Abstract
The gender of cercarial larvae can only be determined using molecular methods. End point PCR methods that amplify repetitive markers on the W chromosome of the female (ZW) parasites have been developed, but sometimes results are ambiguous or incorrect. To more effectively distinguish sexes, and to determine why end point PCR can be incorrect, we quantified the W6 repeat sequence and a specific Z chromosome gene using real-time PCR. The ratio between copy number of W6 and a Z chromosome marker unambiguously identifies gender: females have higher ratios (421-4371) than males (0–21). However, some males have low numbers of W6 elements in their genome, and qPCR demonstrated significantly higher W6/Z marker ratios for male genotypes giving ambiguous end point PCR results compared with males giving clear end point results. The quantitative PCR sexing method developed will be particularly useful where reliable sexing of cercariae is critical, for example when staging genetic crosses.
Keywords: Schistosome, qPCR, sexing methods, cercariae
Schistosomes have separate sexes, which is a unique feature among trematodes. Gender is genetically determined (ZZ males, ZW females) but males and females can be only distinguished morphologically when they are adults. However, no obvious morphological dimorphism is observable during the larval stages (miracidia and cercariae) [1,2], so efficient larval sexing methods are critical for experimental genetics and genomics [3–5], and for studies of sex ratio dynamics in different parasite populations and life-stages [6,7]. Several methods have been developed to sex cercariae. Liberatos et al. [2] developed a cytological approach based on C-band staining of W chromosome, which was validated for the three major species of schistosomes (Schistosoma mansoni, S. haematobium and S. japonicum) and for all larval stages of S. mansoni (miracidia, sporocysts and cercariae). A second molecular approach was made possible in S. mansoni by the discovery of repetitive elements on the W chromosomes which could be detected by Southern blot [8–10]. Later the W1 element was PCR amplified [11] providing a high throughput end point assay for sexing cercariae.
End point PCR amplification sometimes provides incorrect sexing results in S. mansoni populations kept in the laboratory [12,13] (personal observations). This inconsistency results from the fact that the W1 element is not restricted to the W chromosome, and may be found at low copy in male (ZZ) genomes [13]. Hence PCR amplification of W1 markers from males leads them to be incorrectly identified as females. These sexing errors can be extremely costly: while establishing genetic crosses between schistosomes [4,5], we infected hamsters with cercariae identified as male or female using end point PCR assays. Forty-five days later, we discovered that 29% (67/229) were homosexual crosses that contained two male genotypes, due to incorrect sexing results. To reduce incorrect results, more accurate sex identification is required. Several attempts have been made to improve end point PCR approaches. These include sexing using multiple independent PCRs of the same population of cercariae [1] or identification of more female specific W elements [14]. These laborious methods tend to demonstrate the unreliability of end point methods. Unfortunately, none of these end point methods provides a completely reliable solution.
We have developed a quantitative PCR method to determine the proportion of the W6 elements (another repetitive element found on W chromosomes [15]) relative to Z chromosome marker. The ratio of the two marker copy number measures allows efficient identification of female and male cercariae. This quantitative PCR approach also describes variation in copy number in different schistosome lines or individuals and determines the reasons underlying spurious sexing results in end point assays.
Determination of W/Z marker ratio using quantitative PCR
We extracted DNA from cercariae from different parasite populations from the Old and New World (SmLE, Belo Horizonte, Brazil; SmLE-PZQ-R, derived from SmLE population; SmEG, Cairo, Egypt). We chose the parasite lines that we maintain in the laboratory to determine that the methods developed work consistently across several different parasite populations with different geographical origins. We used parents and progeny of genetic crosses (SmLE × SmHR, Puerto Rico; SmLE-PZQ-R × SmEG). We included F1 progeny, because we have often observed misscoring of males as females which is often seen in the first generations of genetic crosses (personal observations) using end point PCR methods.
We recovered all the cercariae (10 to >1000) produced by snails infected with single miracidia in microtubes. These were spun down at 300 × g for 5 minutes, and DNA extracted using a Chelex protocol [4]. The same cercariae were also used to infect hamsters or mice in order to check the adult worm’s gender after 45 days. Adult worms were recovered using classical perfusion methods [16] (IACUC protocols: UTHSCSA 08093X/11087X, TBRI 1419-MA-0/1420-MU-0).
