Abstract
Malaria surveillance is critical for control efforts, but diagnostic methods frequently disagree. Here we compare microscopy, PCR, and a Rapid Diagnostic Test in 7,137 samples from children in the Democratic Republic of the Congo using Latent Class Analysis. PCR had the highest sensitivity (94.6%) and microscopy had the lowest (76.7%).
Keywords: malaria, surveillance, diagnostics, microscopy, PCR, rapid diagnostic test, RDT, latent class analysis, LCA
Malaria is a particularly severe problem in the Democratic Republic of the Congo (DRC) (WHO, 2015). Both surveillance and case management of malaria depend on accurate diagnosis, but different methods of parasite detection vary in ease of use, reliability, and cost.
Three commonly used methods of parasite detection are light microscopy, PCR, and Rapid Diagnostic Tests (RDTs). Microscopy, the traditional diagnostic method, has a limit of detection of about 50 parasites/μL of whole blood (Kilian et al., 2000; Mwingira et al., 2014). PCR can detect as few as 0.002 parasites/μL (Kamau et al., 2011), but is more costly and often unavailable in endemic areas. RDTs are inexpensive and easy to use but perform variably below 200 parasites/μL (WHO, 2014).
Here, in an effort to improve malaria surveillance, we compare microscopy, PCR, and RDT on a large common set of samples. We determined the sensitivity and specificity of microscopy and RDT compared to PCR. Additionally, we used Latent Class Analysis (LCA) to estimate an alloyed gold standard and then computed the sensitivity and specificity of each method.
Samples from children under five years old were obtained during a Demographic and Health Survey (DHS), a population-representative survey conducted between November 2013 and February 2014 (MPSMRM, 2014). Finger- or heel-prick blood was obtained from each participant. Giemsa-stained slides were read by two independent microscopists who conferred upon discordances. RDTs targeting the Plasmodium falciparum histidine-rich protein II (PfHRP2) (SD BIOLINE Malaria Ag P.f, Standard Diagnostics, Gyeonggi-do, Republic of Korea) were performed on-site. Blood was also applied to filter papers as dried blood spots (DBS). These DBS were transported to the University of North Carolina, where from each DBS one ¼” diameter hole was punched into a well of a 96-well deep-well plate (Life Technologies, Carlsbad, CA).
From these DBS, genomic DNA (gDNA) was extracted as previously described (Plowe et al., 1995) and stored at −20°C. Each gDNA sample was tested in duplicate in a duplex real-time PCR assay targeting the P. falciparum lactate dehydrogenase gene (pfldh) and, as a control, the human β-tubulin gene. Primer and probe sequences used to amplify pfldh (Taylor et al., 2010) and human β-tubulin (Beshir et al., 2010) were previously published, with the exception that a VIC-TAMRA probe was used for the human β-tubulin assay. Primer and probe sequences, reaction components, and cycling conditions are given in Table S1.
Cycle threshold (CT) values were imported into Microsoft Excel 2007 (Microsoft, Redmond, WA) and were compiled and analyzed in R (version 3.0.3, www.R-project.org). Samples in which human β-tubulin did not amplify or had a CT value above 35 were discarded. Samples were considered positive for pfldh if both replicates amplified or if one amplified with a CT value of 38 or lower. This assay had a limit of detection between 10 and 50 parasites/μL.
DBS were collected from 9,790 children; of these, 978 were unavailable for parasite testing. From the remaining 8,812 samples, four samples did not amplify human DNA in any replicates, 1,558 were from children aged five or older, and 113 samples lacked data for microscopy and/or RDT; these were excluded. The remaining 7,137 samples constitute the analyzable population.
The overall prevalence of P. falciparum parasites was 38.6% by PCR, 26.8% by microscopy, and 36.2% by RDT. The malaria prevalence data and maps are available in the DHS Final Report (MPSMRM, 2014). Of 6,301 children for whom responses were available, 2,094 (33.2%) were reported to have had a fever in the previous two weeks (40.9% of microscopy-positive, 42.4% of RDT-positive, and 39.4% of PCR-positive children).
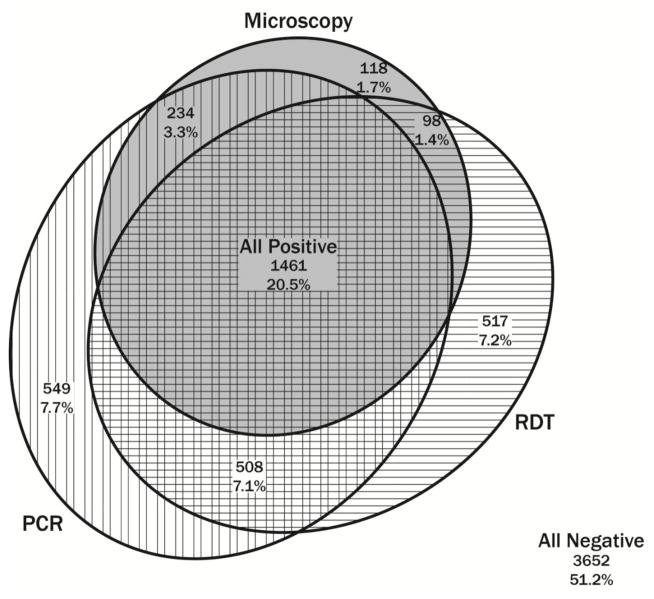
Of the 7,137 samples analyzed here, 3,652 (51.2%) were negative by all three parasite detection methods and 1,461 (20.5%) were positive by all three tests. 234 (3.3%) were positive by PCR and microscopy only, 508 (7.1%) by PCR and RDT only, and 98 (1.4%) by microscopy and RDT only. Additionally, 549 (7.7%) were positive by PCR only, 118 (1.7%) by microscopy only, and 517 (7.2%) by RDT only. The distribution is illustrated in Figure 1.
