Abstract
The role of cyclins and their catalytic partners, the cyclin-dependent kinases (CDKs), as core components of the machinery that drives cell cycle progression is well established. Increasing evidence indicates that mammalian cyclins and CDKs also carry out important roles in other cellular processes such as transcription, DNA damage repair, the control of cell death, differentiation, the immune response and metabolism. Some of these non-canonical functions are performed by cyclins or by CDKs, independent of their respective cell cycle partners, suggesting a substantial divergence in the function of these proteins during evolution.
Introduction
Progression through the cell division cycle is driven by cyclins, which bind and activate their catalytic partners, the cyclin-dependent kinases (CDKs). Specific heterodimeric cyclin–CDK complexes phosphorylate a plethora of cellular proteins to promote entry into the cell cycle and G1 phase, regulate DNA synthesis (during the S phase) and trigger segregation of the newly duplicated chromosomes to the daughter cells during mitosis, thereby ensuring that the cell cycle progresses in an ordered manner1, 2.
Mammalian cyclin and CDK families each contain more than 20 members3, but only a few cyclin–CDK complexes are known to directly participate in the cell division cycle (Figure 1). Growth factors induce the expression of D-type cyclins (D1, D2 and D3), which are therefore regarded as molecular links between the cell environment and the core cell cycle machinery. Once induced, D-cyclins interact with CDK4 or CDK6, and phosphorylate the retinoblastoma protein (pRB), p107 and p130 pocket proteins, which bind and regulate E2F transcription factors during the G1 phase of the cell cycle. Later during G1, E-type cyclins (cyclins E1 and E2) become upregulated and activate CDK2 (and, to a lesser extent, CDK1 and CDK3), resulting in phosphorylation of a broader range of cell cycle-related proteins. The subsequent induction of cyclin A2 in S phase, which partners with CDK2 and CDK1, and activation of cyclin B1-CDK1 at the onset of mitosis drives the progression of cells through the remainder of the cell cycle through phosphorylation of a large number of proteins involved in DNA replication, as well as centrosome and chromosome function1, 2, 4, 5.
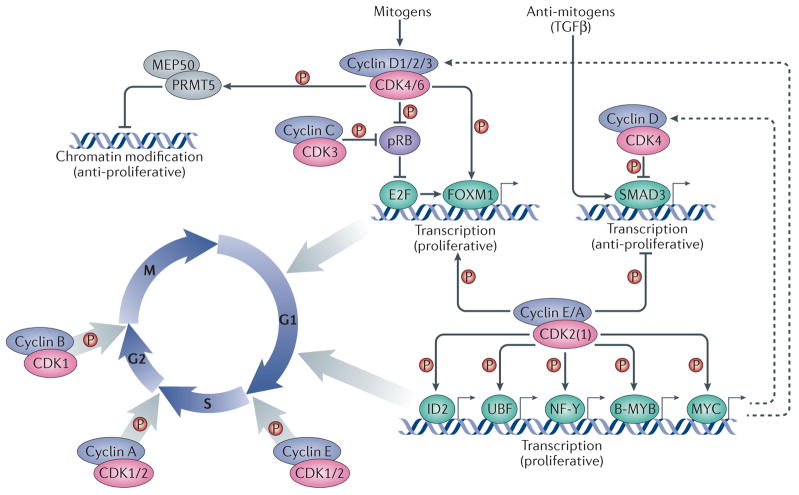
Figure 1. Canonical roles of cyclins and CDKs in the cell division cycle.
Cell cycle progression is driven by heterodimeric complexes formed by cyclin D, cyclin E, cyclin A or cyclin B with cyclin-dependent kinase (CDK) 4, CDK6, CDK2 or CDK1. (Cyclin C–CDK3 complexes also participate in G0 to G1 phase progression although their physiological relevance in this process is less well established). Cell cycle entry is driven by the formation of cyclin D–CDK4/6 complexes in response to mitogenic stimulation. These complexes phosphorylate and partially inactivate the retinoblastoma protein, pRB, thereby releasing its inhibition of the transcription factor E2F. E2F subsequently promotes the expression of multiple cell cycle genes, among which are E-type cyclins, which bind to and activate CDK2 (and, to a lesser extent, CDK1). Activation of CDK2 (and CDK1) by E-cyclins and A-cyclins leads to the phosphorylation of multiple transcription factors, such as the helix-loop-helix (HLH) protein ID2 as well as UBF, NF-Y, B-MYB and MYC, which contribute to cell cycle progression at different levels. Cyclin D1–CDK4 complexes also phosphorylate chromatin modifiers, such as the protein arginine N-methyltransferase 5 (PRMT5) cofactor MEP50, to promote the repression of specific genes with anti-proliferative properties. Multiple cyclin–CDK complexes can phosphorylate the transcription factors SMAD3, which is activated by antimitogens such as TGFb, and FOXM1 to promote cell cycle progression. Entry into mitosis is specifically driven by cyclin B–CDK1 complexes. Dashed arrows indicate indirect connections or connections with multiple steps. Abbreviations: MEP50, methylosome protein 50; NF-Y, nuclear factor Y; UBF, upstream binding factor.
Consistent with a role for cyclin–CDK complexes in promoting cell cycle progression, amplification or genetic rearrangements of cyclin or CDK genes, and/or their overexpression have been documented in nearly all human tumour types6–9. In particular, the cyclin D1 gene (CCND1) represents the second most frequently amplified locus across all types of human cancers. For these reasons cyclin–CDK complexes are considered promising targets for cancer therapy.
Although the canonical role of cyclins and CDKs as essential drivers of cell cycle entry and progression has been firmly established (Figure 1), research carried out over the past two decades provides increasing evidence for additional functions of these proteins in gene transcription, DNA damage repair, cell death, cell differentiation and metabolism. In this Review, we focus on the main biological areas where such non-canonical functions of cyclins and CDKs have been documented. We restricted our review to interphase cyclins and CDKs, that is, proteins operating in the G1, S and G2 phases. Non-cell cycle CDKs such as CDK5, are only discussed in the context of their interactions with interphase cyclins, such as cyclin E-CDK5 function in postmitotic neurons.
Kinase-dependent roles in transcription
In the current model of cell cycle regulation, the main function of cyclin D-CDK4/6 complexes is to phosphorylate and inactivate pocket proteins, thereby enabling the transcription of genes required for cell cycle progression2, 5, 10. In addition, cyclin D-CDKs were shown to phosphorylate several transcription factors, thereby linking cell cycle and transcription (Table 1). Cyclin D–CDK4 complexes phosphorylate SMAD3, which inhibits the anti-proliferative effect of signalling through the transforming growth factor β pathway11 (Figure 1). Cyclin D–CDK4 and cyclin D–CDK6 complexes activate the transcription factor forkhead box protein M1 (FOXM1), which regulates the expression of multiple cell cycle regulators, including proteins that govern the G2–M transition12. Cyclin D1–CDK4-mediated phosphorylation of methylosome protein 50 increases the activity of the chromatin modifier PRMT5 (protein arginine N-methyltransferase 5), which is thought to mediate key events associated with cyclin D1–CDK-dependent neoplastic growth13.
Table 1.
Representative transcription factors regulated by interphase cyclins and CDKs*
| Transcription factor | Cyclin | CDK | Kinase dependent? | Activation or Repression | Biological function(s) | Reference |
|---|---|---|---|---|---|---|
| Regulation by cyclin--CDK complexes | ||||||
| B-Myb | A2 | CDK2 | YES | Activation | Promoting cell cycle progression | 21 |
| c-Myc | E1 | CDK2 | YES | Activation | Repression of cellular senescence | 22 |
| FOXM1C | D1, E1, A2 | CDK2, CDK4 | YES | Activation | Promoting cell cycle progression, repression of cellular senescence | 12, 14 |
| FOXO1 | E | CDK2 | YES | Repression | Mediating genotoxic responses | 93 |
| FOXP3 | E | CDK2 | YES | Repression | Development of regulatory T-cells | 128 |
| GATA4 | D1 | CDK4 | YES | Repression | Preventing cardiomyocyte differentiation | 103 |
| ID2 | E1, A2 | CDK2 | YES | Repression | Promoting cell cycle progression | 15 |
| MEF2 | D3 (D1) | CDK4 | YES | Repression | Preventing muscle differentiation | 102 |
| NF-Y | A2 | CDK2 | YES | Activation | Promoting cell cycle progression | 17,18 |
| NGN2 | A- | CDK2, CDK1 | YES | Repression | Inhibition of neuronal differentiation | 113 |
| NRF1 | D1 | CDK4 | YES | Repression | Inhibition of mitochondrial function | 141 |
| PPARγ | D3 | CDK4 | YES | Repression | Induction of adipogenesis | 109, 154 |
| RUNX2 | D1 | CDK4 | YES | Repression | Inhibition of bone differentiation | 98 |
| SMAD3 | - | CDK2, CDK4 | YES | Repression | Preventing TGFβ signaling | 11 |
| UBF | E1, A2 | CDK2 | YES | Activation | Ribosomal DNA transcription | 16 |
| Regulation by cyclins | ||||||
| RUNX1 (AML1) | D3 | - | NO | Repression | Inhibition of myeloid differentiation | 46 |
| AR | D1, D3 | - | NO | Repression | Inhibition of hormone-dependent gene expression | 33, 36 |
| NeuroD | D1 | - | NO | Repression | Inhibition of neuronal differentiation | 39 |
| C/EBPβ | D1 | - | NO | Activation | Promoting mammary epithelial differentiation | 172 |
| CBP | D1 | - | NO | Activation | Promoting Notch-dependent retinal development | 36,48 |
| DMP1 | D2 (D1, D3) | - | NO | Repression | Modulation of the p53 pathway | 42 |
| ER | D1 | - | NO | Activation | Activation of hormone-dependent gene expression | 30–32 |
| FOXO1, O3 | D1 | - | NO | Repression | Inhibition of anoikis | 97 |
| hATF5 | D3 | - | NO | Activation | Multiple cellular processes | 45 |
| HNF4a | D1 | - | NO | Repression | Inhibition of lipogenesis in the liver | 152 |
| MYOD | D1 | CDK4 | NO | Repression | Blocking myoblast differentiation | 24, 25, 27 |
| PPARγ | D1 | - | NO | Repression | Blocking adipogenesis | 38 |
| RUNX3 | D1 | - | NO | Repression | Cell proliferation | 40 |
| SP1 | D1 | - | NO | Repression | Multiple cellular processes | 28 |
| STAT3 | D1 | - | NO | Repression | Cytokine signaling | 173 |
| TR | D1 | - | NO | Repression | Hormone-dependent gene expression | 41 |
| VDR | D3 | - | NO | Activation | Vitamin-dependent gene expression | 44 |
| Regulation by CDKs | ||||||
| c-JUN | - | CDK6 | NO | Activation | Stimulation of angiogenesis | 54 |
| RUNX1 | - | CDK6 | NO | Repression | Inhibition of myeloid differentiation | 53 |
| STAT3 | D1,2,3 | CDK6 | NO | Activation | Cell cycle inhibition | 54 |
pRB, p107 and 130 proteins are not included.
