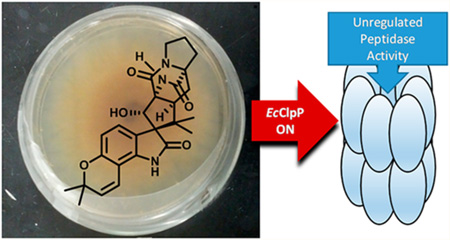
Abstract
Caseinolytic protease P (ClpP) maintains essential roles in bacterial homeostasis. As such, both the inhibition and activation of this enzyme result in bactericidal activity, making ClpP a promising target for antibacterial drug development. Herein, we report the results of a fluorescence-based screen of ~450 structurally diverse fungal and bacterial secondary metabolites. Sclerotiamide (1), a paraherquamide-related indolinone, was identified as the first non-peptide-based natural product activator of ClpP. Structure-activity relationships arising from the initial screen, preliminary biochemical evaluation of 1, and rationale for the exploitation of this chemotype to develop novel ClpP activators are presented.
Graphical abstract
Despite the effectiveness of modern day antimicrobials, the ever-increasing number of multi-drug-resistant pathogens urgently requires the development of alternative treatment strategies.1–3 Of particular importance is the development of therapeutics that operate through clinically unexploited mechanisms of action.2,4 Such therapies present new challenges to bacteria that require the evolution of new resistance regimes and thus lengthen the potential duration of therapeutic efficacy. One approach that deviates definitively from the status quo of traditional antibiotics is to develop compounds capable of imparting bactericidal activity via an activation mechanism, rather than an inhibitory one. Traditional antibiotics shut down or inhibit essential cellular processes (cell wall construction, protein synthesis, replication, etc.) and thus, generally rely upon a normal bacterial growth phase and/or an active cellular target. This dependency of antibiotics on microbial cooperativity often renders dormant persister cells and biofilms unresponsive to inhibitory therapeutics.5 Differentiated therapies that operate through the activation of cellular processes would provide a clinically unexploited approach, potentially independent of growth mode or target activity, and would therefore fill a prevalent void in the current antimicrobial arsenal.
One target, which produces bactericidal activity upon activation, is caseinolytic protease P (ClpP). ClpP is a cylindrical serine protease composed of two stacked heptameric rings, which form a tetradecameric complex that encompasses a large proteolytic core.6–12 The primary physiological role of the ClpP system is to maintain homeostasis through the controlled degradation of defective and obsolete intracellular polypeptides.13–16 Under normal physiological conditions, ClpP proteolytic activity is highly regulated through interactions with ATP-dependent AAA+ co-chaperones17–19 (e.g., ClpA and ClpX), which are responsible for the denaturation, delivery, and translocation of substrates into the proteolytic core of ClpP.12,20–24 In the absence of ATP or co-chaperones, ClpP activity is limited to the degradation of small peptides (<6 amino acids).25,26 Natural product or small-molecule chemoactivation of ClpP, however, triggers unregulated proteolysis of large polypeptides and unstructured proteins.27–29 This indiscriminant degradation results in bacterial self-digestion and eventual cellular death, including microbes in dormant or biofilm phases.30,31 Thus, activation of ClpP is an attractive therapeutic strategy worthy of detailed interrogation and medicinal chemistry efforts.
To date, the acyldepsipeptides (ADEPs, Figure 1) represent the only natural product class identified to activate ClpP.32,33 While elegant structure-activity relationship (SAR) studies and biochemical analyses have structurally and mechanistically characterized ADEP activation of ClpP and provided extremely potent derivatives,25,31,34–37 the utility of this natural product family has been limited. Much of this limitation arises from lack of activity against Gram-negative pathogens and the susceptibility of ADEPs to active efflux.27 Structural alteration of ADEPs to address these detriments has been relatively unsuccessful thus far, in part because the N-acylated phenylalanine pharmacophore38 is the recognized motif for efflux pumps.39 Efforts to diversify the arsenal of ClpP-activating molecules through screening or structure-based approaches have been extremely limited. In fact, other than the ADEP natural products and related congeners, only four chemically distinct small-molecule activators have been discovered, all of which were identified within a single screen of a commercially available library.40,41 In order to allow for the continued exploration of ClpP activation as a differentiated antibacterial strategy, chemically distinct activators are needed. Identification of structurally divergent activating molecules would expand the chemical space upon which SAR studies can be executed and potentially provide scaffolds amenable to development as broad-spectrum antibiotics.
