, et al. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-Vibrio symbiosis. mBio. 2013;4(2):e00167–13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu X, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwartzman JA, et al. The chemistry of negotiation: rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci U S A. 2015;112:566–571. doi: 10.1073/pnas.1418580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherji A, et al. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 88.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geva-Zatorsky N, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med. 2015;21:1091–1100. doi: 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rath CM, et al. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal Chem. 2012;84:9259–9267. doi: 10.1021/ac302039u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nikolakakis K, et al. Using hybridization chain-reaction fluorescent in situ hybridization (HCR-FISH) to track gene expression by both partners during initiation of symbiosis. Appl Environ Microbiol. 2015;81:4728–4735. doi: 10.1128/AEM.00890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bouslimani A, et al. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci U S A. 2015;112:E2120–E2129. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alivisatos A, et al. A unified initiative to harness Earth’s microbiomes. Science. 2015;350:507–508. doi: 10.1126/science.aac8480. [DOI] [PubMed] [Google Scholar]
- 95.Dubilier N, et al. Microbiology: Create a global microbiome effort. Nature. 2015;526:631. doi: 10.1038/526631a. [DOI] [PubMed] [Google Scholar]
- 96.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 97.Mukherjee S, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103–107. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wesener DA, et al. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol. 2015;22:603–610. doi: 10.1038/nsmb.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matamouros S, Miller SI. S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. Biochim Biophys Acta - Biomembranes. 2015;1848:3021–3025. doi: 10.1016/j.bbamem.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saito HE, et al. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol. 2014;80:6527–6538. doi: 10.1128/AEM.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wier AM, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giles DK, et al. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol. 2011;79:716–728. doi: 10.1111/j.1365-2958.2010.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Parsons JB, et al. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol. 2012;194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim JK, et al. Insect gut symbiont susceptibility to host antimicrobial peptides caused by alteration of the bacterial cell envelope. J Biol Chem. 2015;290:21042–21053. doi: 10.1074/jbc.M115.651158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Horváth B, et al. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1500777112. 201500777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heath-Heckman EA, et al. Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid Vibrio symbiosis. Environ Microbiol. 2014;16(12):3669–3682. doi: 10.1111/1462-2920.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]