Abstract
Appreciation for the role of epigenetic modifications in the diagnosis and treatment of diseases is fast gaining attention. Treatment of chronic kidney disease stemming from diabetes or hypertension as well as Wilm’s tumor will all profit from knowledge of the changes in the epigenomic landscapes. To do so it is essential to characterize the epigenomic modifiers and their modifications under normal physiological conditions. The transcription factor Pax2 was identified as a major epigenetic player in the early specification of the kidney. Notably, the progenitors of all nephrons that reside in the cap mesenchyme display a unique bivalent histone signature (expressing repressive epigenetic marks alongside activation marks) on lineage-specific genes. These cells are deemed poised for differentiation and commitment to the nephrogenic lineage. In response to the appropriate inducing signal, these genes lose their repressive histone marks, which allow for their expression in nascent nephron precursors. Such knowledge of the epigenetic landscape and the resultant cell fate or behavior in the developing kidney will greatly improve the overall success in designing regenerative strategies and tissue reprogramming methodologies from pluripotent cells.
Keywords: kidney development, histone code, epigenetics, nephrogenesis, nephron progenitors
Epigenetics
Epigenetics refers to mechanisms that prevail above and beyond the information encoded in the DNA sequence (the genetic code) in regulating gene expression. This is brought about by covalent modifications, not just to the nucleotides in the DNA, but also to the histone proteins that are closely associated with them. These modifications are heritable - although non-genetic in nature – and can be passed on during cellular divisions [1]. Epigenetic mechanisms are paramount to the development of metazoan organisms. Although the cells in these organisms have identical genotype, by virtue of epigenetic mechanisms, they diversify during the course of development to form highly specialized tissues and organ systems.
Despite the heritable nature of epigenetic modifications they are reversible [2]. Covalent modifications such as methylation, acetylation, phosphorylation, sumoylation and ubiquitination together with the enzyme mediators of these modifications constitute the major players. The DNA within the nucleus is organized into a complex structure called chromatin. The chromatin is formed of a repeating structural unit: the nucleosome. Each nucleosome has a protein core formed of histone protein dimers (H2A, H2B, H3, and H4) wrapped by 146bp of double-stranded DNA. The tails of the histone proteins that extend out of the core are a hot bed for covalent modifications especially on the lysine (K) residues. Segments of chromatin bearing different modifications dynamically assume a “closed” (transcriptionally silent) or “open” (transcriptionally active) conformation, which allows the expression of lineage-specific genes in a spatial and temporal manner. The tightly packed, transcriptionally silent chromatin is called heterochromatin while the loosely packed, transcriptionally active chromatin is called euchromatin. Gene regulation at euchromatin regions is guided by epigenetic modifications to modular, cis-acting gene elements, e.g., promoters, enhancers, silencers, insulators, locus control regions. Apart from histone modifications, the methylation of cytosine nucleotides in the DNA itself, as well as non-coding RNAs is included as part and parcel of the epigenetic regulatory signature.
Epigenetic DNA modifiers
Several of the genetic polymorphisms related to complex traits (e.g. blood cell phenotypes) and various disease outcomes (e.g. diabetic kidney disease development) reside within the non-coding regions of the genome [3]. Although non-coding, these regions comprise cell-type specific gene regulatory components, such as enhancers. The consortium ENCODE (ENCylcopedia Of DNA Elements) was created to identify cell type-specific gene regulatory elements including new transcript variants and transcriptional units using various human cell lines [4, 5].
The promoters of genes have regions enriched in cytosine and guanine that are referred to as CpG islands (CGI). The CGIs are 300–3000bp stretches that may be methylated on cytosines. Methylation of cytosine nucleotides can also occur outside of the CGI [6]. It is estimated that there are 27,000 CGIs in the human genome [7]. Despite having a high G+C content, the vast majority of the promoter-associated CGIs are unmethylated, as is characteristic of the promoters of constitutively expressed genes and 40 % of tissue-restricted genes [8, 9]. The lack of methylation renders the chromatin transcriptionally permissive. On the other hand, the proportion of hypermethylated CGIs is estimated at 9 – 25 % and localizes to regions distal to promoters. Hypermethylated CGIs are a hallmark of inactivated X-chromosomes, imprinted genomic regions and improperly silenced genes in neoplastic tissues. Cytosine methylation typically conforms to a transcriptionally repressed state [10] and is mediated by DNA methyl-transferases (DNMTs). With the exception of imprinted DNA segments, fertilization removes all pre-existing cytosine methylation signatures [11]. In humans, DNMT3 (A and B) mediates de novo methylation, while DNMT1 is engaged in the perpetuation of established methylation states during cell divisions. Elevated levels of cytosine methylation correlate with a more differentiated state. Therefore, CGIs normally unmethylated in the germline are methylated in somatic tissues. In embryonic stem cells methylated CGI-promoters have been found on developmental genes such as Rhox2 [12].