Figure 1.
General structure of peptide-based ADEP antibiotics.
To this end, we screened a focused but structurally diverse subset of fungal and bacterial secondary metabolites, housed within the Institute for Natural Products Applications and Research Technologies (INPART) at the University of Oklahoma. The complete library has been established by the Cichewicz Laboratory over nearly a decade and comprises ~450 pure metabolites and >20 000 extracts isolated from fungal and bacterial organisms collected within the United States.42 This continually growing collection includes a rich source of structural diversity (depsipeptides, alkaloids, carbocycles, terpenes, polyketides, nonribosomal peptides, etc.) and has provided leads in a number of targeted phenotypic screens and functional assays.43–46 Thus, we believed this library represented a promising untapped source to screen for novel natural product ClpP activators.
For the initial screen, a fluorescence-based protease assay that utilizes a commercially available internally quenched fluorogenic ClpP substrate, FITC-β-casein, was employed.41 We commenced our screening campaign with only the pure metabolite portion of the available natural product library, and thus ~450 natural products were screened at a single concentration (50 µM) for the activation of recombinant Escherichia coli ClpP (EcClpP). Enopeptin A33,47 (25 µM) and ACP541 (50 µM), a natural ADEP analogue and a small molecule, respectively, were implemented as positive controls. In the presence of an activating molecule, ClpP-mediated cleavage of FITC-β-casein relieves internal quenching of the substrate and yields a fluorescent output of protease activity that can be quantified with a fluorimeter (excitation/emission: 485/538 nm). In the absence of an activating molecule, ClpP is unable to proteolytically cleave FITC-β-casein and fluorescence remains quenched.
As shown in Figure 2A, a threshold of 22 500 raw relative fluorescent units (RFUs) was set, and any compound exhibiting an end-point fluorescence greater than this threshold was analyzed further. In total, 11 hits were identified, two of which were the positive controls, enopeptin A (A, Figure 2A)33,47 and ACP5 (B, Figure 2A).41 The remaining nine hits were resubjected to the assay conditions to confirm the observed activity. All nine demonstrated reproducibility, which provided a respectable initial hit rate of 3.6%. However, when the raw screening data were replotted using the B-score algorithm,48,49 only a single true hit provided a score above three standard deviations from the mean of the general sample population (Figure S2). Thus, to eliminate false positives and confirm the B-score analysis, the original hits were evaluated for intrinsic fluorescence, which disqualified eight of the nine hits (Figure 2B). To measure intrinsic fluorescence, 538 nm emission for each compound (50 µM) in assay buffer was measured following 485 nm excitation. To ensure the only remaining hit was not inducing substrate degradation through inherent proteolytic activity, compound 1 (50 µM) was incubated with FITC-β-casein (1.92 µM) in assay buffer. Fluorescence readings were taken, and no significant time-dependent inherent proteolytic activity was observed. As such, sclerotiamide (1, Figure 2B),50 a paraherquamide-related indolinone natural product, was the only hit to pass both validation filters and B-score analysis and was evaluated further.
Figure 2.
(A) Raw FITC-β-casein degradation screening results. Threshold for analysis is indicated by the red line (ΔRFU ≥ 22 500). Inset: Enlarged area of threshold exceeding “hits”. (B) Fluorescence data, validation of initial “hits”, and structure of sclerotiamide (1). ΔRFU = change in relative fluorescence units, IF = intrinsic compound fluorescence, PA = inherent compound proteolytic activity, ND = not determined.