To perform the quantitative PCR, we designed primers to amplify the W6 element (accession n°HQ880215) and a low copy gene on the Z chromosome (Smp_011570.1). We chose Smp_011570.1 because this gene is present on chromosome Z of the S. mansoni genome assembly (v5.0), was found only on this chromosome following a blastn (v2.2.29) and was annotated as ribosomal protein S4 which is a conserved gene. We determined Smp_011570.1 copy number by qPCR using the single copy autosomal gene α-tubulin 2 (accession n°S79195) as a reference [17]. The average ratio ± sd of Smp_011570.1 copy number/α-tubulin 2 copy number was 1.36 ± 0.50 for females (ZW) and 1.82 ± 0.60 for males (ZZ). The observed ratios confirm that Smp_011570.1 is a low copy number gene.
We quantified copy number of the W6 element and the Z marker (Smp_011570.1) as follows. Reactions were performed in duplicate using AB1 prism 7900HT Sequence Detection system (Applied Biosystems) using the program: 95°C for 10 minutes, [95°C for 15 seconds, 60°C for 1 minute] × 40 cycles. Duplicate reactions showing a difference in Ct greater than 1 were rerun. A melting curve (60°C to 95°C) was recorded at the end of each reaction in order to check for uniqueness of the PCR product. The reaction mixture consisted of 5 μL of SYBR Green MasterMix (Applied Biosystems), 0.3 μL of each forward and reverse primers (10 μM; Table 1), amplifying 195 bp of the W6 element and 404 bp of Smp_011570.1 on Z chromosome, 3.4 μL of sterile water and 1 μL of total DNA template (5–10 ng). As we normalized the W6 copy number by the Z marker copy number for a given sample (and not by the quantity of DNA we added in the well), it is not necessary to standardize the ng amounts of DNA template between the samples. A standard curve was plotted using seven dilutions of a purified W6 element PCR product (W6 copies.μL−1: 1.60×101, 1.60×102, 1.60×103, 1.60×104, 1.60×105, 1.60×106, 1.60×107) and of a purified Smp_011570 PCR product (Smp_011570.1 copies.μL−1: 1.64×101, 1.64×102, 1.64×103, 1.64×104, 1.64×105, 1.64×106, 1.64×107), showing an amplification efficiency of 1.92 and 1.90, respectively. PCR products for standard curves were generated using TaKaRa Taq R001 AM kit (Clonetech) and the manufacturer’s protocol (PCR cycles: 95°C for 5 minutes, [95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds] × 35 cycles, 72°C for 10 minutes), purified using SigmaSpin Sequencing Reaction Clean-Up kit (Sigma) following the manufacturer’s protocol, and quantified using Qubit dsDNA BR Assay kit (Invitrogen). We estimated the number of copies in the PCR products as follows: PCR product length × (average molecular mass of nucleotides (330 g.mole−1) × 2 strands) × Avogadro constant. Following the qPCR experiment, we estimated the number of W6 element and Smp_011570.1 copies for a given sample according to the standard curve.
Table 1.
Primer sequences to amplify W and Z markers.