Epigenetic non-coding RNA modifiers
Long and short non-coding RNAs (ncRNA) are also believed to be significant in the epigenetic regulation of chromatin states [13–16]. This is best exemplified by the ncRNA, Xist, which is involved in random X-chromosome inactivation. It does so by recruiting Polycomb group proteins like Ezh2 which mediates methylation of lysine 27 residue on histone3 tails (H3K27me3) causing epigenetic silencing of the affected Xchromosome [16, 17].
Epigenetic Histone marks and modifiers
Chromatin organization and regulation of gene function rely on a unique combination of post-translational modifications to histone tails - the “Histone code” [18]. The “histone code” serves as a form of encrypted information about the nature and position of specific post-translational modifications within the regulatory elements in a genome. In an effort to understand the epigenetic signature of the genome as a whole (epigenomics), several consortia have been established. Among them are ENCODE (mentioned earlier), Roadmap Epigenomic Mapping Consortium (REMC), and International Human Epigenome Consortium; all conceived to provide a sweeping profile of the epigenomic landscape from several primary and cultured human cells and tissues [19–21]. Synthesis of data from these large-scale epigenome projects have provided the chromatin “fingerprints” of active, poised and repressed gene regulatory elements, such as proximal or distal promoters and enhancers [3, 22]. Access to these datasets is available through the following web links: http://www.encode-roadmap.org or http://genome.ucsc.edu/ENCODE
It is important to appreciate that the chromatin landscape, although stable between cell divisions, is liable to change in response to environmental and metabolic stresses. Chromatin modifying enzymes are reliant on co-factors and substrates that are intermediates of cellular metabolism. Changes in cell metabolism can therefore impact the chromatin modifiers which in turn alter chromatin dynamics. Intra-uterine nutrient restriction was reported to alter the cytosine methylation profile and the expression of the Pdx1 gene of beta-pancreatic cells [21, 23]. Consequently, insulin secretion becomes impaired in these experimental models. Re-organization of the histone signature during in-vitro culture conditions has been reported by Zhu et al [21].
Histone modification patterns
Typically, H3K9me3 (tri-methylation of lysine 9 on histone 3) is a repressive mark found on heterochromatin. It renders the chromatin refractory to reprogramming during induced pluripotent stem cell (iPSCs) generation [24]. By contrast, active promoters feature tri-methylation of H3 Lysine 4 (H3K4me3) and acetylation of H3 Lysine 9 (H3K9Ac). Tri-methylation of H3 Lysine 36 (H3K36me3) is associated with the gene body of actively transcribed genes. Active enhancers have H3K4me1 and H3K27Ac marks while “poised” enhancers bear only the H3K4me1 mark [21, 25, 26].
Chromatin modifications direct cell lineage restriction
The establishment of the metazoan body plan is complex, requiring the correct spatial and temporal specification of cellular identities [27]. In the fly, the body segments are specified by the homeotic gene cluster whose regionalized expression is epigenetically regulated by two major complexes - Polycomb (PcG, repressive) and Trithorax (trxG, activating). The Polycomb complex methylates lysines 9 and 27 on histone H3, while the Trithorax complex methylates lysines 4 on histone H3. Thus, lineage commitment and the acquisition of positional identity are driven by epigenetic modifications that regulate chromatin structure, DNA accessibility and ultimately gene expression at developmental loci. Burney et al [28] compared the epigenetic status of lineage-regulated genes in pluripotent embryonic stem cells (ESCs) as they progressed toward multi-potency (neural stem cells) and finally committed to a neural cell fate. In the pluripotent state, the promoters of lineage-specific genes carry both H3K4me3 (activating) and H3K27me3 (repressive) modifications. This “bivalent” epigenetic status is believed to keep the genes transcriptionally silent although poised for expression in response to the appropriate differentiation cues. Therefore, in the neural progenitors while both epigenetic marks are retained on the promoters of neural developmental genes they soon resolve into “monovalent” H3K4me3 status in the early, immature neurons. However, following neuronal commitment the genes specifying non-neuronal fates are repressed either by the loss of “bivalency” or the retention of the repressive mark H3K27me3 on their promoters.
Maintenance of the adult neural stem cell niche in rodents and non-human primates (in baboons) relies on the chromatin repressive mark H3K9me3 [29]. Enrichment of H3K9me3 in the germinal niche of adult brain ventricles helps maintain cellular integrity by silencing genes that promote neuronal, hepatic and immunological fates. A study by Wen et al [30] found that broad domains of H3K9 di-methylation (H3K9me2) distinguish differentiated cells from ES cells. Typically, the percentage of the genome with H3K9me2 modification increases in differentiated tissues. In mammals about 30 % of the genome carries H3K9me2 modifications. These large organized chromatin K9 modifications (LOCKs) are essential to tissue-specific gene expression and are largely lost in human cancer cell lines.
Nephric lineage epigenetic landscape
The paired domain transcription factor, