Abstract
The CASP experiment is a biannual benchmark for assessing protein structure prediction methods. In CASP11, RBO Aleph ranked as one of the top-performing automated servers in the free modeling category. This category consists of targets for which structural templates are not easily retrievable. We analyze the performance of RBO Aleph and show that its success in CASP was a result of its ab initio structure prediction protocol. A detailed analysis of this protocol demonstrates that two components unique to our method greatly contributed to prediction quality: residue–residue contact prediction by EPC-map and contact–guided conformational space search by model-based search (MBS). Interestingly, our analysis also points to a possible fundamental problem in evaluating the performance of protein structure prediction methods: Improvements in components of the method do not necessarily lead to improvements of the entire method. This points to the fact that these components interact in ways that are poorly understood. This problem, if indeed true, represents a significant obstacle to community-wide progress.
Keywords: CASP, free modeling, ab initio structure prediction, conformational space search, contact prediction, structure prediction pipeline
Introduction
In this paper, we report on the performance of RBO Aleph [1] in the free modeling category of CASP11 (held in 2014). CASP is a biannual experiment to evaluate the state of the art in the field of protein structure prediction. RBO Aleph ranked third among all automatic methods in the free modeling category. The focus of the free-modeling category are difficult protein targets for which homologous structures (templates) are hard to find with fold recognition methods or do not exist. Researchers often approach these difficult modeling targets with ab initio prediction methods that do not depend on such templates.
Ab initio protein structure prediction approaches assume that the native structure coincides with the global minimum of an energy landscape. Finding this minimum can be cast as a search problem. This search problem is extremely difficult, because the energy landscape is rugged and the high-dimensional conformational space is far too large for exhaustive search. Hence, state-of-the-art ab initio prediction methods use information from structural fragments to reduce the dimensionality of the search space and guide the search towards relevant regions (Rosetta [2], I-Tasser [3], QUARK [4], MULTICOM [5], FRAGFOLD [6]). Since information is highly valuable to guide search towards the most promising regions of conformational space, many methods focus on exploiting information sources, such as contacts (EV-Fold [7]). Other approaches leverage information from search space samples themselves, by analyzing the distribution of candidate protein structures (decoys) (MBS [8], NEFILIM [9]).
RBO Aleph includes two unique features that extract information from previously sampled decoys: EPC-map and model-based search. EPC-map [10] predicts residue–residue contacts from decoy structures. It leverages physico-chemical information about native-like properties in local contact networks and combines it with evolutionary information. Model-based search (MBS) [8] guides conformational space search by exploiting the funnel-like structure of the energy landscape. MBS builds a model of the energy landscape by analyzing the distribution of decoy structures. Our ab initio prediction protocol uses these two methods and employs ProSA [11] for decoy selection.
In our analysis of RBO Aleph, we compare our performance to other participating servers in the free-modeling category and investigate three above-average and three below-average predictions (measured by z-score) as examples of successes and failures of our method. Our analysis shows that our above-average predictions (z-scores > 0) originate from the RBO Aleph ab initio structure prediction protocol. To investigate which components of the ab initio contribute to our performance, we analyze conformational space search, predicted residue-residue contacts, and decoy selection.
Our analysis shows that MBS finds lower energy structures than Rosetta’s Monte-Carlo-based conformational space search algorithm [2], which translates into better structure predictions (higher GDT TS) in nine of of 18 cases. We show that using residue–residue contacts in combination with MBS increases the sum z-score of RBO Aleph by 28.8%. This suggests that MBS effectively leverages contact information.
Since our contact prediction (EPC-map) is an important component in RBO Aleph, we compare it with other contact prediction methods. For contact prediction, EPC-map (RBO Aleph) ranks second for medium+long-range contacts (sequence separation > 11) and fifth for long-range contacts (sequence separation > 23).
Finally, we analyze the improvement of the final prediction outcome (GDT TS of resulting first and best of five models) of the individual ab initio components—residue-residue contacts, conformational space search, and decoy selection—in the context of the entire RBO Aleph pipeline. Our data suggests that the performance of the pipeline depends on the interactions and dependencies between the components. These suggest that an entire pipeline cannot be improved by improving its components independently. To counteract this effect, early integration and continuous testing seem to be key factors for the development of effective protein structure prediction pipelines.
Methods
Pipeline Overview
Overview of RBO Aleph
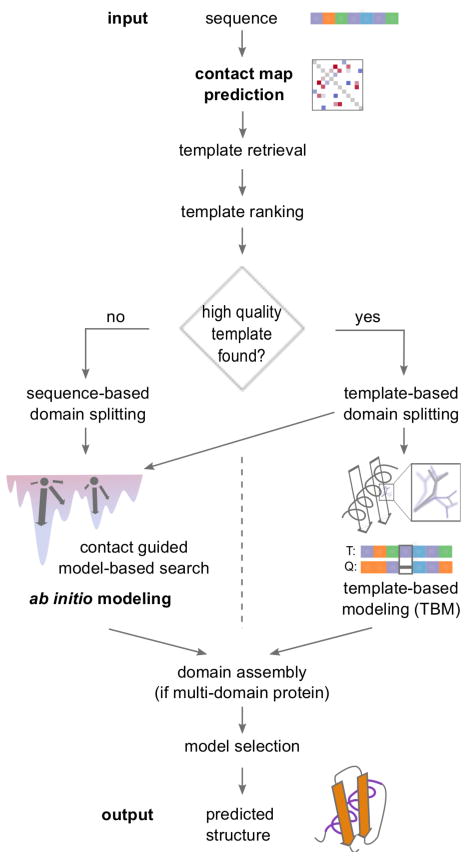
RBO Aleph[1] is a protein structure prediction pipeline that includes template-based modeling, contact prediction, ab initio prediction, domain boundary prediction and domain assembly. Figure 1 provides an overview of the pipeline. First, we describe the pipeline and then discuss the ab initio components in detail.
Figure 1.
Overview of RBO Aleph’s structure prediction pipeline used in CASP11. After predicting contacts with EPC-map, the server retrieves templates with HHsearch, LOMETS, RaptorX, and SPARKS-X. These templates get re-scored with a random-forest classifier. Depending on the existence of suitable templates, RBO Aleph runs template-based modeling with MODELLER or ab initio modeling with MBS guided by predicted contacts. The server ranks decoys sampled by MBS with ProSA and selects the five structures with highest rank as best predictions. RBO Aleph infers domain boundaries of longer sequences from templates or from sequence-based domain prediction with PPRODO and DomPro in the absence of suitable templates. After predicting the individual domains the server assembles them into a whole structure and submits the highest scoring models based on Rosetta energy.
Contact Prediction
We predict residue–residue contacts with EPC-map. Later in the pipeline, we use contacts to re-rank templates and to guide ab initio structure prediction (see section Contact Prediction by EPC-map for details).
Template Retrieval
We retrieve structural templates using the following threading methods: HHPred [12], LOMETS [13], RaptorX [14], and SPARKS-X [15]. Next, we re-score the templates using a random forest classifier [16, 17]. The classifier is trained on features that capture the consensus between the templates, scores of the threading algorithms, and the match between the predicted contacts and templates. Last, we select the best non-overlapping templates.
Domain Boundary Prediction
We split the target sequence into domains using the template alignments. Template-covered domains are assigned to template-based modeling. Uncovered domains, longer than 50 residues are submitted to ab initio prediction. If ab initio domains are larger than 250 amino acids, we split them recursively with a consensus score from PPRODO [18] and DomPro [19].
Modeling
We model template-based domains with MODELLER [20] and any uncovered terminal segments with RosettaRemodel [21]. Ab initio domains are modeled using a contact-guided model-based search (MBS) [8] (see Section Ab Initio Modeling of RBO Aleph).
Decoy Selection and Error Prediction
We score template-based models with the DOPE energy function [22] and ab initio decoys using Rosetta energy [23], decoy clustering [24] and ProSA’s knowledge-based potential [11] (see Section Decoy Selection in Ab initio Structure Prediction). We estimate the prediction error along the backbone with ModFOLD [25].
Domain Assembly
We generate around 200 permutations out of the selected decoys from each domain for assembly. We assemble the domains using Rosetta’s domain assembly protocol [26]. Finally, we select the five top scoring predictions by Rosetta energy[23]. Note that in the case of single domain targets, RBO Aleph submits the highest scoring decoys based on the DOPE energy or ProSA (see above).
Ab Initio Modeling of RBO Aleph
Ab initio prediction searches the native structure in a rugged energy landscape. Because of its size, the conformational space cannot be searched exhaustively [27]. Hence, exploiting knowledge about the search space is key to guide search towards regions likely to contain the native conformation. We exploit two information sources: First, we leverage knowledge about the physicochemical propensities in proteins to predict contacts. Second, we exploit knowledge about the funnel-like structure of the energy landscape to improve search.
In the following sections, we describe the three core components of RBO Aleph’s ab initio modeling pipeline: contact prediction by EPC-map, contact-guided model-based search (MBS), and decoy selection.
Contact Prediction by EPC-map
Residue–residue contact prediction identifies residue pairs that are close in space in the protein structure. Contacts can successfully guide the conformational space search if they are accurate enough [28].
Evolutionary algorithms identify co-evolution signals from multiple-sequence alignments to predict contacts, but require many homologous sequences [29, 30, 31]. However, proteins in the free-modeling (FM) category usually lack deep sequence alignments [32].
EPC-map [10] complements evolutionary information [33] with physicochemical information. While the evolutionary approach is accurate for proteins with deep alignments, the physicochemical approach can predict consistent contact maps, even for proteins without deep sequence alignments. This renders EPC-map an ideal contact prediction for FM targets.
EPC-map leverages physicochemical information encoded in the energy function: Low-energy decoys often contain native-like contacts even if their overall fold is wrong. EPC-map extracts local contact networks from low-energy decoy and encodes them with a graph. Graph-based features are used to differentiate native from non-native contacts using a trained support vector machine (SVM). EPC-map then combines the resulting contacts with the contacts predicted by GREMLIN [33].
RBO Aleph uses the contacts predicted by EPC-map to guide conformational space search towards biologically relevant regions (see Leveraging Contact Information to Guide Search).
Model-Based Search
Model-based search (MBS) [8] is a conformational search algorithm that actively guides search towards low-energy regions. MBS incrementally builds a model of the energy landscape, consisting of energy funnels derived from previously sampled decoys. MBS allocates the computational resources towards the most promising funnels, thereby guiding search towards low-energy regions.
The current implementation of MBS performs Monte-Carlo simulated annealing in six stages. The stages use different fragment libraries (9-mer fragments in the first five stages, and 3-mer fragments in stage six) and increasingly complex energy functions (similar to Rosetta’s ab initio protocol [2]).
In each stage, MBS performs the following steps: First, MBS clusters the sampled decoys into funnels with a heuristic clustering algorithm. Second, it refines the five lowest-energy conformations in each funnel with an all-atom energy function. Next, it scores each funnel using a minimization of the all-atom energy of the lowest energy decoy. Finally, MBS discards half of the lowest-scoring funnels, then allocates the computational resources to the remaining funnels in accordance with their energy scores. Decoys from these funnels are then used as starting points to start new minimization runs, leading to the decoys used for the next stage of MBS.
In CASP11, we used MBS to sample different numbers of decoys for each target depending on the target size and the available computational resources at the time (see Table S2 for a detailed listing of the number of sampled decoys).
Leveraging Contact Information to Guide Search
RBO Aleph guides conformational search by leveraging contacts in the form of distance restraints. The contact restraints are incorporated into the energy function as additional terms, thereby reducing the ruggedness of the energy landscape. RBO Aleph uses the top scoring 1.5L contacts ranked by EPC-map to guide MBS, where L refers to the length of the target domain sequence.
FM targets in CASP are generally difficult, which results in a significant number of false-positive contact predictions. In order for false positives not to negative affect structure prediction, we include the contact-based distances restraints into the energy function using a modified Lorentz potential. This potential does not penalize contact violations. Instead, it maximizes the number of satisfied contacts while allowing some constraints to remain violated without penalty [10]:
where di j denotes the distance between residues i and j in the decoy; the parameters l, u, and w refer to lower bound, upper bound, and half width, respectively. ci j is the weight for the contact between residues i and j and controls the maximum energy that the algorithm rewards to a satisfied contact. In CASP11, RBO Aleph used l = 1.5 Å, u = 8 Å and w = 1.0. We use a uniform contact weight of ci j =1.5 for all 1.5L contacts. Satisfied restraints get full reward, mildly violated ones still get small energy bonus, while strongly violated ones do not affect the energy. We use the same parameters (including the Lorentz potential) when using contacts with Rosetta in our analysis in this paper.
Decoy Selection in Ab initio Structure Prediction
During CASP11 we changed our decoy selection method from Rosetta energy [23] to decoy clustering [24], and finally to ProSA’s quality score [11]. The ProSA score was used for most of the CASP11 experiment. We compare the effect of all three decoy selection methods on our pipeline in this paper. We briefly describe herein the different methods.
ProSA is a knowledge-based potential used to assess the quality of structural models. ProSA uses statistics of the structures in the Protein Data bank (PDB) to identify errors in structural models. The Rosetta energy function is a physical/knowledge-based hybrid composite potential. Decoy clustering is a decoy selection technique based on the assumption that highly populated low-energy regions are more likely to contain near-native folds than small ones [34]. In our pipeline, we use the decoy clustering method Durandal [24] to group decoys by structural similarity. We then select the lowest-energy decoys (using Rosetta Energy) from the three largest clusters.
Data set
Our analysis is based on domains which a) have a native structure available at the time of writing this manuscript, and b) were modeled with the ab initio prediction protocol of RBO Aleph in CASP11. Since the focus of our paper is the analysis of our ab initio structure prediction protocol, we exclude from consideration the seven free-modeling (FM) domains that were modeled by RBO Aleph’s comparative modeling protocol. Furthermore, we excluded target T0831 from our analysis, because we did not submit a prediction for this target in CASP11. This results in 17 target proteins, split into 18 DIFadd FM domains according to RBO Aleph’s definition of domains and split into 20 FM domains according to the official CASP11 domain definition (see Table S1 in Supporting Information).
We use the 18 RBO Aleph assigned domains when analyzing the decoy sets predicted by MBS (Section Evaluation of Conformational Search with Model-Based Search) and the effect of contacts on the decoy ensembles (Section Impact of Predicted Contacts on ab initio Predictions). We use the official CASP11 FM domains when analyzing the final predictions of the full pipeline and in the analyses presented in Sections Performance of Contact Prediction in CASP11 and Evaluation of the Dependencies between Prediction Components of RBO Aleph.
Results and Discussion
We now present the outcome of our analysis of RBO Aleph [1]. We first compare our predictions to the participating servers in CASP11. Then, we select a set of six representative targets that led to above-average and below-average predictions. We discuss these targets because they highlight the successes and challenges of our ab initio protocol. Next, we analyze the ab initio methods unique to our pipeline: conformational search by model-based search (MBS)) [8] and contact prediction by EPC-map [10]. We analyze the performance of the conformational search strategy implemented in MBS [8] and compare it to state-of-the-art methods. Then, we analyze the performance of contact prediction by EPC-map [10] and its impact on our performance in structure prediction. Last, we analyze the interrelation between the components of the pipeline (conformational search, use of contacts, decoy selection) by comparing different pipeline configurations.
RBO Aleph’s Performance in CASP11
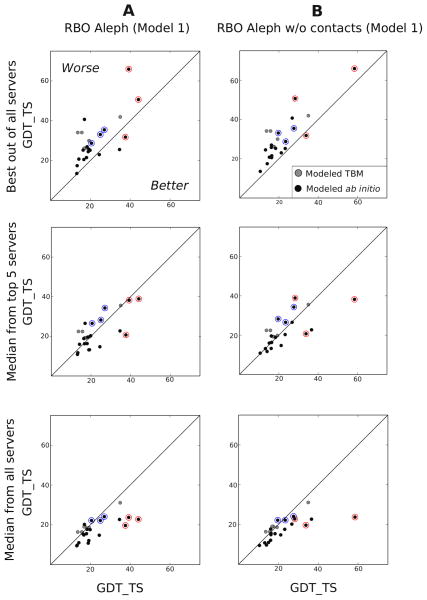
RBO Aleph ranked third among all automated CASP11 methods in the free-modeling category (as ranked by the assessor’s formula by the GDT TS z-scores criterion, computed on first submitted model, called model1 below). Figure 2A compares the performance of RBO Aleph to the other participating servers. We compare the quality (GDT TS) of model1 submissions by RBO Aleph to 1) the best submitted model1 2) median GDT TS of model1 submissions by the top 5 servers (excluding RBO Aleph) 3) median GDT TS of all model1 submissions. To study the effect of contacts on our predictions, we rerun our pipeline without the use of contacts and perform the same comparison as for RBO Aleph (Figure 2B, refer to Table S6 in the Supplementary Information for a complete list of targets and GDT TS values).
Figure 2.