Figure 1.
Proportional Venn diagram showing discordance of malaria parasite detection by PCR, microscopy, and RDT (eulerAPE, version 3.0.0, http://www.eulerdiagrams.org/eulerAPE/). Samples falling within a given circle indicate positivity by that method of detection.
PCR detected the greatest number of infections overall, most likely owing to its lower limit of detection and a high prevalence of low-density infections. Using PCR as the reference standard, the sensitivity of microscopy was 61.6%, similar to sensitivities reported in other studies [reviewed in Okell et al. (2009)]. The specificity of microscopy compared to PCR was 95.1% (Table 1). The DHS microscopists did not report different parasite species, so the “false positive” microscopy diagnoses could have been infections with P. ovale or P. malariae, which are endemic at a low prevalence across the DRC (Taylor et al., 2011).
Table 1.
Sensitivity and specificity of diagnostic methods vs. PCR and LCA.
| vs. PCR | vs. LCA | |||
|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | |
| PCR | - | - | 94.6 (93.3–95.8) | 88.3 (87.2–89.4) |
| Microscopy | 61.6 (59.7–63.4) | 95.1 (94.4–95.7) | 76.7 (74.6–78.8) | 97.2 (96.6–97.8) |
| RDT | 71.6 (69.8–73.2) | 86.0 (84.9–87.0) | 86.9 (85.2–88.6) | 88.1 (87.1–89.2) |
The sensitivity of RDT was 71.6% (Table 1), similar or slightly lower than studies done in pregnant women (Kattenberg et al., 2012) and febrile patients (Faye et al., 2013; Nicastri et al., 2009; Shakely et al., 2013) but higher than a study in Zambia which found an RDT sensitivity of only 17% compared to nested PCR (Laban et al., 2015). The specificity of RDT was 86.0% (Table 1). The lower specificity found here compared to previous studies, especially those in febrile patients, may be attributed to the well-known persistence of HRP2 antigen after parasite clearance (Abeku et al., 2008).
Because each parasite detection method is imperfect, we performed Latent Class Analysis (LCA) in order to estimate an “alloyed” reference standard against which to estimate the operating characteristics of each detection method (Hui & Walter, 1980). This was calculated using the poLCA package for R (version 1.4.1, https://cran.r-project.org/package=poLCA), and was corroborated using the SAS Proc LCA procedure (version 1.3.2, http://methodology.psu.edu/). Using the LCA-generated alloyed gold standard we computed the sensitivities of PCR (94.6%), microscopy (76.7%), and RDT (86.9%), as well as the specificities of PCR (88.3%), microscopy (97.2%), and RDT (88.1%) (Table 1). Thus, using LCA, PCR was the most sensitive method and microscopy was the most specific. This is generally consistent with the findings of an LCA study of febrile patients from Tanzania (Schachterle et al., 2011), although different protocols for PCR and microscopy were used.
This study demonstrates considerable discordances between malaria surveillance methods. While PCR is consistently the most sensitive, concerns remain about its utility in field settings where resources are limited. There has been a large shift toward RDT use for diagnosis and surveillance. Here, RDT missed 13% of infections. However, newer and more sensitive RDTs are under development (http://sites.path.org/dx/malaria/). Improved field-forward methods of malaria parasite detection will greatly facilitate surveillance and control programs.
Supplementary Material
Highlights.