Sclerotiamide was subjected to dose-dependent peptide (Abz-DFAPKMALVPYNO2)21,38 and protein (FITC-β-casein) degradation analysis. As shown in Figure 3 and Table 1, the potency of 1 in the decapeptide degradation assay is similar to that of ACP5 and exhibits an apparent activation constant (Kapp) of 39.6 ± 4.9 µM. In the FITC-β-casein degradation assay, however, ACP5 maintains the same level of potency, whereas 1 exhibits a Kapp of 87.5 ± 5.0 µM, a ~2.2-fold decrease in activity. It is worth noting that the consistent potency of ACP5 in these two assays differs from the trend observed for 1 and ADEP1, which also shows an increase (~10-fold) in Kapp from decapeptide to FITC-β-casein degradation (Table 1). This unique substrate-independent activity exhibited by ACP5 may represent a useful observation in regard to a hypothesis set forth by Houry and colleagues that ACP analogues may interact with more than one binding pocket on EcClpP.41 Additionally, this may suggest that ACP5 and related analogues induce an alternative activation state of EcClpP, which is less sensitive to the size and/or shape of substrates. In agreement with both ADEP1 and ACP5, Hill plot analysis of 1 demonstrates modest positive cooperativity for ClpP binding (Hill coefficient > 1.0). Although the potency is modest, especially in comparison to ADEP1, sclerotiamide is the first non-peptide-based natural product ClpP activator discovered and thus provides unique chemical space to evaluate for SAR studies. As a member of the paraherquamide family, 1 possesses an interesting three-dimensional bicyclo[2.2.2]-diazaoctane motif that has garnered significant attention from the synthetic community51,52 and has recently been reported in closely related compounds that exhibit a range of biological activities.53,54
Figure 3.
Dose-dependent EcClpP activation by ACP5 and 1. (A) Substrate = Abz-DFAPKMALVPYNO2. (B) Substrate = FITC-β-casein.
Table 1.
Dose–Response Comparison of ADEP1, ACP5, and 1 for the Decapeptide (DFAPKMALVPYNO2) and Unstructured Protein (FITC-β-casein) Substrates
| EC50 (µM) | |||
|---|---|---|---|
| compound | DFAP | FITC-casein | Hill slope |
| ADEP1 | 0.1 ± 0.02 | 1.2 ± 0.1 | 1.6/1.5 |
| ACP5 | 25.4 ± 3.6 | 22.2 ± 2.3 | 1.4/1.4 |
| 1 | 39.6 ± 4.9 | 87.5 ± 5.0 | 2.6/1.9 |
Further validation of EcClpP activation by 1 was confirmed by SDS-PAGE analysis (Figure 4), which demonstrates time-dependent activation of EcClpP, resulting in subsequent degradation of unlabeled β-casein. The inability of 1 to completely degrade the substrate has been noted in other ClpP-activating scaffolds with similar potency.41 Interestingly, when 1 was evaluated for its propensity to activate Bacillus subtilis ClpP (BsClpP), the homologue typically utilized to evaluate the activity of ADEP analogues, no significant activation was observed (Figure 4). This lack of BsClpP activation was noted in both the SDS-PAGE analysis and fluorescence-based degradation assays. Surprisingly, similar homologue selective activation was noted with ACP5, albeit at a lower concentration (25 µM) (Figure S3). To the best of our knowledge, this observation provides the first report of ClpP activators that demonstrate noticeable profiles of selectivity between bacterial homologues, and we are currently probing the utility of this observation.
Figure 4.
SDS-PAGE analysis of ClpP homologue-dependent degradation of unlabeled β-casein by 1 (50 µM) in comparison to ADEP1 (1 µM).
Upon reinvestigation of the screened compound set, we noted that six additional members of this family were included within the screen (2–7, Figure 5A). Interestingly, however, all six of these analogues fail to elicit EcClpP (Figure 5B) or BsClpP activation to any significant extent. Most notable from this group is notoamide B (2),55 which lacks only the C-10 α-oriented secondary hydroxy group. This lack of activity by 2 identifies the C-10 α-hydroxy motif as a necessary fixture in this natural product family for ClpP activation. In addition, the three-dimensional character imparted by the spiroindolinone moiety seems to be important, as all other congeners exhibit a fused architecture and thus an altered geometry that fails to activate EcClpP.
Figure 5.
(A) Structures of paraherquamide-related compounds present in the initial FITC-β-casein degradation screen. (B) Relative EcClpP activation of 1 and related compounds. Substrate = Abz-DFAPKMALVPYNO2.
Indicative of its relatively modest Kapp, concentrations up to 100 µM of 1 failed to inhibit the growth of efflux pump deficient Escherichia coli M5418 (ΔacrB, ΔacrD, ΔacrEF::spc ΔemrB ΔemrY ΔentS::cam ΔmacB ΔmdtC ΔmdtF)56 and Pseudomonas aeruginosa PAO1116 ((ΔmexAB-oprM), (ΔmexCD-oprJ), (ΔmexEF-oprN), (ΔmexJK), (ΔmexXY), and (ΔtriABC)).57 These results suggest that although 1 is capable of activating recombinant EcClpP, improvement of potency is required for cellular target engagement and efficacy. Molecular dissection to identify the pharmacophore for this class of ClpP-activating natural products and X-ray cocrystallization studies are currently ongoing in our laboratory. Results from these studies will be communicated in due course and will contribute to structure-guided approaches aimed at improving upon the ClpP activation potency of this chemotype. Furthermore, the process outlined here will be expanded upon to enable the screening of natural product extracts, allowing for the interrogation of much larger chemical diversity.
EXPERIMENTAL SECTION
General Experimental Procedures
Positive controls were purchased from commercial sources and include enopeptin A (Santa Cruz Biotechnology, #sc-397311A), ADEP1 (Cayman Chemical, #15305), and ACP5 (Chembridge #5107473). ClpP substrates were purchased from commercial vendors and include FITC-β-casein (Sigma-Aldrich, C3777, 20–50 µg FITC per mg solid) and Abz-DFAPKMALVPYNO2 (Biomatik). Fluorescence readings were recorded on an i-TECAN Infinite M200 plate reader. Gel images were acquired on a Bio-Rad ChemiDoc MP CCD imaging system. All data analyses were performed with GraphPad Prism.
Protein Purification
Two different E. coli cell strains were used for overexpression. EcClpP was overexpressed in BLR (DE3) from Novagen and BsClpP in BL21 (DE3) from New England Biolabs. Overexpression and purification conditions for both proteins were similar. Cell strains transformed with the relevant plasmids were grown at 37 °C to an OD600 of 0.7 followed by induction with 1 mM isopropyl β-d-thiogalactopyranoside and overnight overexpression at 25 °C. Cell pellets were resuspended in 20 mM Tris and 150 mM NaCl (pH 8.0) and lysed using an Avestin C3 Emulsiflex. Lysates were clarified by centrifugation at 12 000 rpm for 30 min and incubated with Ni-NTA (Qiagen) resin pre-equilibrated with 20 mM Tris and 150 mM NaCl (pH 8.0). Protein was eluted from Ni-NTA resin with 20 mM Tris, 150 mM NaCl, and 500 mM imidazole (pH 8.0) and concentrated. Gel filtration, using a Superdex 200 Increase size exclusion column (GE Healthcare), was used as a final purification step for both proteins. Gel filtration buffer conditions were 25 mM Tris, 100 mM KCl, and 10% glycerol (pH 7.5)58 for EcClpP and 25 mM HEPES, 100 mM KCl, 20 mM MgCl2, 1 mM EDTA, and 10% glycerol (pH 7.6)19 for BsClpP. Protein concentrations were determined using the calculated molar extinction coefficient in 6 M guanidinium hydrochloride.59
FITC-Casein Screening Assay
A 300 nM tetradecameric EcClpP in buffer A (25 mM Tris-HCl, pH 7.5, and 100 mM KCl) was incubated with compounds (50 µM) at 37 °C for 15 min in flat bottom, nonbinding, nonsterile, white polystyrene 96-well plates (Corning 3990). After the preincubation period, 1 µL of a 192 µM FITC-β-casein solution in buffer A was added to each assay well to give a final assay concentration of 1.92 µM FITC-β-casein and final assay volume of 100 µL. Assay plates were incubated at 37 °C, and hydrolysis of the fluorogenic substrate was monitored via an i-TECAN Infinite M200 plate reader (excitation: 485 nm; emission: 538 nm). Readings were taken every 30 min for 6 h. All compounds, including enopeptin A (25 µM, positive control), ACP5 (50 µM, positive control), and DMSO (1%, negative control) were normalized relative to background FITC-β-casein fluorescence (ΔRFU = sample dosed well fluorescence − DMSO dosed well fluorescence).