| Primer name | Primer sequence (5′ to 3′ orientation) | Expected amplicon size (bp) | qPCR efficiency |
|---|---|---|---|
| Sm_W6 F | TGTGAAGCAAAGTGTTCACTG | 195 | 1.92 |
| Sm_W6 R | TTCATCAAGTCAATCACAGCTC | ||
| Smp_011570.1 F | TGGATGTTGGATAAGCTGGG | 404 | 1.90 |
| Smp_011570.1.R | TGGATCTGGATATCGAATGGTC |
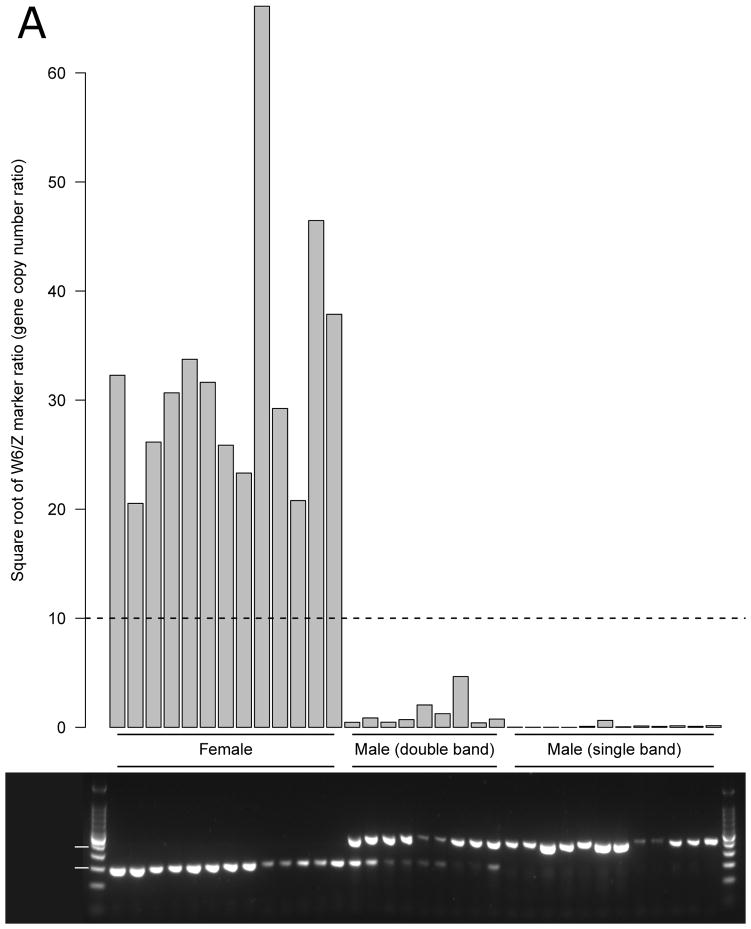
Females contain between 421 and 4371 times more W6 elements than Z markers, while males have between 0 and 21 more W6 elements than Z markers (Figure 1A; Table S1). No distinct differences were observed between the different schistosome populations used. We use the W6/Z marker ratio to determine sex of individuals: this is square root transformed in figure 1 to better visualize the low amplification observed in male parasites. Females have a square root ratio above 10 while males have a square root ratio below this arbitrary threshold. These results are consistent for cercariae from the different parasite populations as well as for F1 and F2 individuals generated from crosses.
Figure 1. Copy number ratio of W marker and Z marker.
A: Square root of the ratio between W marker (W6) and Z marker (Smp_011570.1). Males and females are clearly differentiated from the ratios observed: Females have a high copy number of W marker compared to the Z marker, while in males W markers are absent or rare. B: end point characterization of PCR products. We electrophoresed multiplexed PCR products generated using W and Z marker primers on 2% agarose gels. The size standard is a 100bp ladder (Promega). Females have a 195 bp W6 specific band, and males have a 404 bp Z-specific band. “Double band” individuals are genotypically male but have elevated numbers of W markers relative to most males.
This dramatic difference in the W6/Z marker ratio between females (WZ) and males (ZZ) (at least 20 times) is expected since the W6 element is abundant on the W chromosome [15] but rare or absent elsewhere in the genome. However we observed a low number of W6 elements in males (ZZ) as previously observed for the W1 elements [13]. This result cannot be explained by contamination because we observed no contamination in our qPCR negative controls. Therefore the presence of these elements outside the W chromosome may be a common feature of W elements. Movement of W elements may be due to retrotransposition to other chromosomes (these elements are annotated as transposable elements and showed transcription activity [15]) or to recombination events between W and Z chromosomes (some W6 elements may be present on the pseudo-autosomal region of the Z/W chromosome which is known to recombine [4,18]), or to extrachromosomal DNA, as observed for repetitive elements in Leishmania and human cancer cells [19,20]. However we cannot distinguish these two explanations. Further investigation of the precise genomic locations of W elements will be needed to understand the dynamics of these elements, which might play a major role in sex chromosome evolution as seen in other organisms [21].