Abbreviations: AR, androgen receptor; ER, estrogen receptor; TR, thyroid hormone receptor; VDR, vitamin D receptor; B-Myb, V-Myb Avian Myeloblastosis Viral Oncogene Homolog-Like 2; c-Myc, V-Myc Avian Myelocytomatosis Viral Oncogene Homolog; FOXM1C, Forkhead Box Protein M1C; FOXO1, Forkhead Box Protein O; FOXP3, Forkhead Box Protein P3; GATA4, GATA Binding Protein 4; ID2, Inhibitor Of DNA Binding 2, Dominant Negative Helix-Loop-Helix Protein; MEF2, Myocyte-Specific Enhancer Factor 2; NF-Y, Nuclear Transcription Factor Y; NGN2, Neurogenin 2; NRF1, Nuclear Respiratory Factor 1; PPARγ, Peroxisome Proliferator-Activated Receptor Gamma; RUNX2, Runt-Related Transcription Factor 2; SMAD3, SMAD Family Member 3; UBF, Upstream Binding Transcription Factor; RUNX1, Runt-Related Transcription Factor 1; NeuroD, Neuronal Differentiation; C/EBPβ, CCAAT/Enhancer Binding Protein Beta; CBP, CREB Binding Protein; DMP1, Dentin Matrix Acidic Phosphoprotein 1; FOXO3, Forkhead Box Protein O3; hATF5, human Activating Transcription Factor 5; HNF4α, Hepatocyte Nuclear Factor 4 Alpha; MYOD, Myogenic Differentiation; RUNX3, Runt-Related Transcription Factor 3; SP1, Specificity Protein 1; STAT3, Signal Transducer And Activator Of Transcription 3; c-JUN, V-Jun Avian Sarcoma Virus 17 Oncogene Homolog.
Also E-type and A-type cyclins have been implicated in directly modulating transcription through CDK-dependent phosphorylation of several DNA-binding factors, including SMAD3 and FOXM111, 14, ID2, upstream binding factor (UBF), and nuclear factor Y (NF-Y)15–20 (Figure 1). Cyclin E/A–CDK2 complexes also phosphorylate the transcription factors MYC and B-MYB, thereby enhancing the ability of these proteins to activate target genes during the cell cycle21, 22. In tumours driven by c-MYC overexpression, CDK2 activity seems to be essential for preventing senescence and allowing immortalization of cancer cells23. However, while the functional link between cyclin–CDK kinases and pocket proteins has been firmly established, the contribution of these additional phosphorylation events to normal cell cycle progression remains unknown.
Kinase-independent transcriptional roles
In addition to the role of cyclin-CDK complexes in phosphorylating transcriptional regulators, there is substantial evidence for transcriptional functions of cyclins not involving kinase activity. Interestingly, reports of cell cycle-independent transcriptional roles of cyclins and CDKs have so far been vastly dominated by D-type cyclins.
D-type cyclins have direct roles in transcription
The first indication of a pRB-independent role for cyclins in transcriptional control came from the demonstration that ectopic expression of cyclin D1 abrogated the ability of the myogenic master transcription factor MyoD to activate the muscle creatine kinase (MCK) promoter24. In skeletal myoblasts, cyclin D1 recruits CDK4 to MyoD and thereby blocks MyoD-dependent transcription independently of CDK4 catalytic activity25–27. Cyclin D1 was also shown to repress Sp1-mediated transcription by binding to the TATA box binding protein (TBP)-associated factor TAFII250 in a pRB-independent manner28, 29. A direct interaction between cyclin D1 and the estrogen receptor has been reported to augment estrogen receptor -driven transcription in a CDK kinase-independent manner. Specifically, cyclin D1 was shown to function as a scaffold between the estrogen receptor and members of the steroid receptor coactivator family30, 31, as well as between the estrogen receptor and the chromatin modifier histone acetyltransferase KAT2B (also known as P/CAF)32. In contrast, cyclin D1 and cyclin D3 inhibit the transactivation activity of the androgen receptor, another member of the nuclear steroid hormone receptor family, in a kinase-independent manner, by competing with it for P/CAF binding33–35,36.
Numerous reports have subsequently described interactions of D-type cyclins with many transcription factors (Table 1). Several groups have demonstrated, mostly using protein overexpression systems, that D-type cyclins interact with sequence-specific transcription factors and activate or repress transcription by recruiting chromatin modifiers to gene promoters (or, in some cases, by blocking their access). Cyclin D1 directly binds and inhibits histone acetyltransferases such as p300 and CREB-binding protein (CBP) (in addition to P/CAF) and represses their ability to activate multiple promoters including that of the gene encoding peroxisome proliferator-activated receptor γ (PPARγ), a master regulator of adipogenesis32, 37. Consistent with these data, mouse embryonic fibroblasts lacking cyclin D1 display increased PPARγ activity and are prone to adipogenic differentiation38. Cyclin D1 can also repress transcription by neurogenic differentiation factor 1 (NeuroD) or runt-related transcription factor 3 (RUNX3), independently of CDK activity, by preventing the interaction of these transcription factors with p30039, 40. Furthermore, cyclin D1 can inhibit the transcriptional activity of thyroid hormone receptors by recruiting histone deacetylases to these transcription factors and by promoting the formation of transcriptional repressive complexes41. Cyclin D2 and cyclin D3 can also modulate the activity of several transcription factors by interacting directly with them. A yeast two-hybrid screen identified the Myb-like transcription factor DMP1 as a cyclin D2 interactor42. Enforced expression of any D-type cyclin inhibits the transcriptional activity of DMP1 in a CDK-independent manner42, 43. Intriguingly, DMP1 regulates the expression of p19ARF, an inhibitor of MDM2, a negative regulator of p53, raising the possibility that D-cyclins can modulate both the pRB pathway (through CDK activity) and the p53 pathway (through CDK-independent activity).
Cyclin D3 was reported to interact with and enhance the transactivation of the vitamin D receptor, and this effect was diminished by overexpression of CDK4 or CDK644. Additionally, cyclin D3 interacts with human activating transcription factor 5 (hATF5) and enhances its activity45. On the other hand, cyclin D3 competes with core binding factor β for binding RUNX1 (also known as AML1), thereby reducing the ability of RUNX1 to bind DNA and activate transcription46.
Collectively, these reports suggest that D-cyclins might exert transcriptional effects independent of any associated kinase activity. However, the biological significance of these findings remained unclear. To address this point, human cancer cells were engineered to overexpress wild type cyclin D1 or a mutant cyclin D1 that is unable to activate CDK4 and CDK647. Surprisingly, the set of target genes that were induced was identical in both cases and was lacking E2F target genes, indicating that cyclin D1 plays a kinase- and E2F-independent role in transcription. Further computational analysis implicated the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) as the effector of cyclin D1 in regulating gene expression. Proteomic studies using a knock-in mouse strain expressing a tagged version of cyclin D1 in place of the wild-type protein revealed that cyclin D1 interacts in vivo with several proteins involved in transcription48. Furthermore, chromatin immunoprecipitation coupled to microarray (ChIP-on-chip) analysis revealed an extensive interaction of cyclin D1 with the mouse genome48. In particular, within developing mouse retinas, cyclin D1 physically occupies the Notch1 gene enhancer, to which it recruits CBP acetyltransferase, thereby activating gene expression. Genetic ablation of cyclin D1 decreased recruitment of CBP to the Notch1 gene enhancer, and diminished histone acetylation and expression of Notch1 in the retina. Given an essential role for Notch1 in retinal development49, these findings likely explain, at least in part, retinal developmental abnormalities seen in cyclin D1-null animals50, 51. A direct interaction of cyclin D1 with chromatin was also demonstrated by a ChIP-sequencing study in which tagged cyclin D1 was ectopically expressed in cyclin D1-null mouse embryonic fibroblasts. The authors concluded that cyclin D1 plays a role in chromosomal stability through binding to genes that govern chromosomal integrity52.
Direct transcriptional functions of CDK6
Although the majority of reports have highlighted a CDK4/6-independent function for D-cyclins in regulating transcription, some intriguing studies revealed a kinase-independent role for CDK6 itself in this process. CDK6 was shown to physically interact with and inhibit transcriptional activity of RUNX1 in a kinase-independent fashion, and by doing so block myeloid differentiation53. CDK6, but not CDK4, activates c-JUN and signal transducer and activator of transcription 3 (STAT3) to induce transcription of cyclin-dependent kinase inhibitor 2a (Cdkn2a; encoding the CDK4/6 inhibitor p16INK4A), and Vegfa (encoding vascular endothelial growth factor A, VEGF-A). Whereas the action of CDK6 on p16INK4A expression requires the presence of a D-type cyclin, the effect on VEGF-A expression is independent of cyclin D54. Interestingly, cyclin D1 has also been postulated to contribute to VEGFA gene transcription55, suggesting that both CDK6 and cyclin D1 might regulate angiogenesis through different routes. The observation that CDK6 upregulates its own inhibitor p16INK4a indicates the presence of a negative feedback loop, by which CDK6 safeguards against uncontrolled proliferation triggered by CDK6 overexpression54. CDK6 can exert its pro-proliferative role only upon silencing of the p16INK4a gene, an event frequently seen in human tumors6. Additional data suggest that the transcriptional activity of CDK6 might also be crucial in regulating the balance between quiescence and proliferation in haematopoietic and leukaemic stem cells by modulating the activity of the transcription factor early growth response protein 1 (EGR1) 56, although whether CDK6 binds it directly is currently unknown.
Cyclins and CDKs in DNA damage repair
DNA double-strand breaks (DSBs) are repaired by two distinct mechanisms: the high fidelity homologous recombination, which uses the sister chromatid as a major template for repair, and non-homologous end joining (NHEJ), which directly joins the broken DNA and is therefore error-prone. Work in several systems indicates that components of the core cell cycle machinery have important and direct roles in DSB repair.