Comparison of the GDT TS of first models by our pipeline (with and without the use of contacts) to the first models submitted by other servers. We compare our first models to (from above) the best first model submitted by all servers, the median GDT TS of the first models submitted by top 5 servers in the FM category (according to the website ranking and excluding RBO Aleph), the median GDT TS of the first models submitted by all servers (excluding RBO Aleph). FM Targets that we modeled using ab initio are in black, targets we modeled using template-based modeling are in grey. The targets discussed in the paper are in red (good predictions) and blue (bad predictions).
Our server consistently provides above-average predictions for the majority of the targets: twenty out of the 27 domains modeled by our server (See section Data set) had positive z-scores. We modeled 20 out of the 27 domains using ab initio modeling; the rest has been modeled using template-based modeling. Strikingly, 18 of the ab initio modeled domains had positive z-scores. In contrast, only two of the seven domains modeled with template-based modeling obtained positive z-scores (see Figure S1 in Supporting Information). This indicates that the success of our server in the free modeling category is grounded on its ab initio pipeline, hence we limit the analysis in this paper to targets modeled using ab initio.
We compare in Figure 2A the performance of RBO Aleph to the performance of other servers. The accuracy of our model1 predictions (GDT TS) is higher than the median GDT TS of all model1 submissions for 20 out of 27 domains, whereas it is higher than the median GDT TS of top five servers model1 submissions for eleven domains. Despite consistently providing above-average predictions, we only predict the best model1 for three domains. We discuss one of these targets (T0834 D1) in detail in the next section.
A unique feature of RBO Aleph is the use of contacts predicted by EPC-map in ab initio prediction (see Section Contact Prediction by EPC-map). We compare our prediction with and without the use of contacts to the other servers (Figure 2). Using contacts improves the prediction slightly, increasing the sum of GDT TS of all model1 submissions by 21 points. The use of predicted contacts increases the number of targets for which we rank first from one to three. Nevertheless, we note that even without using contacts, our pipeline performs considerably well compared to the top five predictors: higher GDT TS of model1 submissions than the median GDT TS of top five servers in eleven out of 27 cases. We hypothesize that the reason behind this performance lies in our conformational search algorithm Model-Based Search (MBS). We analyze this further in Sections Evaluation of Conformational Search with Model-Based Search and Evaluation of the Dependencies between Prediction Components of RBO Aleph.
To summarize, Figure 2 gives an overview on the strong performance of our pipeline in CASP11 and offer two preliminary insights: i) contacts predicted by EPC-map improves slightly the ab initio prediction of our pipeline ii) our conformational search algorithm MBS plays an important role in our performance in CASP11. In the remainder of this paper, we analyze the effect of residue-residue contacts, conformational search, and decoy selection on our performance and probe the interrelation between them.
Evaluation of Selected Free Modeling Targets
In the following section, we describe and discuss the prediction of targets that highlight the achievements and problems of our protein structure prediction pipeline. We select a set of six representative targets that led to above-average and below-average predictions (Colored in red and blue respectively in Figure 2). We compare our predictions to other server submissions in CASP11 and analyze the effect of decoy sampling (MBS), contact prediction (EPC-Map) and decoy selection (ProSA) on the overall prediction quality. We focus our analysis on these components because they are the main components of our ab inito prediction protocol and, in the case of MBS and EPC-map, are the unique features of our pipeline.
High quality predictions
We analyze three targets for which RBO Aleph produced models with high GDT TS and z-score, compared to the other servers (Figure 3).
Figure 3.
High quality prediction produced by RBO Aleph. RBO Aleph submitted better models for the targets T0834 D1, T0837 D1, and T0855 D1 than most of the other servers. The arrows on the right side indicate the performance of the individual components (such as decoy, contacts prediction, and decoy selection). An upwards pointing arrow states good performance whereas a downwards pointing arrow indicates bad performance. Although the individual components did not always perform well, the overall prediction quality was high.
T0834 D1
T0834 is a two-domain target with the non-sequential domain T0834 D1. RBO Aleph incorrectly modeled the full-length target as a single domain. Nevertheless, our algorithm accurately predicted the first half of the first domain containing three helices and a beta-sheet motif. The GDT TS score of the submitted model1 is 37.9. Our five submissions rank at positions 1–3 and 5–6. Overall, all ab initio components (decoy sampling, contact prediction, and decoy selection) performed well for this target. The sampled decoy set had a median GDT TS of 30.2, which is higher than the GDT TS of model1 of all other servers except for RaptorX-FM (GDT TS: 32.07). The contact prediction accuracy of 45% for L/5 long+median range contacts only improved the quality of the best decoys (median GDT TS without/with contacts: 31.6/30.2; median GDT TS without/with contacts of top 20% decoys: 34.6/38.1). All five submitted models have higher GDT TS values than the median GDT TS of the decoy set indicating that the decoy selection was successful. In this example the high performance of the individual components results in a high quality prediction despite the incorrect domain parsing.
T0837 D1
RBO Aleph has predicted the fourth best model1 for the single domain target T0837 D1 (GDT TS: 39.7). The gap between the best submitted model (QUARK, GDT TS: 65.7) and the second best submitted model (RaptorX-FM, GDT TS: 45.45) is high (~20 GDT TS values). The top 20% submissions were able to identify and model the seven helices of the target. However, the winning model (QUARK, model1) has the best arrangement and packing of these helices. The median GDT TS of RBO Aleph’s decoy set (GDT TS: 43.6) is higher than the overall performance of the other servers (median GDT TS: 24.8, median GDT TS of the top 20% submissions: 40.2). The best decoy in our decoy set (GDT TS: 64.9) is of similar quality to the best submission for this target (QUARK, GDT TS: 65.7). In addition, the high accuracy of predicted long+medium range contacts (91.7) improved the quality of the decoy set (median GDT TS without/with contacts: 39.1/40.6). Among the five submitted models of RBO Aleph, only model5 has an higher GDT TS value than the median GDT TS of the decoy set. This means that the decoy selection method was not able to produce models with higher quality than expected by a random selection. To summarize, the quality of the sampled decoys together with the accuracy of predicted contacts produced a high quality decoy set that helped to compensate the low performance of decoy selection for this target.
T0855 D1
Model1 for the single domain target T0855 D1 has an GDT TS score of 38.9 and is ranked seventh among the servers. Contact prediction performed well (L/5 long+medium range contact accuracy 91.3%) but did not have a positive effect on the quality of the decoys set (median GDT TS without/with contacts: 37.4/36.3). However, using contacts slightly improved the quality of the top 20% decoys (median GDT TS without/with contacts: 41.0/41.7). The performance of the decoy sampling was of medium quality; the median GDT TS of decoy set (36.3) is higher than the median GDT TS of all servers (23.0). Selected decoys have higher GDT TS than the median GDT TS of the predicted decoy ensemble (median GDT TS of the five submitted models/decoy ensemble: 37.83/36.3), however no model from the top 20% sampled decoys was submitted. The GDT TS of our model1 (GDT TS: 38.91) is 10 units lower than the best submitted model1 (MULTICOM-NOVEL, GDT TS: 51.09). To conclude, contact prediction had a high performance but decoy sampling and selection only showed a medium performance.
Low quality predictions
We analyzed three targets for which RBO Aleph yields models with low GDT TS and z-score in comparison to the other servers (see Figure 4).
Figure 4.
Low quality prediction produced by RBO Aleph. RBO Aleph has submitted worse models for the targets T0761 D2, T0824 D1 and T0834 D2 than most of the other servers. The arrows on the right side indicate the performance of the individual components such as decoy sampling, contacts prediction and decoy selection. An upwards pointing arrow states good performance whereas a downwards pointing arrow indicates bad performance. Although the individual components often performed well, the overall prediction quality of the models is weak.
T0761 D2
T0761 D2 is part of a target consisting of two free modeling domains (see Table S1 in Supplementary Information) which we modeled as a single ab initio domain. Our pipeline predicts a badly packed structure for T0761 D2. Overall, none of the components of the ab initio protocol had a high performance for this target. The predicted long-range contacts have a low accuracy (13.04%). The predicted decoy ensemble is of a higher quality (median GDT TS: 24.5) than the median of server submissions (median GDT TS: 21.9) but worse than the median submissions by the top 20% predictors (median GDT TS: 26.3). The selected decoys have lower GDT TS than the median GDT TS of the predicted decoy ensemble (model1 GDT TS 23.7 / median GDT TS decoy ensemble 24.5). The two best servers for model1 predictions for this target (SAM-T08-server (29.65), ZHOU-SPARKS-X (GDT TS: 28.98) are template-based prediction methods. Furthermore, the server with the best submission for this target used templates to model the structure (BAKER-ROSETTASERVER, GDT TS: 38.27, model4). This might suggests that modeling this target using TBM might have improved our predictions.
T0824 D1
Contact prediction (L/5 long+medium range contact accuracy: 9.1), decoy sampling and decoy selection for the single-domain target T0824 D1 performed poorly resulting in a model that is not able to capture the beta-sheet arrangement of the native structure. The median GDT TS of the decoy set is 23.8 and hence slightly higher than the median GDT TS of the server predictions (22.6) but worse than the median of the 20% best server submissions for model1 (27.6). Decoy selection was not able to select a decoy among the 20% best decoys, but selected a decoy as model1 that is better than the median of the decoy set. One reason for the bad performance is the wrong predicted helix on position 65 to 71 and the missing beta-sheet motif in the core of the structure. Overall, the submissions for this target are of medium quality and the best server submission has a GDT TS value of 33.0 (MULTICOM-CONSTRUCT).
T0834 D2
T0834 is a two domain protein target with a non-sequential domain T0834 D1. RBO Aleph incorrectly modeled the full-length target as one single domain. While RBO Aleph predicted part of the first domain (T0834 D1) with high quality (see above), the prediction quality (model1) for target T0834 D2 is low. For this target the decoy selection performed badly. Although the accuracy of the long range contact prediction was high (long range accuracy: 29.4) and decoy sampling produced a high quality decoy set (median GDT TS of decoy set 31.1), the submitted decoys had far worse quality (median GDT TS of submitted decoys: 30.5; GDT TS of model1: 27.0) than expected by chance (median GDT TS of decoy set 31.1). Overall, the median GDT TS of the decoy set is higher than the median GDT TS of the top 20% predictors for this target (30.8). To sum up, contact prediction and decoy sampling performed well, but decoy selection submitted models with worse quality than expected by chance.
Summary of single targets analysis
Analyzing the target predictions separately provides some insight that might explain the performance of our server. However, the pipeline’s complexity and the interplay of its components makes any conclusion uncertain. With this in mind, we summarize here our observations from this analysis. In the remainder of the paper, we test in more details some of the arising hypothesis.
Our main observation is that the most contributing factor for the success of a prediction is the quality of the sampled decoy set. This in turn is determined by the method used for conformational search and the quality of the guiding contacts. However, to our best knowledge, it is still unclear which properties makes contacts useful in structural prediction; accuracy alone is not a indicator for the effect of distance constraints on ab initio prediction performance [35].
Secondly, we observe that decoy selection can be decisive for the success of a prediction. The case of T0834 D2 provides a very good demonstration of its effect: we predicted accurate contacts and sampled decoys of good quality (relative to the median GDT TS of other servers submissions), yet decoy selection failed by selecting models with lower GDT TS than the median GDT TS of sampled decoys.
In the remainder of the paper, we analyze thoroughly the prediction components we have discussed here. First, we analyze the properties of our conformational search method MBS and compare it to the widely used Monte Carlo search. Then, we attempt to measure the effect of predicted residue-residue contacts on our prediction. Last, we analyze the effects of the different components on a pipeline level and investigate their interrelation.
Evaluation of Conformational Search with Model-Based Search
Model-Based Search (MBS) is a conformational space search algorithm used in our ab initio prediction protocol. MBS’s approach leverages information accumulated during search about the energy landscape. The accumulated information is used to steer search towards low-energy regions. The algorithm incrementally builds a model of the energy landscape using the sampled decoys. This model groups decoys into energy funnels and uses them to guide search towards low-energy regions. For method details please refer to section Model-Based Search.
RBO Aleph’s ab initio prediction protocol uses predicted contacts to guide the conformational search in MBS. In this section, we attempt to analyze the performance of our conformational search in isolation, therefore, we evaluate the performance of MBS without the use of contacts.
In CASP11, we used MBS to predict decoy ensembles with sizes ranging from 1000 to 5000 decoys depending on the target length and the available computational resources (see Table S2 for a detailed list). We used ProSA to select five models for final submissions. We evaluate in this section both the quality of the decoy ensembles predicted by MBS and the quality of the final predictions selected by ProSA.
Evaluation of MBS’s Search Strategy
We evaluate here the performance of the conformational search strategy of MBS by analyzing the quality of the predicted decoy ensembles. Ideally, we would like to compare the decoys predicted by MBS with the decoy ensembles from all other servers. Unfortunately, we do not have access to the predicted decoy ensembles for other servers. Therefore, we evaluate our conformational search strategy by comparing it to the Monte Carlo (MC) strategy, as implemented in Rosetta 3.4. Please note that the predictions by Rosetta 3.4 are different to the BAKER-ROSETTASERVER, which uses a more sophisticated protocol. We evaluate next predictions made with MBS and Rosetta (both without use of contacts) and use the same parameters when running both methods: The same energy function, fragment library, and a corresponding amount of computational resources (see Table S2 for a detailed list of number of generated decoys). By using the same parameters, we ensure that the two methods only differ in their conformational search strategies.
We compare the performance of MBS and Rosetta to sample decoys in terms of 1) the median energy and GDT TS of the decoy set, 2) the median of the 10% lowest-energy decoys, and 3) the highest GDT TS value in the decoy sets. Our aim is to determine if the MBS search strategy succeeds in producing lower energy decoys than Rosetta, and if the MBS search strategy succeeds in producing more accurate decoy ensembles than Rosetta (measured in mean GDT TS and best GDT TS in predicted decoys).
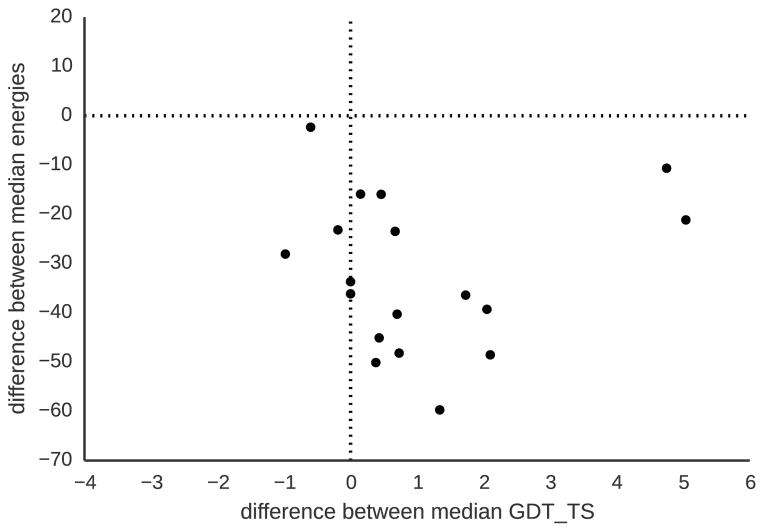
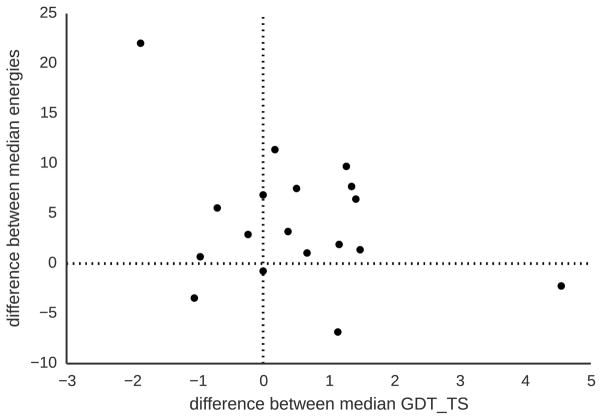
Figure 5 compares the median GDT TS and energies of decoy sets predicted by both methods (See Figure S4 for a detailed plot for all targets decoy sets). MBS finds lower-energy decoys for all domains. The median energy difference over all ensembles between both methods is −35 Rosetta energy units. These findings indicate that MBS successfully allocates resources towards low-energy regions and effectively minimizes the energy.
Figure 5.
Comparing energies and GDT TS of decoys obtained by MBS and Rosetta’s Monte Carlo-based search. The median energy and GDT TS were calculated for each free-modeling domain using all decoys. Negative numbers at the y-axis indicate how much lower the energy of MBS-modeled decoys is compared with Rosetta-modeled decoys. The x-axis shows the difference of the median GDT TS of MBS compared to Rosetta obtained decoys. MBS consistently finds lower energy decoys (−35 lower median energy over all in the ensembles) and better quality decoys (0.565 higher median over all decoys).
We investigate whether the reduced energy translates into decoys with higher GDT TS. We find that, with the exception of five targets, MBS decoy sets have higher median GDT TS than Rosettas Monte Carlo-based search. Over all decoys, MBS produces predictions with median GDT TS that is slightly higher than Rosetta’s Monte Carlo-based search (0.565 median GDT TS difference). These results show that MBS predicts decoy ensembles that have higher median GDT TS and lower median energy than Rosetta. However, this does not mean that MBS is a better search strategy than Rosetta: usually, only the lowest-energy decoys are considered for final model submissions. Therefore, we compare next the performance of the two methods on the lowest-energy decoys.
We analyze the GDT TS and the energy of the 10% lowest-energy decoys (See Figure S2 for a comparison of GDT TS for different energy thresholds). We find that for half of the domains (9/18), MBS produces low-energy decoys with better median GDT TS values than Rosetta. The median GDT TS of MBS and Rosetta are virtually identical (median difference between GDT TS: 0.035). This is in contrast to the analysis of the decoy ensembles which showed that MBS finds better decoys. We compare the best decoys (highest GDT TS) predicted for each target (without regard to energy). We find that Rosetta predicts at least one decoy with better or similar GDT TS than MBS for all targets. On the other hand, we find that for 17 out of 18 domains, MBS finds at least one decoy with lower energy than Rosetta.
These results show that MBS predicts decoy sets with lower energy and higher GDT TS in median than Rosetta. However, when considering the top 10% lowest energy decoys, both method performs similarly in terms of GDT TS. These results suggest that there might be a discrepancy between the energy function and the quality of the decoys: finding lower energy decoys does not always translate into finding decoys with a higher GDT TS. We believe that the reason for this behavior lies in the exploitative strategy employed by the algorithm: MBS focuses search on only a small number of regions of the conformational space. We hypothesize that the addition of new information sources, for instance residue-residue contacts could help steer search from non-native low-energy regions and thereby improve prediction. We evaluate the effect of contacts in section Impact of Predicted Contacts on ab initio Predictions
Impact of Model-Based Search on Final Predictions
The results from the last section show that MBS succeeds in predicting decoys ensembles with similar quality to Rosetta, one of the state-of-the-art methods for ab initio prediction. We evaluate here the models selected by ProSA (without use of contacts) and compare them to other servers in CASP11. We find that the sum of GDT TS z-scores for model1 decreases from 29.5 (with contacts) to 22.9 (without contacts) but this does not impact the ranking of our server. We explain this by the large difference between the z-score of our server and the preceding (Zhang-server, sum of z-scores: 37.9) and the following server (MULTICOM-CLUSTER, sum of z-scores: 20.9) in the ranking. These results confirm that the conformational search strategy of MBS performs comparably to other state-of-the-art prediction methods in the field.
Evaluation of Contact Prediction with EPC-map
Predicted residue-residue contacts by EPC-map are an integral component of RBO Aleph. RBO Aleph uses contacts in two ways: 1) The match between contacts and templates is used as a feature in template selection; 2) The resulting distance constraints are used to direct conformational search in ab initio prediction. In this section, we analyze the results of our contact prediction and its impact on ab initio structure prediction with model-based search (MBS).
Performance of Contact Prediction in CASP11
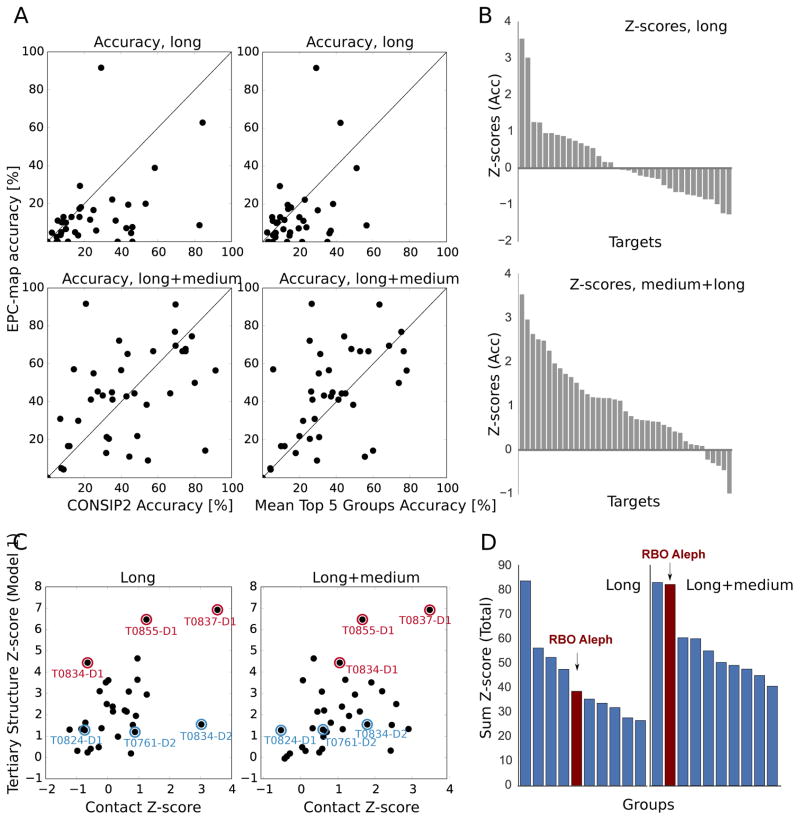
Figure 6 shows a detailed analysis of contact prediction by RBO Aleph in CASP11, which is performed with EPC-map [10]. The analysis in this section is based on official data from CASP11 (data taken from http://www.prediction-center.org/). Thus, we report here the L/5 ( L is the length of the domain) contact prediction performance on all free-modeling domains that were processed by RBO Aleph.
Figure 6.
Overview of residue-residue contact prediction results of EPC-map, the contact prediction method employed by RBO Aleph. All results in this Figure refer to the top L/5 predicted contacts. All targets in this Figure are free-modeling targets. A: Comparison of long (sequence separation > 23) residues and long+medium (sequence separation > 11) range contact prediction accuracy of EPC-map with CONSIP2, the winning method in CASP11, and the per-target mean accuracy of the top 5 contact predictors of CASP11. B: z-scores for individual targets of RBO Aleph. C: Correlation between contact prediction z-scores (Acc) with tertiary structure prediction z-scores (average of MAMMOTH and Dali scores, model 1) of RBO Aleph. The z-scores weakly correlate with a pearson/spearman rank correlation coefficients are 0.51/0.47 and 0.45/0.48, for long and long-medium range contacts, respectively. This indicates that EPC-map contacts at least partially contribute to the performance of RBO Aleph in the FM category. Targets that we discuss later in this paper are marked red (RBO Aleph submitted good predictions, compared to the other groups) and blue (RBO Aleph submitted poor predictions, compared to the other groups). D: Ranking of the top ten contact prediction methods by the sum of z-scores (total, i.e. Acc+Xd). RBO Aleph (EPC-map) ranks second for medium+long range contacts and fifth for long-range contacts.
First, we compare contact prediction accuracy by RBO Aleph with the leading methods that participated in CASP11. In this analysis, we compare RBO Aleph with CONSIP2 (the best contact prediction method in CASP11), and with the mean accuracy obtained by the other top five groups excluding RBO Aleph (see Figure 6A). Since we only include free-modeling targets that have been submitted by RBO Aleph and the compared method, the averages might slightly differ from the results on the CASP11 website.
Overall, the average contact prediction accuracy of RBO Aleph for medium+long range contacts (sequence separation >11 amino acids) is 43.2% for the L/5 top scoring contacts (39 evaluated domains). RBO Aleph’s medium+long range contact prediction accuracy is comparable with CONSIP2 (mean accuracy 43.0% for 39 evaluated domains) and higher than the mean medium+long contact prediction accuracy of the top 5 methods (37.3%). On the other hand, RBO Aleph’s long-range contacts are much less accurate than the contacts from CONSIP2 and lower than the mean contact accuracy of the top 5 methods (mean accuracy 13.7/25.5/18.8 for RBO Aleph/CONSIP2/mean top 5 group performance).
Next, we analyze the performance of RBO Aleph, compared with all other groups in CASP11. This is quantified by per-target z-scores. Intuitively, a positive z-score indicates that RBO Aleph performed better in contact prediction than the average CASP11 predictor. Figure 6B shows the z-scores for all target domains. Tables S4 and S5 show accuracy, Xd values, and z-scores for each target domain. RBO Aleph reaches positive z-scores for 17/36 domains (47.2%) for long-range contacts and 34/39 (87.1%) for medium+long-range contacts.
We analyze the correlation between RBO Aleph’s contact prediction performance and RBO Aleph’s ab initio structure prediction performance (see Figure 6C). Contact prediction and tertiary structure prediction z-scores correlate moderately (pearson/spearman rank correlation coefficients are 0.51/0.47 and 0.45/0.48, for long and long-medium range contacts, respectively). This indicates that EPC-map contacts contribute, at least partially, to RBO-Aleph’s tertiary structure prediction performance of CASP11 free-modeling targets.
Overall, RBO Aleph ranks 5th for long-range contacts and 2nd for medium+long range contacts in CASP11 (Figure 6D).
In summary, medium+long range contacts from RBO Aleph are comparable in accuracy with the top groups in CASP11. Long range contacts are less accurate than from the best CASP11 groups, but we find a moderate correlation between long-range contact z-score and tertiary structure prediction z-score, which indicates some contribution of EPC-map to the free-modeling performance of RBO Aleph.
Impact of Predicted Contacts on ab initio Predictions
In this section, we evaluate whether EPC-map contacts improve tertiary structure prediction. We evaluate first the impact of incorporating contacts on the median GDT TS of the predicted decoy ensembles. Second, we evaluate the effect of contacts on the final predictions submitted by the pipeline.
RBO Aleph uses contact information from EPC-map to guide MBS runs. In contrast to evolutionary contact prediction methods, EPC-map is specifically designed to have low dependency on available homologous sequences [10]. We suspect that this could provide an advantage in the context of CASP since deep alignments are often not available for hard CASP targets [36].
To assess the influence of contact prediction on ab initio structure prediction, we rerun structure prediction with MBS, albeit without EPC-map contacts. We implement EPC-map contacts in MBS as described in section Leveraging Contact Information to Guide Search.
The results of this analysis are summarized in Figure 7. Overall, the impact of contact information on the decoy quality is modest (median GDT TS improvement by 0.45). The improvement is more pronounced in the 10% low-energy ensemble (median GDT TS improvement of 1.07). Refer to Figure S3 in Supporting Information for detailed energy-GDT TS scatter plots.
Figure 7.
Analysis of improvements in ab initio structure prediction by comparing MBS with and without the help of contacts. The median energy and GDT TS were calculated for each free-modeling domain using all decoys. Negative numbers at the y-axis indicate how much lower the energy of decoys from contact-guided MBS is compared to MBS without contact guidance. The x-axis shows the difference of the median GDT TS of MBS with contacts and MBS without contacts. Contact-guided prediction runs produce decoys with slightly higher quality (0.45 higher median GDT TS over all decoy ensembles).
One remarkable example of contact-guided search is T0837 for which the median GDT TS improves by 4.55 (3.94 for low-energy ensemble). We also have precise contacts for T0806 but this did not result in pronounced sampling improvements (median GDT TS improvement: 1.27), presumably due to the size (256 residues) and the complex topology of the target. We could not identify the reasons why low contact precision decreases decoy quality in some cases (T0761), while improving the decoy quality in other cases (T0777 and T0832 D1). We analyze next the effect of the contacts on the submitted predictions to CASP11. We find that incorporating contacts into MBS increases the sum GDT TS of the model1 submissions by 21. This result in an increase of the GDT TS z-score of RBO Aleph from 22.9 to 29.5 (relative increase by 28.8%, absolute increase by 6.6%). However, the increase in z-score do not result in change of ranking in the FM-category.
To summarize, these results shows that contacts improves decoy sampling, however this effect is weak when averaged over all targets. Still, a small number of targets seems to benefit significantly from predicted contacts. Most importantly, the small correlation between the z-scores of contact and protein structure prediction (Figure 6C) seems to imply that contacts may provide an edge over other groups. Consequently, contacts may be most important in the context of CASP which rewards the relative and not the absolute performance in prediction.
Impact of Domain Boundary Prediction
Our pipeline predicts domain boundaries using an algorithm based on template alignments and a consensus of state-of-the art prediction methods (see Domain Boundary Prediction). In the course of CASP11, we only split three free modeling targets into multiple domains. In comparison, the official assignments counts seven free modeling targets containing multiple domains (see Table S1 in Supporting Information). We predicted accurate domain boundaries (within 40 residues from the official assignment) for only two of these: T0794 and T0808. The model1 prediction for these domains ranked fourth and seventh respectively. We modeled the remnant five multi-domain targets as single-domain targets. Despite the incorrect domain parsing, our predictions had positive z-scores (GDT TS) for all but one (T0834 D2) of these FM-domains (see Table S1 and Figure S1 in Supplementary Information). In summary, domain boundary prediction has been used scarcely in our predictions, we believe this suggests it has only played a minor role in our performance in CASP11.
Evaluation of the Dependencies between Prediction Components of RBO Aleph
The analysis of individual pipeline components (conformational search, contact prediction, decoy selection, etc) in separation is necessary to understand their potential of improving the outcome of prediction. But does a ”better” individual component always translate into an improved pipeline? Or do pipeline components interact in unexpected ways with each other?
We set out to investigate the interaction between individual components and quantify their net effect on the prediction quality of the final pipeline. We measure the pipeline quality by the GDT TS of the first or best-of-five selected models. In this analysis, we rerun all predictions using the different pipeline configurations and assess the changes in prediction quality. In particular, we are interested in the following three questions: What is the effect of contact restraints on performance? Does MBS search improve performance? Which decoy selection method is the best?
Recall that RBO Aleph uses the following component combination: Contact restraints from EPC-map, MBS as a search method, and ProSA for decoy selection. We compare all combination of pipelines made of the following con-figurations: Contacts/no contacts, MBS/Monte Carlo (Rosetta 3.4) as search methods and ProSA/Clustering/Rosetta Energy for decoy selection. We note here that we use the same paramters for the use of contacts in Rosetta (see Leveraging Contact Information to Guide Search for more details).
We start our analysis by running a head-to-head bootstrap analysis to compare the different configurations. This analysis provides robust comparisons between the different configuration pairs. Then, we inspect how the different configurations would have ranked in CASP11. Lastly, we discuss how the different components interrelate, what is their effect on the pipeline, and what lessons we learned.
How Do the Different Pipeline Configurations Compare to Each Other?
On a sufficiently large set of proteins, it would suffice to simply compare the outcome (sum of GDT TS, mean GDT TS and median GDT TS) of different pipeline combinations. However, since we have a small data set of few free-modeling domains (20 official CASP11 domains, see Section Data set), the results could strongly depend on the used data set. To mitigate the effect of small data set size, we compare the different configurations by a head-to-head bootstrap analysis. In this analysis, we randomly remove five target domains for a pair of pipeline configurations and note which configuration has a higher mean GDT TS over the modified target set. This process is repeated 1000 times. The fraction of ”wins” indicates the relative performance of a configuration over another. This analysis reduces the effect of small data set size by sampling different data set compositions.
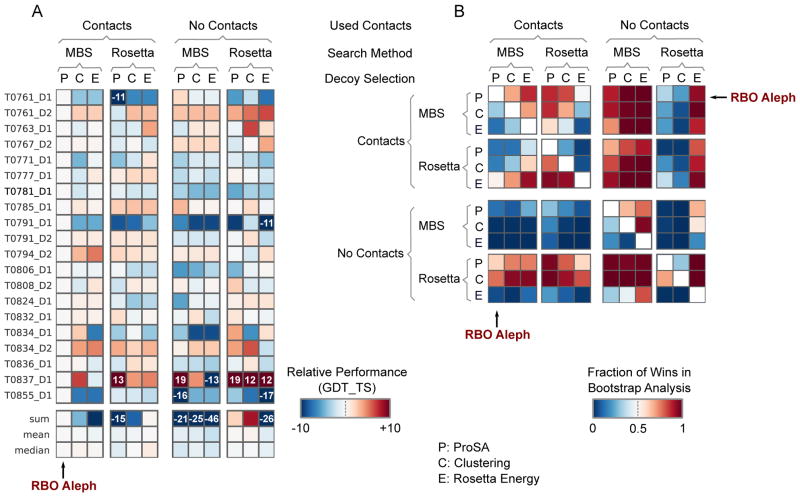
The results of the head-to-head bootstrap analysis are shown in Figure 8B. The target based performance of each pipeline relative to RBO Aleph is shown in Figure 8A.
Figure 8.
Effect of component combinations on the performance of the RBO Aleph pipeline (model 1). A: Target-based analysis of various component combinations. The colors of the heat map cells encode the difference in performance of a particular component combination, relative to RBO Aleph. B: Head-to-head bootstrap analysis of the different component combinations. In this analysis, we randomly remove five target domains for a pair of component combinations. The method with higher mean GDT TS scores a ”win”. We repeat this process 1000 times for each component combination pair. The colors of the heat map cells encode the fraction of ”wins” of a particular method combination over another one.
The head-to-head comparison indicates that RBO Aleph is an effective pipeline that is only slightly outperformed by No Contacts/Rosetta/ProSA (RBO Aleph fraction of wins: 0.36) and No Contacts/Rosetta/Clustering (RBO Aleph fraction of wins: 0.25). Notably, RBO Aleph is more effective than any configuration using contacts. However, we find that differences in performance are generally small. The worst performing contact-guided combination is Rosetta with ProSA for decoy selection, with a sum/mean/median ΔGDT TS of −14.7/−0.7/−1.3, relative to RBO Aleph (Figure 8A).
We also compare RBO Aleph to pipeline variants without contact restraints. Contact restraints improve the quality of the first models that result from MBS search (for example, sum/mean/median ΔGDT TS of −21.2/−1.1/−1.8 for MBS with ProSA). We also find that Rosetta without contacts and either ProSA (sum/mean/median ΔGDT TS of 2.3/0.1/−0.3) or Clustering (sum/mean/median GDT TS of 8.4/0.4/−0.1) as a decoy selection method perform slightly better than the RBO Aleph configuration. However, Rosetta does not perform better if Rosetta Energy is used to select decoys.
How Would Have the Different Pipelines Ranked in CASP11?
After comparing the performance of different pipeline compositions, we investigate whether different configurations would have changed our rank in CASP11. We perform a rank stability analysis to measure the robustness of a particular pipeline composition on the final CASP11 ranking. In this analysis, we randomly remove five target domains and compute the resulting rank. We rank the servers by the sum of z-scores (based on GDT TS) of the first submitted model (model 1). We repeat this process 1000 times. We also include the submitted models of FM targets that RBO Aleph modeled by template-based modeling, but should have been modeled by ab initio modeling. This results in 27 analyzed, official target domains.
Table 1 summarizes the results of this analysis. Most component combinations do not impact the rank of RBO Aleph. All configurations with a lower rank involve not using contacts. These include i) all configurations based on MBS without contact restraints (mean rank 3.5–6.0) and ii) the configuration Rosetta no contacts and decoy selection using the Rosetta energy (mean rank 3.8). This result indicates again that the predicted contacts by EPC-map are helpful.
Table 1.
Impact of component combinations on CASP11 ranking of RBO Aleph. For each component combination, we randomly remove five target domains and compute the rank among server groups on FM targets by the sum of z-scores (GDT TS, model 1). We repeat this process 1000 times and report the mean, best, and worst rank. MBS without contacts results in a lower rank, so does No Contacts/Rosetta and Energy for decoy selection. For all other methods, the rank is remarkably stable. This indicates that contact restraints benefit MBS calculations.
| Used Contacts | Search Method | Decoy Selection | Mean Rank | Best Rank | Worst Rank |
|---|---|---|---|---|---|
| Contacts | MBS | ProSA | 3.0 | 2 | 6 |
| Contacts | MBS | Clustering | 3.0 | 2 | 6 |
| Contacts | MBS | Energy | 3.0 | 2 | 6 |
| Contacts | Rosetta | ProSA | 3.1 | 3 | 8 |
| Contacts | Rosetta | Clustering | 3.0 | 2 | 7 |
| Contacts | Rosetta | Energy | 3.0 | 2 | 5 |
| No Contacts | MBS | ProSA | 3.5 | 3 | 9 |
| No Contacts | MBS | Clustering | 4.1 | 2 | 9 |
| No Contacts | MBS | Energy | 6.0 | 3 | 10 |
| No Contacts | Rosetta | ProSA | 3.0 | 2 | 8 |
| No Contacts | Rosetta | Clustering | 3.0 | 2 | 4 |
| No Contacts | Rosetta | Energy | 3.8 | 3 | 10 |
Across all other method combinations, the rank of RBO Aleph is remarkably stable. This might be explained by the relatively high z-score difference between RBO Aleph (sum z-score: 29.5) and the next higher ranked server (Zhang-Server[37] sum z-score: 37.9), and the next lower ranked server (MULTICOM-CLUSTER[38], sum z-score: 20.9).
We repeat the analysis with the best out of five models (see Table S3 in Supporting Information). In this case, Rosetta with contact restraints and ProSA is the winning component combination (mean rank 5.0). We attribute this finding to Rosetta’s exploratory sampling strategy that infrequently produces higher GDT TS decoys that are then selected by decoy selection. In this analysis too, using contact restraints improve the ranking of the pipelines: The best combination using contacts ranks 5.0 while the best configuration not using contacts only ranks 5.5 (mean rank over 1000 trials).
Discussion of Pipeline-Level Analysis
Overall, the rank of RBO Aleph remains remarkably stable along the investigated dimensions: Contact restraints, search methods, and decoy selection. Certainly, there are more components in RBO Aleph that might have contributed to our CASP11 rank, such as domain boundary prediction and assembly, decision whether or not to model a protein with ab initio methods, secondary structure prediction, and fragment selection for ab initio structure prediction. It is beyond the scope of this paper to analyze all of these elements.
We discuss in this section the three main findings on the interaction of the components on the pipeline level. Please recall that this analysis is based on 20 official CASP11 free-modeling domains (because not all structures where available to us at the time of the analysis), which is about half of the domains that have been officially evaluated in CASP11. Therefore, our findings might not hold for the entire CASP11 free-modeling experiment and do not perfectly explain the performance of RBO Aleph. Nevertheless, this analysis represents our best effort to understand the intricacies of structure prediction pipelines. However, our results are at least true for the analyzed data set and our findings might be similar when other target sets are investigated.
RBO Aleph leverages contacts effectively
We find that contact restraints clearly improves the performance when MBS is used as a search method (Figure 8). A possible explanation is that contact restraints complement the search strategy of MBS.
Recall that MBS leverages observations from the energy landscape to steer search towards promising regions. Briefly, MBS leverages sampled decoys to build a model of the energy landscape, then directs the search towards the most promising regions in that model (see Model-Based Search). This strategy succeeds in finding lower energy decoys (see Figure 5) but can lead to poor results when we exploit inaccurate information. In this case, MBS ends up steering the search towards the wrong regions of the space. As a result, despite having low energy, the accuracy of the predicted models will be low. This might be especially true for difficult modeling targets in CASP, where search might easily mislead by local minima or errors in the energy function. EPC-map contacts include additional information about the native state and guide MBS towards regions containing native-like conformations. Thus, contact restraints effectively counteract the weakness of MBS to overly exploit information from the energy landscape. Contact information effectively smooths the energy landscape and in this more ”funnel-like” energy landscape, the MBS search heuristic performs much better. Since MBS seems to effectively leverage contact restraints effectively, it might particularly benefit from future improvements in contact prediction.
Pipeline Analysis Reveals Other Effective Component Configurations
The pipeline analysis reveals configurations that are as effective or slightly better than RBO Aleph. In particular, we find that Monte Carlo sampling (implemented in Rosetta) without contacts and ProSA/Clustering for decoy selection performs slightly better than MBS. We note here that the difference in performance is minimal and does not affect the rank of the pipeline in CASP11.
We suspect that the exploratory sampling used in Rosetta’s Monte Carlo search, coupled with effective decoy selection, provide an advantage in the CASP context: Free-modeling targets in CASP are very difficult modeling cases. For these targets, MBS might not identify the correct funnels and therefore can not correctly guide conformational search. In addition, the accuracy of contact prediction for CASP targets is low in some cases and can therefore guide search towards regions without native-like structures. In these cases, the exploitative behavior of MBS has a detrimental effect. In contrast, broad sampling with Monte Carlo explores the energy landscape without being too much biased by potentially wrong information. On the other hand, Monte Carlo search in Rosetta leverages contacts less effectively than MBS(see Figure 8). We believe that a successful search strategy should effectively balance the exploitation of prior knowledge (energy function information, contacts) in search with the exploration of the conformational space. MBS and Monte Carlo search seem to fall on opposite ends of the exploration-exploitation spectrum and a method that unites the strength of both approaches should result in improved structure prediction.
The Effect of Decoy Selection Strongly Depend on the Pipeline Configuration
Our last finding is that certain combinations of components perform well in a structure prediction pipeline and others do not. Notably, our analysis confirms the finding of Kosciolek and Jones [31] that, for contact-guided predictions, decoy selection based on energy is superior to Clustering.
We believe this has important implications on the general approach of analyzing structure prediction systems. We note that individually assessing single components of our pipeline would have led to different and sometimes contradicting conclusions regarding the performance of our pipeline. For example, had we only used Clustering as a decoy selection method, we would have concluded that contact restraints improve prediction by MBS but hurt predictions made by Rosetta. On the other hand, had we analyzed the components individually, we would have come to the conclusion that contact restraints improve conformational space search for MBS and Rosetta.
Clearly, the systematic analysis of pipeline configurations reveals that improvements in components of the method do not necessarily lead to improvements of the entire method. This counter intuitive finding suggests that the components interact in ways that are currently poorly understood. This points to a possible fundamental problem in evaluating and developing protein structure prediction methods.
Our finding, if true, suggests that a practitioner that aims to build and improve a structure prediction pipeline, cannot simply incorporate a seemingly improved component, because it might negatively interact with other pipeline components. This is a significant obstacle, because individual components might —in the most extreme case— only effective in the context in which they were originally developed. This limited compatibility of structure prediction components might severely hinder community-wide progress and understanding the interactions between components might be a critical to advance the field.
Conclusion
We presented an analysis of the results obtained by our server RBO Aleph in the free-modeling (FM) category during CASP11. We compared the results of our pipeline to all participating servers and investigated which parts of our pipeline have contributed to our performance. We found that the success of RBO Aleph was the result of the ab initio modeling protocol. Hence, we analyzed the performance of the unique methods in this protocol: Conformational space search by model-based search (MBS) and contact prediction by EPC-map.
In our analysis, MBS consistently finds lower-energy decoys than the Monte-Carlo-based sampling algorithm of Rosetta. However, in only half of the cases the MBS conformational space search method also leads to decoy ensembles of higher GDT TS (median GDT TS of all decoys). This means that conformational space search consistently finds lower-energy decoys without increasing the structural accuracy of the sampled decoys. This finding may mean that there are deficiencies in the employed energy functions, hindering progress in protein structure prediction.
We showed that MBS benefits from additional information contained in residue–residue contacts. This additional information may in fact counteract deficiencies in the energy function, leading MBS towards promising regions of conformational space in spite of these. However, Rosetta’s Monte-Carlo-based search, which in contrast to contact-guided MBS relies heavily on exploration during search, infrequently samples higher GDT TS decoys than MBS. This suggests that a central remaining problem in conformational space search is to adequately balance exploration of the search space and the exploitation of available information. We believe that addressing this problem in future research would result in improved conformational space search strategies.
We also analyzed the interactions of the individual ab initio components (conformational search method, use of contacts, decoy selection method) in the context of the entire structure prediction pipeline. We find that components interact in unexpected ways. An improved component does not necessarily lead to improved results of the entire pipeline. If this observation also holds true for pipelines other than RBO Aleph, it would point to a significant impediment of the community-wide effort to advance structure prediction. We believe that understanding the effects of component interactions on the overall performance of a prediction pipeline may be vital to enable progress in the field.
Supplementary Material
Acknowledgments
This work was supported by the Alexander-von-Humboldt Foundation through an Alexander-von-Humboldt professorship sponsored by the German Federal Ministry for Education and Research (BMBF), http://www.humboldt-foundation.de/web/start.html; and by the National Institutes of Health [grant number 1R01GM076706].
Footnotes
Conflict of interest statement.
None declared.
References
- 1.Mabrouk M, Putz I, Werner T, Schneider M, Neeb M, Bartels P, Brock O. RBO Aleph: leveraging novel information sources for protein structure prediction. Nucleic Acids Res. 2015:gkv357. doi: 10.1093/nar/gkv357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Meth Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y. I-TASSER server for protein 3d structure prediction. BMC Bioinf. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins: Structure, Function, and Bioinformatics. 2012;80(7):1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Eickholt J, Cheng J. MULTICOM: A multi-level combination approach to protein structure prediction and its assessments in CASP8. Bioinformatics. 2010;26(7):882–888. doi: 10.1093/bioinformatics/btq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DT. Predicting novel protein folds by using fragfold. Proteins: Structure, Function, and Bioinformatics. 2001;45(S5):127–132. doi: 10.1002/prot.1171. [DOI] [PubMed] [Google Scholar]
- 7.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3d structure computed from evolutionary sequence variation. PloS one. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunette T, Brock O. Guiding conformation space search with an all-atom energy potential. Proteins: Struct, Func, Bioinf. 2008;73(4):958–972. doi: 10.1002/prot.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrestha R, Zhang KY. Improving fragment quality for de novo structure prediction. Proteins: Structure, Function, and Bioinformatics. 2014;82(9):2240–2252. doi: 10.1002/prot.24587. [DOI] [PubMed] [Google Scholar]
- 10.Schneider M, Brock O. Combining physicochemical and evolutionary information for protein contact prediction. PLoS ONE. 2014;9(10):e108438. doi: 10.1371/journal.pone.0108438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins: Struct, Func, Bioinf. 1993;17(4):355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with hhpred. Proteins: Struct, Func, Bioinf. 2009;77(S9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng J, Xu J. RaptorX: Exploiting structure information for protein alignment by statistical inference. Proteins: Struct, Func, Bioinf. 2011;79(S10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011;27(15):2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit–learn: Machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 17.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 18.Sim J, Kim SY, Lee J. PPRODO: Prediction of protein domain boundaries using neural networks. Proteins: Struct, Func, Bioinf. 2005;59(3):627–632. doi: 10.1002/prot.20442. [DOI] [PubMed] [Google Scholar]
- 19.Cheng J, Sweredoski MJ, Baldi P. DOMpro: Protein domain prediction using profiles, secondary structure, relative solvent accessibility, and recursive neural networks. Data Min Knowl Discov. 2006;13(1):1–10. [Google Scholar]
- 20.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 21.Huang PS, Ban YEA, Richter F, Andre I, Vernon R, Schief WR, Baker D. RosettaRemodel: A generalized framework for flexible backbone protein design. PLoS ONE. 2011;6(8):e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen M-y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leaver-Fay A, OMeara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache RA, Lyskov S, et al. Scientific benchmarks for guiding macromolecular energy function improvement. Meth Enzymol. 2013;523:109. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berenger F, Shrestha R, Zhou Y, Simoncini D, Zhang KYJ. Durandal: Fast exact clustering of protein decoys. J Comput Chem. 2012;33(4):471–474. doi: 10.1002/jcc.21988. [DOI] [PubMed] [Google Scholar]
- 25.McGuffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24(4):586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 26.Wollacott AM, Zanghellini A, Murphy P, Baker D. Prediction of structures of multidomain proteins from structures of the individual domains. Protein Sci. 2007;16(2):165–175. doi: 10.1110/ps.062270707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Bio. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 28.Kim DE, DiMaio F, Yu-Ruei Wang R, Song Y, Baker D. One contact for every twelve residues allows robust and accurate topology-level protein structure modeling. Proteins: Struct, Func, Bioinf. 2014;82(S2):208–218. doi: 10.1002/prot.24374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS ONE. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel M, Hayat S, Skwark MJ, Sander C, Marks DS, Elofsson A. PconsFold: Improved contact predictions improve protein models. Bioinformatics. 2014;30(17):i482–488. doi: 10.1093/bioinformatics/btu458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosciolek T, Jones DT. De novo structure prediction of globular proteins aided by sequence variation–derived contacts. PLoS ONE. 2014;9(3):e92197. doi: 10.1371/journal.pone.0092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue–residue contact prediction in CASP10. Proteins: Struct, Func, Bioinf. 2014;82(S2):138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balakrishnan S, Kamisetty H, Carbonell JG, Lee SI, Langmead CJ. Learning generative models for protein fold families. Proteins: Struct, Func, Bioinf. 2011;79(4):1061–1078. doi: 10.1002/prot.22934. [DOI] [PubMed] [Google Scholar]
- 34.Shortle D, Simons KT, Baker D. Clustering of low-energy conformations near the native structures of small proteins. Proc Natl Acad Sci USA. 1998;95(19):11158–11162. doi: 10.1073/pnas.95.19.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DT, Singh T, Kosciolek T, Tetchner S. Metapsicov: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics. 2015;31(7):999–1006. doi: 10.1093/bioinformatics/btu791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monastyrskyy B, D’Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins: Structure, Function, and Bioinformatics. 2014 Feb;82:138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015:gkv342. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng J, Li J, Wang Z, Eickholt J, Deng X. The MULTICOM toolbox for protein structure prediction. BMC Bioinf. 2012;13(1):65. doi: 10.1186/1471-2105-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.