The very low W6/Z marker ratios observed in male (ZZ) parasites also requires explanation. If we suppose the Z marker as a single copy gene, so there should be two copies in male worms. Therefore, a W6/Z marker ratio of ~1 is expected if the larvae inherit a single W6 copy from each parent while a ratio of ~0.5 is expected if the larvae inherit a single W6 copy of one of the two parents. The presence of very low ratios (≤ 0.1) may indicate that not all thecercariae present in the pool used for DNA extraction carry the W6 element. The genetic explanation for this is unclear, given that all cercariae emerging from a single snail are clonally derived from a single sporocyst. However, regardless of the mechanism, the presence of low numbers of W6 elements complicates interpretation of PCR end-point assays, but presents no problem for qPCR approaches.
Sex determination using multiplex end point PCR
We used the same primers to develop an end point multiplexed PCR for sex determination to compare with the qPCR method. Each PCR was performed with the TaKaRa Taq R001 AM kit (Clonetech) and composed of 8.325 μL of sterile water, 1.5 μL of 10× buffer, 1.2 μL of dNTP(2.5 mM each), 0.9 μL of MgCl2 (25 mM), 0.5 μL of each primers of both markers (10 μM; Table 1), 0.075 μL of Taq polymerase (5 U.μL−1) and 1 μL of DNA template. We amplified DNA using a GeneAmp PCR system 9700 thermocycler (Applied Biosystems) with the following program: 95°C for 5 minutes, [95°C for 30 seconds, 55°C for 30 seconds, 72°C for 45 seconds] × 35 cycles, 72°C for 10 minutes. The size of the expected product is 195 bp for the W6 element and 404 bp for the Z marker (as previously mentioned).
We applied the end point method to the same samples to allow direct comparison with the qPCR method. The results obtained showed three distinct band patterns (Figure 1B; Figure S1): (i) a single band at 195 bp for female cercariae, (ii) a single band at 404 bp for most males, and (iii) a double band at 195 and 404bp from some males. The unique W6 band at 195 bp found in females is expected, because of the small size of this amplicon and the much higher number of W6 elements than Z markers. This leads to the preferential amplification of the W marker. The unique Z marker band (404 bp) found in most males is also expected because W markers are outnumbered by Z markers. In some males, we observed a double band pattern in which both 195 bp and 404 bp fragments are amplified. The qPCR results show that these males have a significantly higher number of W6 elements than in males with only the single 404 bp band (Wilcoxon test, p = 9.9.10−4). Given that all males tested contain a W element, it is not surprising that previous end point methods frequently misscored males as females.
In addition to its simple setup, this multiplex end point PCR has two advantages over previous end point methods. Competition between W6 element and Z marker amplification limits amplification of low copy W6 elements in males. Second, this method does not require a second PCR of an autosomal marker (in addition to the W element) to test the presence of amplifiable DNA within the sample [4].
Conclusion
Quantitative PCR provides a reliable approach to determine gender by counting the number of W6 elements relative to the number of Z chromosome markers. This approach avoids misscoring of males as females which can occur when a small number of W elements are present in the male genome. The quantitative PCR sexing method developed will be particularly useful where reliable sexing of cercariae is critical, for example when establishing genetic crosses. We also showed that using the same set of primers in a multiplex end point PCR provides good results for sexing cercariae. We observed an intermediate double band pattern in some males which was explained by a significantly greater number of W6 repeats than in males showing a single 404 bp band.
Supplementary Material
Independent PCR (1) using either the W6 primers (W marker) or the Smp_011570.1 primers (Z marker) showing the expected bands either at 195 bp or 404 bp. This result is the same for both female (ZW) and male (ZZ), because males carry low numbers of W6 copies. When all primer sets are used in multiplex PCR, three patterns are obtained: a band at 195 bp only for female (W6 amplification outcompetes Z marker amplification), a double band at 195 bp and 404 bp (W6 and Z marker are co-amplified) and a band at 404 bp for males (Z marker amplification outcompetes W6 amplification). Competition between primer sets in multiplexed PCR reactions is the key to this end-point PCR and minimizes the chance of spurious sexing results. L: 100 bp ladder (lowest band = 100 bp, brightest band = 500 bp).