Decapeptide Degradation Assay
As described previously,38 25 nM tetradecameric EcClpP in buffer B (25 mM HEPES, pH 7.5, 100 mM KCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT) was incubated with the compound of interest over a range of concentrations at 30 °C for 15 min in flat bottom, nonbinding, nonsterile, white polystyrene 96- well plates (Corning 3990). After the preincubation period 1 µL of a 1.5 mM Abz-DFAPKMALVPYNO2 (Biomatik) solution was added to each assay well to give a final assay concentration of 15 µM fluorogenic decapeptide and final assay volume of 100 µL. Assay plates were incubated at 30 °C, and hydrolysis of the fluorogenic peptide was monitored via an i-TECAN Infinite M200 plate reader (excitation: 320 nm; emission: 420 nm). Readings were taken every 30 min for 2 h. All compounds, including ADEP1 (positive control) and DMSO (1%, negative control), were normalized relative to background Abz-DFAPKMALVPYNO2 fluorescence (ΔRFU = sample dosed well fluorescence − DMSO dosed well fluorescence).
FITC-Casein Degradation Assay
A 257 nM tetradecameric EcClpP in buffer A (25 mM Tris-HCl, pH 7.5, and 100 mM KCl) was incubated with the compound of interest over a range of concentrations at 37 °C for 15 min in flat bottom, nonbinding, nonsterile, white polystyrene 96-well plates (Corning 3990). After the preincubation period 1 µL of a 0.45 mM FITC-β-casein solution in buffer A was added to each assay well to give a final assay concentration of 4.5 µM FITC-β-casein and final assay volume of 100 µL. Assay plates were incubated at 37 °C, and hydrolysis of the fluorogenic substrate was monitored via an i-TECAN Infinite M200 plate reader (excitation: 485 nm; emission: 538 nm). Readings were taken every 30 min for 6 h. All compounds, including ADEP1 (positive control) and DMSO (1%, negative control), were normalized relative to background FITC-β-casein fluorescence (ΔRFU = sample dosed well fluorescence − DMSO dosed well fluorescence).
SDS-PAGE Analysis
A 420 nM tetradecameric EcClpP or BsClpP in buffer A (59 µL) was incubated with the compound of interest at the desired concentration for 15 min at 37 °C in Eppendorf tubes. A 16 µL amount of a 1 mg/mL bovine β-casein (Sigma-Aldrich C6905, Bio-Ultra ≥98%) solution in buffer A was then added for a final substrate concentration of 7.5 µM and a final assay volume of 75 µL. The reaction was quenched with 25 µL of Laemmli loading buffer (Alfa Aesar J60015) at 80 °C. Individual reactions were quenched after 5 min, 30 min, 1 h, and/or 2 h depending on the compound. The samples were then boiled for approximately 1 min prior to loading. Time-dependent degradation of the β-casein band was visualized using SDS-Page on a 10% acrylamide Bis-Tris gel.
MIC Determination
Using the 2-fold dilution technique in 96-well microtiter plates, cells were subcultured (1:100 dilution) in fresh LB media (tryptone 10 g/L, yeast extract 5 g/L, and NaCl 5 g/L) and grown to an OD600 of ~1.0. Cells were inoculated at a density of 105 cells per mL into LB media in the presence of 2-fold increasing concentrations of 1. MIC values were determined visually after incubation of the microtiter plates at 37 °C for 18 h.