7,137 samples from children in the DRC were tested by microscopy, RDT, and PCR
Latent Class Analysis was used to compare the three diagnostic methods
PCR was the most sensitive (94.6%), microscopy was the least sensitive (76.7%)
Microscopy was the most specific (97.2%)
Acknowledgments
NIH (Grant 5R01AI107949 to Steven R. Meshnick) and NSF (Grant BSC-1339949 to Michael Emch). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeku TA, Kristan M, Jones C, Beard J, Mueller DH, Okia M, Rapuoda B, Greenwood B, Cox J. Determinants of the accuracy of rapid diagnostic tests in malaria case management: evidence from low and moderate transmission settings in the East African highlands. Malar J. 2008;7:202. doi: 10.1186/1475-2875-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshir KB, Hallett RL, Eziefula AC, Bailey R, Watson J, Wright SG, Chiodini PL, Polley SD, Sutherland CJ. Measuring the efficacy of anti-malarial drugs in vivo: quantitative PCR measurement of parasite clearance. Malar J. 2010;9:312. doi: 10.1186/1475-2875-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye B, Nath-Chowdhury M, Tine RC, Ndiaye JL, Sylla K, Camargo FW, Martel N, Foly K, Lo AC, Abiola A, Sow D, Ndiaye M, Ndiaye D, Ndao M, Gaye O. Accuracy of HRP2 RDT (Malaria Antigen P.f(R)) compared to microscopy and PCR for malaria diagnosis in Senegal. Pathog Glob Health. 2013;107:273–278. doi: 10.1179/2047773213Y.0000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui SL, Walter SD. Estimating the error rates of diagnostic tests. Biometrics. 1980;36:167–171. [PubMed] [Google Scholar]
- Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF. Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J Clin Microbiol. 2011;49:2946–2953. doi: 10.1128/JCM.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenberg JH, Tahita CM, Versteeg IA, Tinto H, Traore Coulibaly M, D’Alessandro U, Schallig HD, Mens PF. Evaluation of antigen detection tests, microscopy, and polymerase chain reaction for diagnosis of malaria in peripheral blood in asymptomatic pregnant women in Nanoro, Burkina Faso. Am J Trop Med Hyg. 2012;87:251–256. doi: 10.4269/ajtmh.2012.12-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian AH, Metzger WG, Mutschelknauss EJ, Kabagambe G, Langi P, Korte R, von Sonnenburg F. Reliability of malaria microscopy in epidemiological studies: results of quality control. Trop Med Int Health. 2000;5:3–8. doi: 10.1046/j.1365-3156.2000.00509.x. [DOI] [PubMed] [Google Scholar]
- Laban NM, Kobayashi T, Hamapumbu H, Sullivan D, Mharakurwa S, Thuma PE, Shiff CJ, Moss WJ, Research SAICoEfM. Comparison of a PfHRP2-based rapid diagnostic test and PCR for malaria in a low prevalence setting in rural southern Zambia: implications for elimination. Malar J. 2015;14:25. doi: 10.1186/s12936-015-0544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministère du Plan et Suivi de la Mise en oeuvre de la Révolution de la Modernité[MPSMRM], MdlSP, ICF International. Enquête Démographique et de Santé en République Démocratique du Congo 2013–2014. Rockville, MD, USA: MPSMRM, MSP et ICF International; 2014. [Google Scholar]
- Mwingira F, Genton B, Kabanywanyi AN, Felger I. Comparison of detection methods to estimate asexual Plasmodium falciparum parasite prevalence and gametocyte carriage in a community survey in Tanzania. Malar J. 2014;13:433. doi: 10.1186/1475-2875-13-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastri E, Bevilacqua N, Sane Schepisi M, Paglia MG, Meschi S, Ame SM, Mohamed JA, Mangi S, Fumakule R, Di Caro A, Capobianchi MR, Kitua A, Molteni F, Racalbuto V, Ippolito G. Accuracy of malaria diagnosis by microscopy, rapid diagnostic test, and PCR methods and evidence of antimalarial overprescription in non-severe febrile patients in two Tanzanian hospitals. Am J Trop Med Hyg. 2009;80:712–717. [PubMed] [Google Scholar]
- Okell LC, Ghani AC, Lyons E, Drakeley CJ. Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis. 2009;200:1509–1517. doi: 10.1086/644781. [DOI] [PubMed] [Google Scholar]
- Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- Schachterle SE, Mtove G, Levens JP, Clemens EG, Shi L, Raj A, Munoz B, Reller ME, West S, Dumler JS, Sullivan D, Team PP. Prevalence and density-related concordance of three diagnostic tests for malaria in a region of Tanzania with hypoendemic malaria. J Clin Microbiol. 2011;49:3885–3891. doi: 10.1128/JCM.01157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakely D, Elfving K, Aydin-Schmidt B, Msellem MI, Morris U, Omar R, Weiping X, Petzold M, Greenhouse B, Baltzell KA, Ali AS, Bjorkman A, Martensson A. The usefulness of rapid diagnostic tests in the new context of low malaria transmission in Zanzibar. PLoS One. 2013;8:e72912. doi: 10.1371/journal.pone.0072912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Juliano JJ, Trottman PA, Griffin JB, Landis SH, Kitsa P, Tshefu AK, Meshnick SR. High-throughput pooling and real-time PCR-based strategy for malaria detection. J Clin Microbiol. 2010;48:512–519. doi: 10.1128/JCM.01800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Messina JP, Hand CC, Juliano JJ, Muwonga J, Tshefu AK, Atua B, Emch M, Meshnick SR. Molecular malaria epidemiology: mapping and burden estimates for the Democratic Republic of the Congo, 2007. PLoS One. 2011;6:e16420. doi: 10.1371/journal.pone.0016420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Malaria Rapid Diagnostic Test Performance: Results of WHO product testing of malaria RDTs: Round 5 (2013) World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- World Health Organization. World Malaria Report 2015. World Health Organization; Geneva, Switzerland: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.