This table contains, for each sample tested: the parasite stage used (cercariae), the S. mansoni (Sm) population (LE: SmLE, from Brazil; HR: SmHR, from Puerto Rico; EG: SmEG, from Egypt; PZQR: SmLE-PZQ-R, from Brazil, a population derived from SmLE and showing praziquantel resistance), the genetic cross from where the parasite was sampled, the generation of the cross (F0: parent populations, F1: F1 population, F2: F2 population), the PCR profile obtained (lower, double or upper band, see Figure 1B and Figure S1), the corresponding sex (female or male), the copy number data for the W6 and Z-specific markers, and the ratio computation.
Acknowledgments
This study was supported by NIH grants [R01-AI097576 (T.J.C.A.), 1R01AI115691 and P50 AI098507 (P.T.L.)], World Health Organization [HQNTD1206356 (P.T.L.)], and the UTHSCSA Presidents Collaborative Research Fund (P.T.L.). Biomphalaria glabrata snails were provided by the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) through NIH-NIAID Contract HHSN272201000005I for distribution through BEI Resources. The molecular work at TBRI was conducted in facilities constructed with support from Research Facilities Improvement Program Grant (C06 RR013556) from the National Center for Research Resources (NIH). W.L was supported by a Cowles fellowship from Texas Biomedical Research Institute. A.C.A.M received support from CAPES (17766/12-5). We thank the Theodor Bilharz Research Institute (Egypt) for providing the SmEG parasites.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Frédéric D. Chevalier, Email: fcheval@txbiomed.org.
Winka Le Clec’h, Email: winkal@txbiomed.org.
Ana Carolina Alves de Mattos, Email: anademattos@cpqrr.fiocruz.br.
Philip T. LoVerde, Email: loverde@uthscsa.edu.
Timothy J.C. Anderson, Email: tanderso@txbiomed.org.
References
- 1.Boissier J, Durand P, Moné H. PCR effectiveness for sexing Schistosoma mansoni cercariae: application for sexing clonal cercarial populations. Mol Biochem Parasitol. 2001;112:139–141. doi: 10.1016/S0166-6851(00)00344-3. [DOI] [PubMed] [Google Scholar]
- 2.Liberatos JD, Short RB. Identification of sex of schistosome larval stages. J Parasitol. 1983;69:1084–1089. [PubMed] [Google Scholar]
- 3.Pica-Mattoccia L, Dias LC, Cioli D. Genetic complementation analysis of two independently isolated hycanthone-resistant strains of Schistosoma mansoni. Mem Inst Oswaldo Cruz. 1992;87(Suppl 4):211–214. doi: 10.1590/S0074-02761992000800032. [DOI] [PubMed] [Google Scholar]
- 4.Criscione CD, Valentim CLL, Hirai H, LoVerde PT, Anderson TJC. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 2009;10:R71. doi: 10.1186/gb-2009-10-6-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentim CLL, Cioli D, Chevalier FD, Cao X, Taylor AB, Holloway SP, et al. Genetic and molecular basis of drug resistance and species-specific drug action in Schistosome parasites. Science. 2013;342:1385–1389. doi: 10.1126/science.1243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liberatos JD. Schistosoma mansoni: male-biased sex ratios in snails and mice. Exp Parasitol. 1987;64:165–177. doi: 10.1016/0014-4894(87)90140-8. [DOI] [PubMed] [Google Scholar]
- 7.Lepesant JMJ, Boissier J, Climent D, Cosseau C, Grunau C. Female biased sex-ratio in Schistosoma mansoni after exposure to an allopatric intermediate host strain of Biomphalaria glabrata. Exp Parasitol. 2013;135:350–356. doi: 10.1016/j.exppara.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Spotila LD, Rekosh DM, Boucher JM, LoVerde PT. A cloned DNA probe identifies the sex of Schistosoma mansoni cercariae. Mol Biochem Parasitol. 1987;26:17–20. doi: 10.1016/0166-6851(87)90125-3. [DOI] [PubMed] [Google Scholar]
- 9.Drew AC, Brindley PJ. Female-specific sequences isolated from Schistosoma mansoni by representational difference analysis. Mol Biochem Parasitol. 1995;71:173–181. doi: 10.1016/0166-6851(95)00048-6. [DOI] [PubMed] [Google Scholar]
- 10.Webster P, Mansour TE, Bieber D. Isolation of a female-specific, highly repeated Schistosoma mansoni DNA probe and its use in an assay of cercarial sex. Mol Biochem Parasitol. 1989;36:217–222. doi: 10.1016/0166-6851(89)90169-2. [DOI] [PubMed] [Google Scholar]
- 11.Gasser RB, Morahan G, Mitchell GF. Sexing single larval stages of Schistosoma mansoni by polymerase chain reaction. Mol Biochem Parasitol. 1991;47:255–258. doi: 10.1016/0166-6851(91)90187-B. [DOI] [PubMed] [Google Scholar]
- 12.Quack T, Doenhoff M, Kunz W, Grevelding CG. Schistosoma mansoni: the varying occurrence of repetitive elements in different strains shows sex-specific polymorphisms. Exp Parasitol. 1998;89:222–227. doi: 10.1006/expr.1998.4293. [DOI] [PubMed] [Google Scholar]
- 13.Grevelding CG. Genomic instability in Schistosoma mansoni. Mol Biochem Parasitol. 1999;101:207–216. doi: 10.1016/S0166-6851(99)00078-X. [DOI] [PubMed] [Google Scholar]
- 14.Portela J, Grunau C, Cosseau C, Beltran S, Dantec C, Parrinello H, et al. Whole-genome in-silico subtractive hybridization (WISH) - using massive sequencing for the identification of unique and repetitive sex-specific sequences: the example of Schistosoma mansoni. BMC Genomics. 2010;11:387. doi: 10.1186/1471-2164-11-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lepesant JMJ, Cosseau C, Boissier J, Freitag M, Portela J, Climent D, et al. Chromatin structural changes around satellite repeats on the female sex chromosome in Schistosoma mansoni and their possible role in sex chromosome emergence. Genome Biol. 2012;13:R14. doi: 10.1186/gb-2012-13-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvall RH, DeWitt WB. An improved perfusion technique for recovering adult schistosomes from laboratory animals. Am J Trop Med Hyg. 1967;16:483–486. doi: 10.4269/ajtmh.1967.16.483. [DOI] [PubMed] [Google Scholar]
- 17.Duvaux-Miret O, Baratte B, Dissous C, Capron A. Molecular cloning and sequencing of the alpha-tubulin gene from Schistosoma mansoni. Mol Biochem Parasitol. 1991;49:337–340. doi: 10.1016/0166-6851(91)90079-l. [DOI] [PubMed] [Google Scholar]
- 18.Spotila LD, Hirai H, Rekosh DM, Lo Verde PT. A retroposon-like short repetitive DNA element in the genome of the human blood fluke, Schistosoma mansoni. Chromosoma. 1989;97:421–428. doi: 10.1007/BF00295025. [DOI] [PubMed] [Google Scholar]
- 19.Wahl GM. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 20.Ubeda JM, Raymond F, Mukherjee A, Plourde M, Gingras H, Roy G, et al. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol. 2014;12:e1001868. doi: 10.1371/journal.pbio.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Independent PCR (1) using either the W6 primers (W marker) or the Smp_011570.1 primers (Z marker) showing the expected bands either at 195 bp or 404 bp. This result is the same for both female (ZW) and male (ZZ), because males carry low numbers of W6 copies. When all primer sets are used in multiplex PCR, three patterns are obtained: a band at 195 bp only for female (W6 amplification outcompetes Z marker amplification), a double band at 195 bp and 404 bp (W6 and Z marker are co-amplified) and a band at 404 bp for males (Z marker amplification outcompetes W6 amplification). Competition between primer sets in multiplexed PCR reactions is the key to this end-point PCR and minimizes the chance of spurious sexing results. L: 100 bp ladder (lowest band = 100 bp, brightest band = 500 bp).
This table contains, for each sample tested: the parasite stage used (cercariae), the S. mansoni (Sm) population (LE: SmLE, from Brazil; HR: SmHR, from Puerto Rico; EG: SmEG, from Egypt; PZQR: SmLE-PZQ-R, from Brazil, a population derived from SmLE and showing praziquantel resistance), the genetic cross from where the parasite was sampled, the generation of the cross (F0: parent populations, F1: F1 population, F2: F2 population), the PCR profile obtained (lower, double or upper band, see Figure 1B and Figure S1), the corresponding sex (female or male), the copy number data for the W6 and Z-specific markers, and the ratio computation.