Supplementary Material
Acknowledgments
This work was supported by the University of Oklahoma Junior Faculty Fellowship (A.S.D.), Oklahoma Center for the Advancement of Science and Technology (OCAST, HR15-161), and University of Oklahoma start-up funding. The initial screen was partially subsidized by an Undergraduate Research Opportunity Program (UROP) Award (J.A.C.). Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103640. In addition, we thank Prof. R. Sauer and Dr. K. Schmitz (MIT) for the EcClpP and BsClpP plasmids, Prof. R. Cichewicz (OU) and his research team for access to the natural product library, and Prof. H. Zgurskaya (OU) and her research team for preliminary microbiology work.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Structures of enopeptin A and ACP5, B-score analysis of initial screen, ACP5 SDS-PAGE analysis, ADEP1 degradation curves (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Antibiotic Resistance Threats in the United States, 2013. Center for Disease Control and Prevention; 2013. [Google Scholar]
- 2.Fischbach MA, Walsh CT. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright GD. BMC Biol. 2010;8:123–128. doi: 10.1186/1741-7007-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh CT, Wencewicz TA. J. Antibiot. 2014;67:7–22. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. Multidrug Tolerance of Biofilms and Persister Cells. In: Romeo T, editor. Bacterial Biofilms. Springer; 2008. pp. 107–131. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman S, Clark WP, Maurizi MR. J. Biol. Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 7.Hwang BJ, Woo KM, Goldberg AL, Chung CH. J. Biol. Chem. 1988;263:8727–8734. [PubMed] [Google Scholar]
- 8.Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark WP, Maurizi MR. J. Biol. Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 9.Katayama-Fujimura Y, Gottesman S, Maurizi MR. J. Biol. Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- 10.Maurizi MR, Clark WP, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. J. Biol. Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 11.Maurizi MR, Thompson MW, Singh SK, Kim SH. Methods in Enzymology. Vol. 244. Philadelphia, PA: Elsevier; 1994. Endopeptidase Clp: ATP-Dependent Clp Protease from Escherichia coli. p 314.10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Hartling JA, Flanagan JM. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman S, Wickner S, Maurizi MR. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 14.Kruger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. J. Bacteriol. 2000;182:3259–3265. doi: 10.1128/jb.182.11.3259-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raju RM, Goldberg AL, Rubin EJ. Nat. Rev. Drug Discovery. 2012;11:777–789. doi: 10.1038/nrd3846. [DOI] [PubMed] [Google Scholar]
- 16.Yu AY, Houry WA. FEBS Lett. 2007;581:3749–3757. doi: 10.1016/j.febslet.2007.04.076. [DOI] [PubMed] [Google Scholar]
- 17.Joshi SA, Hersch GL, Baker TA, Sauer RT. Nat. Struct. Mol. Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- 18.Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Nat. Struct. Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- 19.Martin A, Baker TA, Sauer RT. Mol. Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson M, Satyendra SK, Maurizi MR. J. Biol. Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 21.Thompson MW, Maurizi MR. J. Biol. Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 22.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordova JC, Olivares AO, Shin Y, Stinson BM, Calmat S, Schmitz KR, Aubin-Tam ME, Baker TA, Lang MJ, Sauer RT. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee ME, Baker TA, Sauer RT. J. Mol. Biol. 2010;399:707–718. doi: 10.1016/j.jmb.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carney DW, Schmitz KR, Truong JV, Sauer RT, Sello JK. J. Am. Chem. Soc. 2014;136:1922–1929. doi: 10.1021/ja410385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brotz-Oesterhelt H, Beyer D, Kroll HP, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, Bandow JE, Sahl HG, Labischinski H. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 28.Kirstein J, Hoffmann A, Lilie H, Schmidt R, Rubsamen-Waigmann H, Brotz-Oesterhelt H, Mogk A, Turgay K. EMBO Mol. Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BG, Park EY, Lee KE, Jeon H, Sung KH, Paulsen H, Rubsamen-Schaeff H, Brotz-Oesterhelt H, Song HK. Nat. Struct. Mol. Biol. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- 30.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis K, Conlon B, Nelson ML, Polastri MP, Dahl TA. WO 2012/135846 A1. 2012 Oct 4;