Yeast
In Saccharomyces cerevisiae, inhibition of the sole cyclin-dependent kinase, Cdc28 (which is homologous to CDK1) after the induction of DSBs cripples end resection of damaged DNA, thereby preventing the loading of the homologous recombination repair proteins Rad51 and replication protein A to sites of DNA damage57, 58; when Cdc28 is inhibited before the induction of DSBs, the incidence of error-prone NHEJ increases substantially, indicating that the kinase activity of Cdc28 is necessary to support homologous recombination 57 (Figure 2a). In Schizosaccharomyces pombe, the CDK1 homologue, Cdc2, promotes the formation of Rad51 foci, resulting in the formation of recombination intermediates at early stages of homologous recombination. At a later stage of repair, Cdc2 and a checkpoint protein Crb2 were shown to control topoisomerase III activity, thereby ensuring complete processing of recombination intermediates59. S. cerevisiae Cdc28 can also regulate DNA damage repair by phosphorylating multiple substrates, such as Sae2, an endonuclease required for the initiation of resection of single-stranded DNA60. Another substrate of the S. cerevisiae Cdc28 is the Srs2 helicase, the phosphorylation of which prevents its sumoylation and targets it to dismantle specific DNA structures, such as D-loops, in a helicase-dependent manner during homologous recombination61. During DNA damage repair Cdc28 also phosphorylates the G2 checkpoint protein Rad962–64 and the resection nuclease Dna265. Collectively, these studies present a strong case for the requirement of CDK activity in the resection of DSB-ends and in mediating the choice between the homologous recombination and NHEJ repair pathways in yeast66.
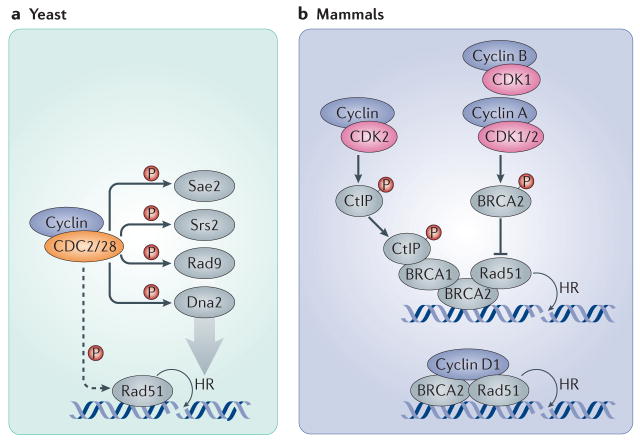
Figure 2. Cyclins and CDKs in DNA double-strand breaks (DSBs) repair.
a| In yeast, the sole CDK (CDC28 in S. cerevisiae and CDC2 in S. pombe) participates in the recruitment of Rad51 to DNA double strand breaks and phosphorylates several proteins involved in the DNA damage response and the repair of DSBs through homologous recombination, such as the endonuclease Sae2, the helicase Srs2, the G2 checkpoint protein Rad9 and the resection nuclease Dna2. b| In mammalian cells, CDK2 and CDK1 (together with A-, B- and potentially also E-type cyclins) modulate homologous recombination by the direct phosphorylation of BRCA2, preventing it from binding and recruiting RAD51. Conversely, CDK2 can support homologous recombination by phosphorylating CtBP-interacting protein (CtIP; the human homologue of Sae2), which promotes the recruitment of BRCA1 to DSBs. In addition, cyclin D1 helps to recruit RAD51 to DSB sites by directly interacting with it in a CDK-independent manner, thereby promoting homologous recombination. Dashed arrows indicate indirect connections or connections with multiple steps.
Mammals
Contrary to yeast, cyclin A- and cyclin B-associated CDK activity was found to inhibit homologous recombination in mammalian cells. Cyclin A–CDK1/2 and cyclin B–CDK1 complexes phosphorylate Ser 3291 of BRCA267, thereby preventing BRCA2 from binding to RAD51 and from recruiting it to sites of DNA damage (Figure 2b). DNA damage leads to reduced levels of Ser3291 phosphorylation and restoration of the interaction between BRCA2 and RAD5167. Phosphorylation of Ser3291 is low in S-phase but increases towards mitosis, suggesting that termination of homologous recombination by CDKs is necessary for normal chromosome segregation.. Consistent with this notion, expression of mutated forms of BRCA2 that cannot be phosphorylated by CDK does not change the rate of homologous recombination, but delays the onset of mitosis68. Conversely, CDK2 was also shown to support homologous recombination by phosphorylating CtBP-interacting protein (CtIP, the human homologue of Sae2), thereby promoting its interaction with BRCA1 and the MRE11 exonuclease and leading to the resection of DSBs 69 (Figure 2b).
As a consequence of DNA damage, cyclin D1 is degraded and cyclin D–CDK4/6 complexes are disrupted, thereby contributing to cell cycle arrest70. Expression of degradation-resistant mutants of cyclin D1 results in increased levels of DNA damage, confirming the importance of preventing cell cycle progression in these conditions71. An unexpected direct role for cyclin D1 in DNA damage repair was first suggested by the observation that targeting cyclin D1 to chromatin led to co-recruitment of RAD5172. Subsequently, it was reported73 that cyclin D1 localizes, in a BRCA2-dependent manner, to irradiation-induced DSBs in human cancer cells, where it helps to recruit RAD51, thereby stimulating homologous recombination (Figure 2b). Depletion of cyclin D1 in human cancer cells reduced the recruitment of RAD51 and decreased homologous recombination, and rendered cells more sensitive to DNA damage. Notably, this function of cyclin D1 was shown to be kinase independent and to take place also in pRB-deficient cells73.
A positive role for cyclin D1 in DNA damage repair might seem at odds with the observation that cyclin D1 becomes rapidly degraded after DNA damage. However, a relatively small pool of cyclin D1 persists after irradiation, and cyclin D1 becomes redistributed to DNA damage sites. This DNA-bound pool of cyclin D1 likely plays a positive role in DNA damage repair73. On the other hand, forced overexpression of cyclin D1 might override cell cycle arrest following DNA damage, leading to DNA replication stress and, ultimately, apoptosis. Consistent with this notion, overexpression of cyclin D1 in cultured tumour cells rendered the cells more sensitive to radiation and triggered apoptosis74, 75. One important and unresolved issue is how overexpression of cyclin D1, which is seen frequently in human cancers, affects the response of tumour cells to DNA damage in vivo.
Balancing cell proliferation and death
The regulation of cell cycle is tightly linked to the control of cell death. Indeed, early studies indicated that the pRB–E2F pathway can modulate the expression of multiple pro- or anti-apoptotic proteins5, 76. Overexpression of cyclins in cultured cells leads to either increased apoptosis or increased survival depending on the cellular setting. For instance, ectopic expression of cyclin D1 induces cell death and increases sensitivity to cytotoxic agents in a variety of cell lines74, 77–81. By contrast, overexpression of cyclin D1 protects fibrosarcoma cells from cell death82, and suppression of cyclin D1 using antisense oligonucleotides sensitizes pancreatic cancer cells to apoptosis induced by cytotoxic agents83. Similarly, ectopic expression of cyclin D3 in leukemic T-cells prevents them from entering apoptosis normally induced by phorbol myristate acetate84, and ablation of cyclin D3 in T-cell acute leukemia cells triggers cell death85. Whether these effects are direct or mediated by the E2F–pRB pathway is unclear in most of the cases. However, D-type cyclins and CDKs can also modulate apoptotic cell death through direct interaction with the apoptotic machinery. In mantle cell lymphoma cyclin D1, which is frequently overexpressed in this tumor type, binds and sequesters the pro-apoptotic protein BAX in the cytoplasm, therefore inhibiting apoptosis86 (Figure 3). CDK4 interacts with the apoptosis inhibitor survivin (also known as baculoviral IAP repeat-containing protein 5). As a result, p21Cip1 is released from its complex with CDK4 and interacts with mitochondrial procaspase 3 to suppress Fas-mediated cell death87.
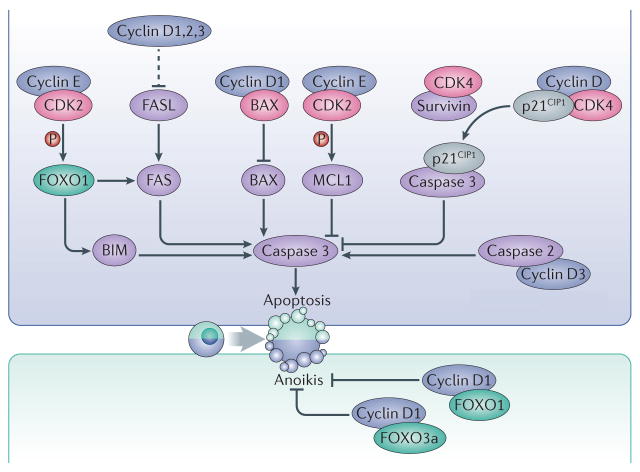
Figure 3. Regulation of cell death by interphase cyclins and CDKs.
Interphase cyclins modulate apoptosis using CDK-dependent and independent mechanisms. Cyclin E–CDK2 complexes phosphorylate the prosurvival factor MCL1 as well as the transcription factor FOXO1, leading to inhibition or activation of apoptosis, respectively. Phosphorylation of MCL1 stabilizes the protein while phosphorylation of FOXO1 leads to upregulated expression of the pro-apoptotic factors FAS and BIM. Binding of survivin to CDK4 causes the release of the CDK inhibitor p21 (CIP1), which binds and inhibits caspase 3. D-type cyclins can also directly interact with the pro-apoptotic proteins BAX or caspase 2. Moreover, the three D-type cyclins have overlapping roles in repressing the expression of the death receptor FAS and its ligand, FASL, thereby preventing apoptosis in haematopoietic cells. Cyclin D1 also interacts with the transcription factors FOXO1 and FOXO3a and prevents anoikis. Dashed arrows indicate indirect connections or connections with multiple steps.
A pro-apoptotic function for cyclin D3 is suggested by the observation that it can physically interact with caspase 2, leading to the stabilization of this protease and promoting its cleavage into an active form88 (Figure 3). Cyclin D1-CDK kinase activity might play a role in mediating cell death under pathological conditions. Increased activity of cyclin D1-CDK4 correlating with neuronal death, has been reported in a rat model of stroke89. Moreover, neuronal cell death triggered by treatment of rats with kainic acid can be prevented by injection of CDK4 or cyclin D1 antisense-oligonucleotides into brain ventricles90. Administration of flavopiridol, a pharmacological inhibitor of CDKs into brain ventricles, prevents neuronal cell death in a model of focal ischemia89.
Analyses of mice lacking D-type cyclins support the pro-survival function of these proteins in select tissues. Genetic ablation of cyclin D1 leads to increased apoptosis of photoreceptor cells in postnatal animals91. Whereas constitutive, germline ablation of cyclin D1 (or cyclin D2 or D3) does not cause obvious cell death in other organs, an acute shutdown of all three D-type cyclins together in bone marrow of adult mice triggers massive cell apoptosis. In this compartment D-cyclins repress the expression of a death receptor Fas and its ligand, FasL (Figure 3); ablation of the three D-cyclins results in upregulation of Fas and FasL proteins leading to hematopoietic cell apoptosis92.
Evidence for a direct role of cyclin E–CDK2 or cyclin A–CDK2 complexes in control of cell death is more limited. Cyclin E–CDK2 affects the apoptotic response through phosphorylation of the transcription factor forkhead box protein O1 (FOXO1), thus upregulating the expression of the pro-apoptotic proteins FAS and BIM (also known as Bcl2-L-11)93. On the other hand, cyclin E–CDK2 complexes were suggested to promote survival by directly phosphorylating the anti-apoptotic factor MCL194 (Figure 3).
In addition to apoptosis, cyclins and CDKs can also modulate other cell death pathways. Cyclin D1 expression suppresses autophagy in the mammary epithelium95, a function that might be mediated by the pRB–E2F pathway96. In prostate cancer cell lines, cyclin D1 was postulated to inhibit anoikis by binding to FOXO1 and FOXO3a, and blocking their ability to induce anoikis in a CDK-independent manner97 (Figure 3).
Control of cell differentiation
Terminal differentiation is usually coupled to permanent exit from the cell cycle. The levels of cyclins typically decline when cells exit the cell cycle and undergo differentiation. Moreover, induction of the expression of CDK inhibitors during cell differentiation prevents activation of cyclin-CDK complexes in terminally differentiated cells (Box 1). On the other hand, expression of cyclin-CDK complexes in proliferating cells inhibits pRB function, thereby promoting proliferation and inhibiting differentiation. Indeed pRB, in addition to its well-established cell cycle role, binds and regulates the activity of several cell-type-specific transcription factors, including MyoD, myocyte-specific enhancer factor 2 (MEF2), RUNX2 and many others, thereby linking cell cycle arrest and differentiation5.
Box 1. CDK inhibitors.
Two different families of proteins can inhibit CDK activity159. Members of the CIP/KIP (CDK interacting protein/Kinase inhibitory protein) family of inhibitors — p21 (CIP1), p27 (KIP1) and p57 (KIP2), encoded by the cyclin-dependent kinase inhibitor 1 (CDKN1) genes — bind most cyclin–CDK complexes and prevent their activation. CDKN2-encoded p16 (INK4A), p15 (INK4B), p18 (INK4C) and p19 (INK4D) proteins, which constitute the INK4 family, bind CDK4 and CDK6 and block their association with the D-type cyclins. Owing to their inhibition of CDKs and cell proliferation, CDK inhibitors are tumour suppressors, and deletion of the CDKN2A–CDKN2B locus, which encodes p16, p15 and another protein, p14 (ARF) involved in the activation of the p53 tumour suppressor, is one of the most frequent mutation in human tumours160.
CDK inhibitors are involved in many cellular processes, including cell cycle control, transcription, cell migration and metabolism; and they can affect cell quiescence, senescence, differentiation or cell death, depending on the cellular setting112, 143, 155, 161–164. In most cases, these functions are mediated by inhibition of specific cyclin–CDK complexes. However, INK4 or CIP/KIP proteins can have additional activities that are unrelated to their function as CDK inhibitors165. For example, p19 (INK4D) has been proposed to participate in DNA damage repair and senescence in a CDK-independent manner through directly interacting with chromatin166, 167. p21 (CIP1) can function as a Rho-kinase (ROCK) inhibitor, and p57 (KIP2) modulates the subcellular localization of LIM kinase (LIMK), a serine/threonine kinase involved in regulation of actin filaments168. Similarly, p27 (KIP1) has an important role in the control of cell cytoskeleton and migration169 by physically interacting with RhoA and by blocking its activation, resulting in impaired stabilization of actin fibres and increased cell motility170,171.
Cyclins and CDKs have also been shown to carry out direct, pRB-independent roles in cell differentiation. For example, cyclin D1–CDK4 complexes phosphorylate and target for degradation RUNX2, a transcription factor that drives osteoblast differentiation, thereby inhibiting bone differentiation98. CDK1 and CDK2 can likely affect cell differentiation through phosphorylation and activation of EZH2, the catalytic subunit of the histone Lys methyltransferase Polycomb repressive complex 2 (PRC2). PRC2 is an important regulator of gene expression by catalyzing the trimethylation of histone H3 Lys 27 (H3K27me3), and it influences differentiation by repressing lineage-specific genes99, 100.
Roles of cyclins and CDKs in muscle differentiation
As mentioned above, cyclin D1 and CDK4 can prevent transcription activation mediated by MyoD, and hence inhibit myoblast differentiation24–27, 101 (Figure 4a). Although this activity is partially kinase-independent, maximal repression of MyoD, as well as repression of another muscle-specific basic helix-loop-helix (bHLH) factor, myogenin, requires CDK kinase activity102. Cyclin D–CDK4 complexes also block the association of MEF2 with glutamate receptor-interacting protein 1(GRIP1), a member of the steroid receptor family of transcription co-activators, thereby inhibiting the ability of MEF2 to induce muscle gene expression102. This block requires CDK activity, although the exact molecular mechanism mediating it is unclear102. Cyclin D1 can also inhibit the differentiation of cardiomyocytes through CDK4-dependent phosphorylation of GATA4, which targets this transcription factor for degradation103 (Figure 4a).
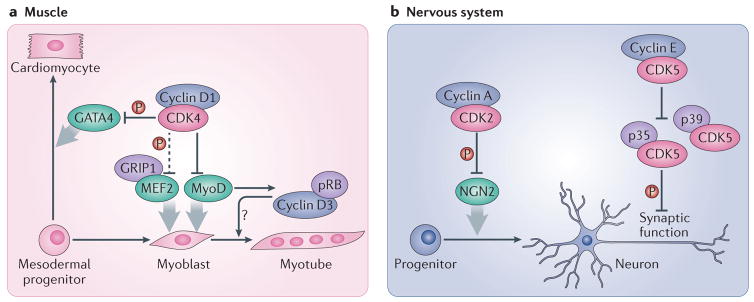
Figure 4. Roles of interphase cyclins and CDKs in cell differentiation.
a| In muscle progenitor cells, cyclin D1 interacts with the transcription factor MyoD and inhibits its activity. This effect requires CDK4 but is partially independent of its kinase function. However, the catalytic activity of CDK4 is required for the inhibition by cyclin D1–CDK4 of GRIP1–MEF2 complexes, which are also involved in muscle differentiation. Cyclin D1–CDK4 complexes also phosphorylate and inactivate the GATA4 transcription factor, and thereby repress the differentiation of cardiomyocytes. On the other hand, cyclin D3, which is stabilized in differentiated muscle cells by its interaction with pRB, seems to promote muscle cell differentiation by an unknown mechanism. b| In the nervous system, the differentiation of neuronal progenitors into neurons is blocked by cyclin A–CDK2 complexes, which phosphorylate and inactivate the NGN2 transcription factor. Cyclin E regulates synaptic plasticity by forming kinase-inactive complexes with CDK5 and sequestering it from its activators p35 and p39, thereby inhibiting the phosphorylation of synaptic CDK5 substrates. Dashed arrows indicate indirect connections or connections with multiple steps.
By contrast, specific D-cyclins are upregulated during terminal differentiation of certain cell types. In differentiating skeletal myoblasts, the expression of cyclin D3 is strongly upregulated (over 20-fold), in part by MyoD101, 104, 105. In differentiated myotubes, cyclin D3 becomes stabilized by binding to pRB106 (Figure 4a). High levels of cyclin D3 have also been observed in several other terminally differentiated cell types, including many types of epithelia, oocytes and ovarian corpora lutea, podocytes within renal glomeruli, adipocytes (see below) and cells of the exocrine pancreas107–110. On the basis of these observations, cyclin D3 has been postulated to carry out cell cycle-independent functions in a variety of terminally differentiated cells. However, these findings seem to be at odds with the observation that mice lacking cyclin D3 develop normal muscle and epithelia111. This might be attributed to functional redundancy between the three D-type cyclins, and conditional knockout approaches involving deletion of all D-cyclins in postmitotic tissues might help to clarify this point.
Cyclin D–CDKs in neuronal development
Cyclin–CDK complexes can inhibit neuronal differentiation, not only through the pRB–E2F pathway112 but also by phosphorylating and inactivating transcription factors that drive neurogenesis. For instance, neurogenin 2 (NGN2), a master regulator of neuronal differentiation, is inhibited by multisite phosphorylation by CDKs (Figure 4b)113, 114, and a non-phosphorylatable NGN2 mutant can drive neuronal differentiation in the presence of CDK activity113. Cyclin D1–CDK4 activity in neural stem cells is also thought to inhibit neurogenesis and expand the population of basal progenitors by shortening the duration of G1, although the exact mechanism remains to be clarified115. In support for a role for cyclin–CDK complexes in inhibiting neuronal differentiation, embryos from Cdk2–/–Cdk4–/– knockout mice lack the intermediate zone and cortical plate in their brains, due to premature neuronal differentiation and depletion of basal progenitor cells116.
Other data support the opposite conclusion — namely, that D-cyclins promote neurogenesis. A cell cycle-independent function for cyclins in the nervous system was first proposed based on the upregulation of cyclin D1-associated kinase activity in rat brains at the onset of neuronal differentiation117, 118. High levels of cyclin D1 persisted in the brains of adult mice, whereas the levels of cyclin D2 decreased, suggesting a specific role for cyclin D1 in neuronal differentiation. By contrast, high levels of cyclin D2 were observed in the brains of adult mice, suggesting that cyclin D2 carries out a distinct role in terminally differentiated neurons119.
Loss-of-function and gain-of-function experiments in embryonic chick spinal cord also supported a role for cyclin D1 in promoting neuronal differentiation120. In vivo knockdown of cyclin D1, but not cyclin D2, significantly reduced differentiation of motoneuron progenitors. Conversely, forced overexpression of cyclin D1 enhanced neurogenesis, whereas cyclins D2 and D3 inhibited this process. Interestingly, overexpression of a cyclin D1 mutant that is unable to activate CDKs promoted neurogenesis to an even higher extent than wild-type cyclin D1120. Collectively these findings suggest a differential role for cyclins D1 and D2 in neurogenesis, but the molecular mechanism involved remains unclear.
Cyclin E in neuronal development
Cell cycle proteins also affect the balance between progenitors and neurons by controlling the regulation of asymmetric cell divisions of neural progenitors121. In Drosophila melanogaster, cyclin E is expressed in the thoracic cell lineage, which undergoes asymmetric divisions giving rise to neuroblasts (central nervous system stem cells in D. melanogaster) and to glial progenitors. Depletion of cyclin E renders the thoracic lineage capable of generating only glial progenitors, whereas ectopic expression of cyclin E in the abdominal lineage forces it into asymmetric divisions122. Mechanistically, cyclin E interacts with and inhibits the activity of Prospero, a transcription factor required for glial cell identity123. The role of E-type cyclins in asymmetric cell divisions of mammalian neural progenitors has not been explored.