- 32.Michael KH, Kastner RE. 4,492,650. U.S. Patent. 1985 Jan 8;
- 33.Osada H, Yano T, Koshino H, Isono K. J. Antibiot. 1991;44:1463–1466. doi: 10.7164/antibiotics.44.1463. [DOI] [PubMed] [Google Scholar]
- 34.Gersch M, Famulla K, Dahmen M, Gobl C, Malik I, Richter K, Korotkov VS, Sass P, Rubsamen-Schaeff H, Madl T, Brotz-Oesterhelt H, Sieber SA. Nat. Commun. 2015;6:6320. doi: 10.1038/ncomms7320. [DOI] [PubMed] [Google Scholar]
- 35.Hinzen B, Raddatz S, Paulsen H, Lampe T, Schumacher A, Habich D, Hellwig V, Benet-Buchholz J, Endermann R, Labischinski H, Brotz-Oesterhelt H. ChemMedChem. 2006;1:689–693. doi: 10.1002/cmdc.200600055. [DOI] [PubMed] [Google Scholar]
- 36.Socha AM, Tan NY, LaPlante KL, Sello JK. Bioorg. Med. Chem. 2010;18:7193–7202. doi: 10.1016/j.bmc.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 37.Lee RE, Zhao Y, Griffith E, Zheng Z, Singh A. WO 2015/031871 A1. 2015 Mar 5;
- 38.Carney DW, Compton CL, Schmitz KR, Stevens JP, Sauer RT, Sello JK. ChemBioChem. 2014;15:2216–2220. doi: 10.1002/cbic.201402358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Compton CL, Carney DW, Groomes PV, Sello JK. ACS Infect. Dis. 2015;1:53–58. doi: 10.1021/id500009f. [DOI] [PubMed] [Google Scholar]
- 40.Houry WA, Leung E, Batey RA, Cossette M, Goodreid J, Datti A, Liu J, Nhieu AJ, Gray-Owen SD, Fai EF, Eger BT. WO 2012/079164 A1. 2012 Jun 21;
- 41.Leung E, Datti A, Cossette M, Goodreid J, McCaw SE, Mah M, Nakhamchik A, Ogata K, El Bakkouri M, Cheng YQ, Wodak SJ, Eger BT, Pai EF, Liu J, Gray-Owen S, Batey RA, Houry WA. Chem. Biol. 2011;18:1167–1178. doi: 10.1016/j.chembiol.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 42.Du L, Robles AJ, King JB, Powell DR, Miller AN, Mooberry SL, Cichewicz RH. Angew. Chem., Int. Ed. 2014;53:804–809. doi: 10.1002/anie.201306549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu G, Wang B, Zhang G, King JB, Cichewicz RH, Li J. Sci. Rep. 2015;5:14694–14711. doi: 10.1038/srep14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B, You J, King JB, Cai S, Park E, Powell DR, Cichewicz RH. J. Nat. Prod. 2014;77 doi: 10.1021/np500531j. 227310.1021/np500531j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du L, Risinger AL, King JB, Powell DR, Cichewicz RH. J. Nat. Prod. 2014;77:1753–1757. doi: 10.1021/np500387h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du L, Robles AJ, King JB, Mooberry SL, Cichewicz RH. J. Nat. Prod. 2014;77:1459–1466. doi: 10.1021/np5002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koshino H, Osada H, Yano T, Uzawa J, Isono K. Tetrahedron Lett. 1991;32:7707–7710. [Google Scholar]
- 48.Brideau C, Gunter B, Pikounis B, Liaw A. J. Biomol. Screening. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 49.Pelz O, Gilsdorf M, Boutros M. BMC Bioinf. 2010;11 doi: 10.1186/1471-2105-11-185. 18510.1186/1471-2105-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whyte AC, Gloer JB. J. Nat. Prod. 1996;59:1093–1095. doi: 10.1021/np960607m. [DOI] [PubMed] [Google Scholar]
- 51.Finefield JM, Kato H, Greshock TJ, Sherman DH, Tsukamoto S, Williams RM. Org. Lett. 2011;13:3802–3805. doi: 10.1021/ol201284y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercado-Marin EV, Garcia-Reynaga P, Romminger S, Pimenta EF, Romney DK, Lodewyk MW, Williams DE, Andersen RJ, Miller SJ, Tantillo DJ, Berlinck RG, Sarpong R. Nature. 2014;509:318–324. doi: 10.1038/nature13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finefield JM, Frisvad JC, Sherman DH, Williams RM. J. Nat. Prod. 2012;75:812–833. doi: 10.1021/np200954v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng J, Zhang XY, Tu ZC, Xu XY, Qi SH. J. Nat. Prod. 2013;76:983–987. doi: 10.1021/np400132m. [DOI] [PubMed] [Google Scholar]
- 55.Kato H, Yoshida T, Tokue T, Nojiri Y, Hirota H, Ohta T, Williams RM, Tsukamoto S. Angew. Chem., Int. Ed. 2007;46:2254–2256. doi: 10.1002/anie.200604381. [DOI] [PubMed] [Google Scholar]
- 56.Rosner JL, Martin RG. J. Bacteriol. 2013;195:1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mima T, Joshi S, Gomez-Escalada M, Schweizer HP. J. Bacteriol. 2007;189:7600–7609. doi: 10.1128/JB.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flanagan JM, Wall JS, Capel MS, Schneider DK, Shanklin J. Biochemistry. 1995;34:10910–10917. doi: 10.1021/bi00034a025. [DOI] [PubMed] [Google Scholar]
- 59.Gill SC, von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.