Interestingly, cyclin E is upregulated in the brains of postnatal animals as neurons undergo terminal differentiation. In terminally differentiated neurons, cyclin E forms a complex with CDK5124 (Figure 4b), which is a well-established regulator of neuronal differentiation125. Cyclin E sequesters CDK5 into kinase-inactive cyclin E–CDK5 complexes, away from the CDK5 activating partners p35 and p39. By this mechanism, cyclin E controls phosphorylation of key synaptic proteins and regulates synaptic plasticity and memory formation. Ablation of cyclin E in the mouse brains decreased synaptic function and rendered animals unable to normally retain memories124. These findings raise a possibility that abnormalities in cyclin E levels might underlie some cases of human learning disabilities124.
Immune system differentiation and function
A role for CDK2 in suppressing immune tolerance was highlighted by the observations that when cardiac allografts were transplanted into Cdk2-null recipient mice, a long-term allograft acceptance was observed, in contrast to wild-type recipients, which rejected the grafts126. Cdk2-deficient regulatory T-cells (Treg) displayed increased suppressive capacity, likely explaining the increased immune tolerance in Cdk2-null mice126, 127. Cyclin E–CDK2 was shown to phosphorylate and negatively regulate forkhead box protein P3 (FOXP3), a transcription factor required for the development and function of Treg128. Furthermore, in developing lymphocytes, cyclin A–CDK2 triggers the periodic degradation of the recombinase RAG-2, thereby restricting V(D)J recombination to G1 phase129. Mutation of CDK2 substrate residues of RAG-2 abrogated cell cycle-dependent degradation of this protein130.
Regulation of cell migration
During keratinocyte differentiation, cyclin D1 co-localizes with SEC6, a subunit of the exocyst complex involved in vesicle trafficking, as well as with β1 integrin and RalA (a GTPase that regulates the exocyst). Ectopic overexpression of cyclin D1 increases β1 integrin recycling upon differentiation, as demonstrated by a pronounced loss of membrane-bound β1 integrin, and leads to reduced keratinocyte adhesion to the extracellular matrix131. Cell migration and the actin cytoskeleton are also modulated by cyclins and CDKs at different levels132. CDK6 was demonstrated to localize to the ruffling edges of spreading fibroblasts prior to the formation of filamentous actin, and to promote migration in a αvβ3 integrin-dependent manner 133. Cyclin D1 was implicated in promoting fibroblast migration by inhibiting the thrombospondin 1 and Rho kinase (ROCK) signalling pathways134. Cyclin D1 can also enhance cell migration by negatively regulating the transcription of the E3 ubiquitin ligase factor S-phase kinase-associated protein 2 (SKP2)135. SKP2 promotes the degradation of the CDK inhibitor p27Kip1, which was implicated in cell migration through control of the RhoA pathway136 (Box 1). Cyclin D1 can also affect cell migration by binding to, and promoting the phosphorylation of filamin A, a member of the actin-binding filamin protein family. Knockdown of cyclin D1 or ectopic expression p16INK4A reduced the phosphorylation of filamin A and of many other actin binding proteins, suggesting a direct role for a cyclin D1-associated kinase in regulating the cytoskeleton137. Furthermore, it was postulated that cyclin D1, in a CDK-independent manner, mediates its effect on cell migration by transcriptionally regulating Dicer138, which is an endoribonuclease required for microRNA processing. In summary, although cyclin D1 has been shown to promote cell migration in multiple cell types, the major mechanism responsible for this function remains unclear. It also remains to be seen whether overexpression of cyclin D1, seen in many cancer types, contributes to the increased invasiveness of tumour cells.
Roles of cyclins and CDKs in metabolism
Multiple cellular functions of cyclins and CDKs converge to control the generation of cellular energy and metabolism at both the cellular and organismal levels.
Cellular energy production and metabolism
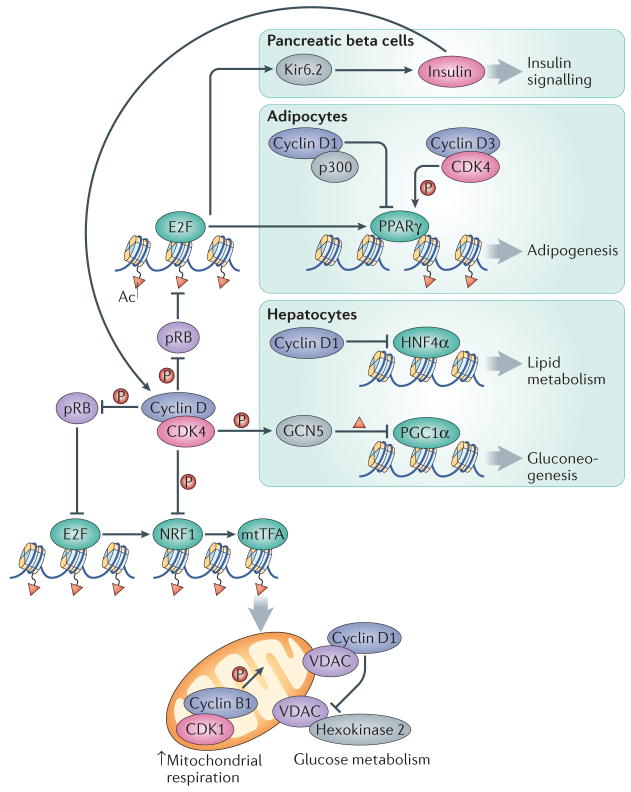
In D. melanogaster and in mammalian cells, cyclin D–CDK4 activity positively regulates mitochondrial biogenesis through the pRB–E2F-dependent control of mitochondrial transcription factors, such as NRF1 and NRF2139 (Figure 5). By contrast, cyclin D1-CDK4 can also inhibit mitochondrial function: depletion of cyclin D1 in mouse breast cancer cells in vivo promoted expression of genes involved in mitochondrial function and increased the number of mitochondria140, and cyclin D1-null mouse mammary epithelial cells had increased mitochondrial size140. The inhibitory effects of cyclin D1 on mitochondrial function were attributed to mediating the repression of NRF1 through CDK-dependent phosphorylation, leading to impaired expression of mitochondrial transcription factor A (mtTFA), a factor essential for mitochondrial respiration141 (Figure 5). Cyclin D1 was also shown to affect mitochondrial function by inhibiting the expression and activity of the glycolytic enzyme hexokinase 2140.
Figure 5. An integrated view of the roles of interphase cyclins and CDKs in the control of cellular energy and physiological metabolism.
Cyclin–CDK complexes display opposing roles in controlling mitochondrial function. Whereas their activity promotes the expression of the NRF1 factor through the pRB–E2F pathway, cyclin D1-CDK4 can also directly phosphorylate and inactivate NRF1, leading to decreased transcription of mitochondrial proteins by mitochondrial transcription factor A (mtTFA). Moreover, although the activity of mitochondria is directly stimulated by cyclin B1–CDK1, cyclin D1 binding to voltage-dependent anion channel protein (VDAC) prevents hexokinase 2 activation and the use of glucose in mitochondria. In pancreatic beta cells, cyclin D–CDK4 complexes promote insulin secretion through the E2F-mediated expression of the potassium ATP channel component Kir6.2. Insulin, in turn, induces the expression of D-type cyclins through a positive feedback loop. In adipocytes, adipogenesis is inhibited by the interaction of cyclin D1 with the histone acetyltransferase p300, which leads to inhibition of the transcription factor peroxisome proliferator-activated receptor γ (PPARγ). Conversely, adipogenesis is activated by cyclin D3–CDK4 -mediated PPARγ phosphorylation. Gluconeogenesis is inhibited in hepatocytes through the cyclin D1–CDK4-dependent phosphorylation and activation of the acetyltransferase GCN5, which acetylates and inactivates the transcription factor PPARγ coactivator-1α (PGC1α). In addition, cyclin D1 can bind and inactivate the transcription factor hepatocyte nuclear factor-4α (HNF4α), resulting in deficient lipid metabolism.
Cyclin D1 can also exert its effect on mitochondria independently of CDK4 by binding to the voltage-dependent anion channel protein (VDAC) in the outer mitochondrial membrane, thereby preventing VDAC from binding to hexokinase 2 and impairing the access of ADP to the inner mitochondrial membrane142. Further observations that cyclin D1 can directly bind additional lipogenic enzymes and mitochondrial proteins48 point to cyclin D1 as an important regulator of metabolic activities through different pathways143. In addition to cyclin D1, also cyclin E has been proposed to inhibit mitochondrial biogenesis and oxidative metabolism in a transcriptional manner, but the exact mechanism is unknown144. Lastly, the cyclin B1–CDK1 complex was shown to localize to the matrix of mitochondria, where it phosphorylates multiple mitochondrial proteins such as the respiratory chain complex 1 subunits, leading to increased mitochondrial respiration. It was postulated that this event coordinates mitochondrial respiration with G2–M phase progression145.
Regulation of metabolism at the organismal level
A physiological role for cell cycle proteins in metabolism was first documented by the observations that mice lacking CDK4 developed diabetes due to the loss of pancreatic islet-beta cells146, 147. Conversely, knock-in mice expressing a constitutively active CDK4 mutant developed pancreatic hyperplasia146. Mice lacking cyclin D2 also displayed substantially reduced beta cell mass and reduced islet size, leading to glucose intolerance148. This phenotype was further exacerbated by the additional heterozygous deletion of cyclin D1, and cyclin D1+/−D2−/− mice developed life-threatening diabetes by 3 months of age149. Ablation of cyclin D3 exacerbated diabetes in non-obese diabetic mice, but this did not affect the proliferation rate of beta cells, raising the possibility that cyclin D3 might have a cell cycle-independent role in this tissue110. Glucose challenge was proposed to promote cyclin D2–CDK4 complex formation and kinase activity, which indirectly induces the expression of Kir6.2, a component of the potassium ATP channel that positively regulates insulin secretion (Figure 5)150. Conversely, inhibition of CDK4 activity leads to lower expression of Kir6.2, decreases insulin secretion and results in glucose intolerance150. The effect of cyclin D2–CDK4 activity on Kir6.2 levels is mediated by E2F1, but it occurs independently of cell cycle progression150. An independent mechanism linking insulin signalling to cyclin D1–CDK4 was provided by the observation that insulin-mediated upregulation of cyclin D1, and subsequent activation of cyclin D1–CDK4 in hepatocytes leads to phosphorylation and activation of the histone acetyltransferase GCN5 (also known as KAT2A). GCN5 is responsible for the acetylation and inhibition of the transcription factor PPARγ coactivator 1α (PGC1α)151, which inhibits the expression of gluconeogenesis genes, thereby suppressing hepatic glucose production151 (Figure 5). Cyclin D1 was shown to inhibit lipid metabolism in the liver, in a kinase-independent manner, through its interaction with and suppression of the transcription factor hepatocyte nuclear factor 4α152.
The functions of cyclins and CDKs in modulating metabolic pathways contribute to the ability of these proteins to affect differentiation. A positive role for cyclin D3 in promoting adipocyte differentiation is supported by the observation that knockdown of cyclin D3 inhibits adipogenesis in vitro153. Consistent with these data, mice lacking cyclin D3 or CDK4 are protected from diet-induced obesity, have smaller adipocytes and reduced expression of adipogenesis genes109, 154. CDK4 — likely in complex with cyclin D3 — also participates in adipocyte differentiation by directly interacting with and activating PPARγ, the master regulator of adipogenesis109, 154 (Figure 5). Furthermore, cyclin D–CDK4 complexes may engage the pRB–E2F pathway to inhibit the pRB-mediated repression of PPARγ155. Cyclin D1, on the other hand, was shown to inhibit adipogenesis by inhibiting p300 and recruiting histone deacetylases to PPAR response element of the lipoprotein lipase promoter, and cyclin D1-null fibroblasts are prone to adipogenic differentiation37, 38.
Conclusions and perspectives
Research carried out over the past two decades indicates that interphase cyclins and CDKs display functions that extend well beyond regulation of cell cycle progression. Yet, further work is needed to reconcile often conflicting models and observations, which are perhaps confounded by the overlapping and/or compensatory roles of the different cyclin and CDK proteins in mammals. Dysregulation of cell cycle proteins is commonly found in several pathological conditions including cancer, neurodegeneration and cardiac disease, but the relevance of non-canonical functions of cell cycle proteins in controlling processes other than cell proliferation — such as the control of transcription, cell death, differentiation and metabolism — remains to be fully explored.
Some of the proposed non-canonical roles might not reflect a normal physiological protein function, but a gain-of-function event that occurs in tumour cells as a consequence of cyclin and/or CDK-overexpression. Given that several inhibitors of CDK4/6 kinase activity are currently in clinical trials156, 157, it will be important to elucidate which of these cell cycle-independent roles of cyclins and CDKs truly contribute to tumorigenesis in vivo. According to the prevailing model, the major role of CDK4 and CDK6 is to inactivate pRB. Indeed, inhibition of CDK4 and CDK6 has no effect on proliferation of tumour cells that lost pRB expression158. Given the increasing number of reported CDK4/6 substrates, it remains to be seen whether CDK4/6 inhibition in pRB-negative tumour cells affects other non-canonical functions of cyclin D-CDK4/6 kinases, and, by doing so, contributes to the therapeutic effect.
Online summary.
In addition to their well-established functions in driving cell proliferation, cell cycle proteins have several non-canonical roles.
D-type cyclins and their partner cyclin-dependent kinase 6 (CDK6) have direct, kinase-independent roles in augmenting or repressing gene expression.
In mammalian cells, cyclin D1 promotes, whereas cyclin A–CDK2 inhibits DNA double-strand break (DSB) repair through homologous recombination. In yeast, CDK activity seems to dictate the choice of DSB repair between non-homologous end joining and homologous recombination.
Cyclins are postulated to regulate apoptosis, autophagy and anoikis. Analyses of mice lacking D-cyclins support pro-survival roles for these proteins in specific tissues.
Cyclin D1, CDK6 and the CDK-inhibitor p27 (KIP1) can affect the actin cytoskeleton and cell migration through several different mechanisms.
Cell cycle proteins have important roles in development and have important functions in the nervous system and in regulating the immune response.
Cyclins and CDKs are shown or postulated to regulate metabolism through different routes, including a direct role in controlling mitochondrial function.
Acknowledgments
This work was supported by R01 CA083688 R01 CA132740 and P01 CA080111 (to P.S.) and grants from the Spanish Ministry of Economy and Competitiveness (SAF2012-38215, SAF2014-57791-REDC and BFU2014-52125-REDT) and Comunidad de Madrid (S2010/BMD-2470) (to M.M.)
Glossary
- Allografts
Transplants from genetically non-identical individuals of the same species
- Anoikis
A special form of programmed cell death induced in anchorage-dependent cells by their detachment from the surrounding matrix
- Apoptosis
Programmed cell death characterized by the activation of specific proteases, termed caspases, which induce chromatin fragmentation and the degradation of multiple cellular components
- Asymmetric cell division
A especial form of cell division in which the daughter cells have different fates, likely as a consequence of asymmetric distribution of cellular components during division
- Autophagy
The degradation and recycling of cellular components and organelles by lysosomes
- Basic Helix–Loop–Helix (bHLH) transcription factor
A protein that contains two helices separated by a loop (the HLH domain), which binds DNA in a sequence-specific manner
- Chromatin modifier
A protein complex that, through catalyzing specific histone or DNA modifications, induces changes in chromatin to make it accessible (open) or inaccessible (closed) to the transcriptional machinery
- D-loop
Displacement loop formed in homologous recombination repair. Two strands of a homologous chromosome are separated for a stretch by an invading broken strand forming a displaced single-strand structure, a D-loop
- Exocyst complex
A highly conserved, octameric protein complex that regulates vesicle delivery to and docking at the cell surface
- Gluconeogenesis
Generation of glucose from non-carbohydrate carbon substrates
- Quiescence
A postmitotic state in which cells do not divide, although they may be induced to re-enter the cell cycle in response to specific stimuli
Biographies
Per Hydbring received his Ph.D. degree from the Swedish University of Agricultural Sciences and Karolinska Institutet in Sweden, before joining the laboratory of Dr. Piotr Sicinski at Dana-Farber Cancer Institute/Harvard Medical School for his postdoctoral training. Per will move on to a position as Assistant Professor at Karolinska Institutet in 2016. His current research is focused on coding- and noncoding RNAs in cancer.
Marcos Malumbres received his Ph.D. degree from the University of Leon in Spain. After a postdoctoral training under the supervision of Angel Pellicer at the New York University Medical Center, he moved back to Spain to join Mariano Barbacid’s lab. Since 2005, he leads the Cell Division and Cancer group at the Spanish National Cancer Research Centre (CNIO), where he is interested in the molecular basis of cell cycle regulation and its implications in cancer therapy.
Piotr (Peter) Sicinski received his M.D. and Ph.D. degrees from the Warsaw Medical School in Warsaw, Poland. He was a visiting scientist at the Medical Research Council in Cambridge, UK before joining the laboratory of Dr. Robert A. Weinberg at the Whitehead Institute for his postdoctoral training. Peter joined the faculty of the Harvard Medical School in 1997, where he is now a Professor of Genetics. His laboratory, located in the Dana-Farber Cancer Institute, studies the roles of cell cycle proteins in physiology and in cancer.
Footnotes
Competing interest statement
The authors declare no competing interests.
References
- 1.Morgan DO. The cell cycle: Principles of control. New Science Press; 2007. [Google Scholar]
- 2.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 3.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33:1890–903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 5.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24:2909–15. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 8.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 9.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–31. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 10.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–28. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–31. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 12.Anders L, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–34. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal P, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–40. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luscher-Firzlaff JM, Lilischkis R, Luscher B. Regulation of the transcription factor FOXM1c by Cyclin E/CDK2. FEBS Lett. 2006;580:1716–22. doi: 10.1016/j.febslet.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Hara E, Hall M, Peters G. Cdk2-dependent phosphorylation of Id2 modulates activity of E2A-related transcription factors. EMBO J. 1997;16:332–42. doi: 10.1093/emboj/16.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–9. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yun J, et al. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J Biol Chem. 2003;278:36966–72. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 18.Chae HD, Yun J, Bang YJ, Shin DY. Cdk2-dependent phosphorylation of the NF-Y transcription factor is essential for the expression of the cell cycle-regulatory genes and cell cycle G1/S and G2/M transitions. Oncogene. 2004;23:4084–8. doi: 10.1038/sj.onc.1207482. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–97. [PMC free article] [PubMed] [Google Scholar]
- 20.Ma T, et al. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saville MK, Watson RJ. The cell-cycle regulated transcription factor B-Myb is phosphorylated by cyclin A/Cdk2 at sites that enhance its transactivation properties. Oncogene. 1998;17:2679–89. doi: 10.1038/sj.onc.1202503. [DOI] [PubMed] [Google Scholar]
- 22.Hydbring P, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci U S A. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campaner S, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–9. 1–14. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 24.Rao SS, Chu C, Kohtz DS. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix regulators. Mol Cell Biol. 1994;14:5259–67. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skapek SX, Rhee J, Spicer DB, Lassar AB. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–4. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 26.Skapek SX, Rhee J, Kim PS, Novitch BG, Lassar AB. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation. Mol Cell Biol. 1996;16:7043–53. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang JM, Wei Q, Zhao X, Paterson BM. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999;18:926–33. doi: 10.1093/emboj/18.4.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao Z, Robbins PD. Differential regulation of E2F and Sp1-mediated transcription by G1 cyclins. Oncogene. 1995;10:221–8. [PubMed] [Google Scholar]
- 29.Adnane J, Shao Z, Robbins PD. Cyclin D1 associates with the TBP-associated factor TAF(II)250 to regulate Sp1-mediated transcription. Oncogene. 1999;18:239–47. doi: 10.1038/sj.onc.1202297. [DOI] [PubMed] [Google Scholar]
- 30.Zwijsen RM, et al. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–15. doi: 10.1016/s0092-8674(00)81879-6. These manuscripts reported a CDK-independent role of cyclin D1 in regulating the transcriptional activity of the estrogen receptor, and an interaction between cyclin D1 and histone acetyltransferases. [DOI] [PubMed] [Google Scholar]
- 31.Neuman E, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–47. doi: 10.1128/mcb.17.9.5338. These manuscripts reported a CDK-independent role of cyclin D1 in regulating the transcriptional activity of the estrogen receptor, and an interaction between cyclin D1 and histone acetyltransferases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon C, Suthiphongchai T, DiRenzo J, Ewen ME. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc Natl Acad Sci U S A. 1999;96:5382–7. doi: 10.1073/pnas.96.10.5382. These manuscripts reported a CDK-independent role of cyclin D1 in regulating the transcriptional activity of the estrogen receptor, and an interaction between cyclin D1 and histone acetyltransferases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knudsen KE, Cavenee WK, Arden KC. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59:2297–301. [PubMed] [Google Scholar]
- 34.Petre CE, Wetherill YB, Danielsen M, Knudsen KE. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J Biol Chem. 2002;277:2207–15. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 35.Petre-Draviam CE, et al. Specificity of cyclin D1 for androgen receptor regulation. Cancer Res. 2003;63:4903–13. [PubMed] [Google Scholar]
- 36.Reutens AT, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 37.Fu M, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280:29728–42. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 38.Fu M, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280:16934–41. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 39.Ratineau C, Petry MW, Mutoh H, Leiter AB. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J Biol Chem. 2002;277:8847–53. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 40.Iwatani K, Fujimoto T, Ito T. Cyclin D1 blocks the anti-proliferative function of RUNX3 by interfering with RUNX3-p300 interaction. Biochem Biophys Res Commun. 2010;400:426–31. doi: 10.1016/j.bbrc.2010.08.094. [DOI] [PubMed] [Google Scholar]
- 41.Lin HM, Zhao L, Cheng SY. Cyclin D1 Is a Ligand-independent Co-repressor for Thyroid Hormone Receptors. J Biol Chem. 2002;277:28733–41. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 42.Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16:6457–67. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jian Y, et al. Cyclin D3 interacts with vitamin D receptor and regulates its transcription activity. Biochem Biophys Res Commun. 2005;335:739–48. doi: 10.1016/j.bbrc.2005.07.141. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, et al. Cyclin D3 interacts with human activating transcription factor 5 and potentiates its transcription activity. Biochem Biophys Res Commun. 2004;321:954–60. doi: 10.1016/j.bbrc.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 46.Peterson LF, et al. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005;25:10205–19. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamb J, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–34. doi: 10.1016/s0092-8674(03)00570-1. This manuscript revealed the relevance of C/EBPβ in the regulation of transcription by cyclin D1 using genome-wide analysis of tumor cells. [DOI] [PubMed] [Google Scholar]
- 48.Bienvenu F, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–8. doi: 10.1038/nature08684. A genetic–proteomic in vivo analysis revealed the transcriptional function of cyclin D1 during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006;133:913–23. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- 50.Sicinski P, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–30. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 51.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–72. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 52.Casimiro MC, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest. 2012;122:833–43. doi: 10.1172/JCI60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujimoto T, Anderson K, Jacobsen SE, Nishikawa SI, Nerlov C. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26:2361–70. doi: 10.1038/sj.emboj.7601675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kollmann K, et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–81. doi: 10.1016/j.ccr.2013.07.012. Demonstration of a transcriptional, kinase-independent role of CDK6 in controlling cell proliferation and angiogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasui M, et al. Antisense to cyclin D1 inhibits vascular endothelial growth factor-stimulated growth of vascular endothelial cells: implication of tumor vascularization. Clin Cancer Res. 2006;12:4720–9. doi: 10.1158/1078-0432.CCR-05-1213. [DOI] [PubMed] [Google Scholar]
- 56.Scheicher R, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125:90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–7. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–75. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saponaro M, et al. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010;6:e1000858. doi: 10.1371/journal.pgen.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granata M, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang G, Tong X, Weng S, Zhou H. Multiple phosphorylation of Rad9 by CDK is required for DNA damage checkpoint activation. Cell Cycle. 2012;11:3792–800. doi: 10.4161/cc.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abreu CM, et al. Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1. PLoS Genet. 2013;9:e1003310. doi: 10.1371/journal.pgen.1003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–9. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferretti LP, Lafranchi L, Sartori AA. Controlling DNA-end resection: a new task for CDKs. Front Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. This manuscript reported that CDK-dependent phosphorylation controls the formation of BRCA2–Rad51 complexes. [DOI] [PubMed] [Google Scholar]
- 68.Ayoub N, et al. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr Biol. 2009;19:1075–85. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–92. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 70.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 71.Aggarwal P, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–22. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, et al. Alternative cyclin D1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70:8802–11. doi: 10.1158/0008-5472.CAN-10-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jirawatnotai S, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–4. doi: 10.1038/nature10155. Proteome-wide analysis of cyclin D1-interacting proteins in human cancer cells revealed a role for cyclin D1 in recruiting Rad51 to DNA damage sites and in promoting homologous recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Coco Martin JM, Balkenende A, Verschoor T, Lallemand F, Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59:1134–40. [PubMed] [Google Scholar]
- 75.Shintani S, Mihara M, Ueyama Y, Matsumura T, Wong DT. Cyclin D1 overexpression associates with radiosensitivity in oral squamous cell carcinoma. Int J Cancer. 2001;96:159–65. doi: 10.1002/ijc.1014. [DOI] [PubMed] [Google Scholar]
- 76.Indovina P, Pentimalli F, Casini N, Vocca I, Giordano A. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget. 2015;6:17873–90. doi: 10.18632/oncotarget.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kranenburg O, van der Eb AJ, Zantema A. Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J. 1996;15:46–54. [PMC free article] [PubMed] [Google Scholar]
- 78.Duquesne F, Florent M, Roue G, Troussard X, Sola B. Ectopic expression of cyclin D1 impairs the proliferation and enhances the apoptosis of a murine lymphoid cell line. Cell Death Differ. 2001;8:51–62. doi: 10.1038/sj.cdd.4400768. [DOI] [PubMed] [Google Scholar]
- 79.Roue G, Pichereau V, Lincet H, Colomer D, Sola B. Cyclin D1 mediates resistance to apoptosis through upregulation of molecular chaperones and consequent redistribution of cell death regulators. Oncogene. 2008;27:4909–20. doi: 10.1038/onc.2008.126. [DOI] [PubMed] [Google Scholar]
- 80.Han EK, et al. Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27kip1, inhibits growth, and enhances apoptosis. Cell Growth Differ. 1996;7:699–710. [PubMed] [Google Scholar]
- 81.Niu MY, Menard M, Reed JC, Krajewski S, Pratt MA. Ectopic expression of cyclin D1 amplifies a retinoic acid-induced mitochondrial death pathway in breast cancer cells. Oncogene. 2001;20:3506–18. doi: 10.1038/sj.onc.1204453. [DOI] [PubMed] [Google Scholar]
- 82.Hochhauser D, et al. Effect of cyclin D1 overexpression on drug sensitivity in a human fibrosarcoma cell line. J Natl Cancer Inst. 1996;88:1269–75. doi: 10.1093/jnci/88.18.1269. [DOI] [PubMed] [Google Scholar]
- 83.Kornmann M, et al. Inhibition of cyclin D1 expression in human pancreatic cancer cells is associated with increased chemosensitivity and decreased expression of multiple chemoresistance genes. Cancer Res. 1999;59:3505–11. [PubMed] [Google Scholar]
- 84.Boonen GJ, et al. Cyclin D3 regulates proliferation and apoptosis of leukemic T cell lines. J Biol Chem. 1999;274:34676–82. doi: 10.1074/jbc.274.49.34676. [DOI] [PubMed] [Google Scholar]
- 85.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–51. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beltran E, et al. A cyclin-D1 interaction with BAX underlies its oncogenic role and potential as a therapeutic target in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2011;108:12461–6. doi: 10.1073/pnas.1018941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suzuki A, et al. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–53. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 88.Mendelsohn AR, Hamer JD, Wang ZB, Brent R. Cyclin D3 activates Caspase 2, connecting cell proliferation with cell death. Proc Natl Acad Sci U S A. 2002;99:6871–6. doi: 10.1073/pnas.072290599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Osuga H, et al. Cyclin-dependent kinases as a therapeutic target for stroke. Proc Natl Acad Sci U S A. 2000;97:10254–9. doi: 10.1073/pnas.170144197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.osuIno H, Chiba T. Cyclin-dependent kinase 4 and cyclin D1 are required for excitotoxin-induced neuronal cell death in vivo. J Neurosci. 2001;21:6086–94. doi: 10.1523/JNEUROSCI.21-16-06086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma C, Papermaster D, Cepko CL. A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proc Natl Acad Sci U S A. 1998;95:9938–43. doi: 10.1073/pnas.95.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choi YJ, et al. D-cyclins repress apoptosis in hematopoietic cells by controlling death receptor Fas and its ligand FasL. Dev Cell. 2014;30:255–67. doi: 10.1016/j.devcel.2014.06.015. This study demonstrated the overlapping roles of D-type cyclins in controlling FAS-mediated apoptosis in hematopoietic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–7. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 94.Choudhary GS, et al. Cyclin E/Cdk2-dependent phosphorylation of Mcl-1 determines its stability and cellular sensitivity to BH3 mimetics. Oncotarget. 2015;6:16912–25. doi: 10.18632/oncotarget.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brown NE, et al. Cyclin D1 activity regulates autophagy and senescence in the mammary epithelium. Cancer Res. 2012;72:6477–89. doi: 10.1158/0008-5472.CAN-11-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang H, et al. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–93. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gan L, et al. Cyclin D1 promotes anchorage-independent cell survival by inhibiting FOXO-mediated anoikis. Cell Death Differ. 2009;16:1408–17. doi: 10.1038/cdd.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen R, et al. Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J Biol Chem. 2006;281:16347–53. doi: 10.1074/jbc.M603439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–14. doi: 10.1038/ncb2116. This study reported a link between CDKs and the epigenetic control of gene expression through phosphorylation of polycomb repressor complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaneko S, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. This study reported a link between CDKs and the epigenetic control of gene expression through phosphorylation of polycomb repressor complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rao SS, Kohtz DS. Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor beta. A role for cyclin D1 in control of myoblast differentiation. J Biol Chem. 1995;270:4093–100. doi: 10.1074/jbc.270.8.4093. [DOI] [PubMed] [Google Scholar]
- 102.Lazaro JB, Bailey PJ, Lassar AB. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 2002;16:1792–805. doi: 10.1101/gad.U-9988R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakajima K, et al. Coordinated regulation of differentiation and proliferation of embryonic cardiomyocytes by a jumonji (Jarid2)-cyclin D1 pathway. Development. 2011;138:1771–82. doi: 10.1242/dev.059295. [DOI] [PubMed] [Google Scholar]
- 104.Kiess M, Gill RM, Hamel PA. Expression of the positive regulator of cell cycle progression, cyclin D3, is induced during differentiation of myoblasts into quiescent myotubes. Oncogene. 1995;10:159–66. [PubMed] [Google Scholar]
- 105.Cenciarelli C, et al. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol Cell Biol. 1999;19:5203–17. doi: 10.1128/mcb.19.7.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Santa F, et al. pRb-dependent cyclin D3 protein stabilization is required for myogenic differentiation. Mol Cell Biol. 2007;27:7248–65. doi: 10.1128/MCB.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17:1027–37. doi: 10.1038/sj.onc.1202016. [DOI] [PubMed] [Google Scholar]
- 108.Doglioni C, et al. Cyclin D3 expression in normal, reactive and neoplastic tissues. J Pathol. 1998;185:159–66. doi: 10.1002/(SICI)1096-9896(199806)185:2<159::AID-PATH73>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 109.Sarruf DA, et al. Cyclin D3 promotes adipogenesis through activation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2005;25:9985–95. doi: 10.1128/MCB.25.22.9985-9995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saavedra-Avila NA, et al. Cyclin D3 promotes pancreatic beta-cell fitness and viability in a cell cycle-independent manner and is targeted in autoimmune diabetes. Proc Natl Acad Sci U S A. 2014;111:E3405–14. doi: 10.1073/pnas.1323236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sicinska E, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–61. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 112.Ajioka I. Coordination of proliferation and neuronal differentiation by the retinoblastoma protein family. Dev Growth Differ. 2014;56:324–34. doi: 10.1111/dgd.12127. [DOI] [PubMed] [Google Scholar]
- 113.Ali F, et al. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–77. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hindley C, et al. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development. 2012;139:1718–23. doi: 10.1242/dev.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–31. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 116.Lim S, Kaldis P. Loss of Cdk2 and Cdk4 induces a switch from proliferation to differentiation in neural stem cells. Stem Cells. 2012;30:1509–20. doi: 10.1002/stem.1114. [DOI] [PubMed] [Google Scholar]
- 117.Tamaru T, Trigun SK, Okada M, Nakagawa H. Identification of cells expressing a D type G1 cyclin in matured brain: implication for its role in neuronal function. Neurosci Lett. 1993;153:169–72. doi: 10.1016/0304-3940(93)90314-b. [DOI] [PubMed] [Google Scholar]
- 118.Tamaru T, Okada M, Nakagawa H. Differential expression of D type cyclins during neuronal maturation. Neurosci Lett. 1994;168:229–32. doi: 10.1016/0304-3940(94)90457-x. [DOI] [PubMed] [Google Scholar]
- 119.Ross ME, Carter ML, Lee JH. MN20, a D2 cyclin, is transiently expressed in selected neural populations during embryogenesis. J Neurosci. 1996;16:210–9. doi: 10.1523/JNEUROSCI.16-01-00210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lukaszewicz AI, Anderson DJ. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc Natl Acad Sci U S A. 2011;108:11632–7. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Malumbres M. Physiological relevance of cell cycle kinases. Physiol Rev. 2011;91:973–1007. doi: 10.1152/physrev.00025.2010. [DOI] [PubMed] [Google Scholar]
- 122.Berger C, Pallavi SK, Prasad M, Shashidhara LS, Technau GM. A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat Cell Biol. 2005;7:56–62. doi: 10.1038/ncb1203. [DOI] [PubMed] [Google Scholar]
- 123.Berger C, et al. Cell cycle independent role of Cyclin E during neural cell fate specification in Drosophila is mediated by its regulation of Prospero function. Dev Biol. 2010;337:415–24. doi: 10.1016/j.ydbio.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 124.Odajima J, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell. 2011;21:655–68. doi: 10.1016/j.devcel.2011.08.009. This manuscript described an unexpected function of cyclin E in inhibiting CDK5 activity in terminally differentiated neurons, and in regulating synaptic plasticity, learning and memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Su SC, Tsai LH. Cyclin-dependent kinases in brain development and disease. Annu Rev Cell Dev Biol. 2011;27:465–91. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- 126.Chunder N, Wang L, Chen C, Hancock WW, Wells AD. Cyclin-dependent kinase 2 controls peripheral immune tolerance. J Immunol. 2012;189:5659–66. doi: 10.4049/jimmunol.1202313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wells AD, Morawski PA. New roles for cyclin-dependent kinases in T cell biology: linking cell division and differentiation. Nat Rev Immunol. 2014;14:261–70. doi: 10.1038/nri3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morawski PA, Mehra P, Chen C, Bhatti T, Wells AD. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J Biol Chem. 2013;288:24494–502. doi: 10.1074/jbc.M113.467704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–81. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- 130.Jiang H, Ross AE, Desiderio S. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J Biol Chem. 2004;279:8478–86. doi: 10.1074/jbc.M311219200. [DOI] [PubMed] [Google Scholar]
- 131.Fernandez-Hernandez R, et al. Cyclin D1 localizes in the cytoplasm of keratinocytes during skin differentiation and regulates cell-matrix adhesion. Cell Cycle. 2013;12:2510–7. doi: 10.4161/cc.25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bendris N, Lemmers B, Blanchard JM. Cell cycle, cytoskeleton dynamics and beyond: the many functions of cyclins and CDK inhibitors. Cell Cycle. 2015;14:1786–98. doi: 10.1080/15384101.2014.998085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fahraeus R, Lane DP. The p16(INK4a) tumour suppressor protein inhibits alphavbeta3 integrin-mediated cell spreading on vitronectin by blocking PKC-dependent localization of alphavbeta3 to focal contacts. EMBO J. 1999;18:2106–18. doi: 10.1093/emboj/18.8.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Z, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26:4240–56. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z, et al. Cyclin D1 induction of cellular migration requires p27(KIP1) Cancer Res. 2006;66:9986–94. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 136.Li Z, et al. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem. 2008;283:7007–15. doi: 10.1074/jbc.M706992200. [DOI] [PubMed] [Google Scholar]
- 137.Zhong Z, et al. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res. 2010;70:2105–14. doi: 10.1158/0008-5472.CAN-08-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu Z, et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812. doi: 10.1038/ncomms3812. This manuscript described control of Dicer by cyclin D1 and its multiple implications for understanding the interaction between cell cycle proteins and non-coding RNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lopez-Mejia IC, Fajas L. Cell cycle regulation of mitochondrial function. Curr Opin Cell Biol. 2015;33:19–25. doi: 10.1016/j.ceb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 140.Sakamaki T, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–69. doi: 10.1128/MCB.02074-05. These reports established a molecular link between cyclin D1 and control of mitochondrial function through the inhibition of nuclear respiratory factor 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang C, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567–72. doi: 10.1073/pnas.0603363103. These reports established a molecular link between cyclin D1 and control of mitochondrial function through the inhibition of nuclear respiratory factor 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tchakarska G, Roussel M, Troussard X, Sola B. Cyclin D1 inhibits mitochondrial activity in B cells. Cancer Res. 2011;71:1690–9. doi: 10.1158/0008-5472.CAN-10-2564. [DOI] [PubMed] [Google Scholar]
- 143.Kaplon J, van Dam L, Peeper D. Two-way communication between the metabolic and cell cycle machineries: the molecular basis. Cell Cycle. 2015;14:2022–32. doi: 10.1080/15384101.2015.1044172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu Y, Swartz KL, Siu KT, Bhattacharyya M, Minella AC. Fbw7-dependent cyclin E regulation ensures terminal maturation of bone marrow erythroid cells by restraining oxidative metabolism. Oncogene. 2014;33:3161–71. doi: 10.1038/onc.2013.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Z, et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell. 2014;29:217–32. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rane SG, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 147.Tsutsui T, et al. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol Cell Biol. 1999;19:7011–9. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114:963–8. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kushner JA, et al. Cyclins D2 and D1 are essential for postnatal pancreatic beta-cell growth. Mol Cell Biol. 2005;25:3752–62. doi: 10.1128/MCB.25.9.3752-3762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Annicotte JS, et al. The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat Cell Biol. 2009;11:1017–23. doi: 10.1038/ncb1915. This study provided the first molecular link between control of the cell cycle and insulin secretion in pancreatic beta cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee Y, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510:547–51. doi: 10.1038/nature13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hanse EA, et al. Cyclin D1 inhibits hepatic lipogenesis via repression of carbohydrate response element binding protein and hepatocyte nuclear factor 4alpha. Cell Cycle. 2012;11:2681–90. doi: 10.4161/cc.21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Phelps DE, Xiong Y. Regulation of cyclin-dependent kinase 4 during adipogenesis involves switching of cyclin D subunits and concurrent binding of p18INK4c and p27Kip1. Cell Growth Differ. 1998;9:595–610. [PubMed] [Google Scholar]
- 154.Abella A, et al. Cdk4 promotes adipogenesis through PPARgamma activation. Cell Metab. 2005;2:239–49. doi: 10.1016/j.cmet.2005.09.003. Unexpected results showing that adipogenesis is induced by a direct interaction between cyclin D3–CDK4 and PPARγ, a master regulator of adipogenesis. [DOI] [PubMed] [Google Scholar]
- 155.Aguilar V, Fajas L. Cycling through metabolism. EMBO Mol Med. 2010;2:338–48. doi: 10.1002/emmm.201000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Turner NC, Huang Bartlett C, Cristofanilli M. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2015;373:1672–3. doi: 10.1056/NEJMc1510345. [DOI] [PubMed] [Google Scholar]
- 157.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–46. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Finn RS, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 160.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–77. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 161.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–69. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 162.Caldon CE, Sutherland RL, Musgrove E. Cell cycle proteins in epithelial cell differentiation: implications for breast cancer. Cell Cycle. 2010;9:1918–28. doi: 10.4161/cc.9.10.11474. [DOI] [PubMed] [Google Scholar]
- 163.Myatt SS, Lam EW. Promiscuous and lineage-specific roles of cell cycle regulators in haematopoiesis. Cell Div. 2007;2:6. doi: 10.1186/1747-1028-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Beukelaers P, et al. Cycling or not cycling: cell cycle regulatory molecules and adult neurogenesis. Cell Mol Life Sci. 2012;69:1493–503. doi: 10.1007/s00018-011-0880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 166.Ceruti JM, Scassa ME, Flo JM, Varone CL, Canepa ET. Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene. 2005;24:4065–80. doi: 10.1038/sj.onc.1208570. [DOI] [PubMed] [Google Scholar]
- 167.Ogara MF, et al. Chromatin relaxation-mediated induction of p19INK4d increases the ability of cells to repair damaged DNA. PLoS One. 2013;8:e61143. doi: 10.1371/journal.pone.0061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18:851–5. doi: 10.1101/gad.1205304. [DOI] [PubMed] [Google Scholar]
- 169.Nagahara H, et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4:1449–52. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 170.McAllister SS, Becker-Hapak M, Pintucci G, Pagano M, Dowdy SF. Novel p27(kip1) C-terminal scatter domain mediates Rac-dependent cell migration independent of cell cycle arrest functions. Mol Cell Biol. 2003;23:216–28. doi: 10.1128/MCB.23.1.216-228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Besson A, Gurian-West M, Schmidt A, Hall A, Roberts JM. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004;18:862–76. doi: 10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu Q, et al. Cyclin D1 and C/EBPbeta LAP1 operate in a common pathway to promote mammary epithelial cell differentiation. Mol Cell Biol. 2014;34:3168–79. doi: 10.1128/MCB.00039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bienvenu F, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J Biol Chem. 2001;276:16840–7. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]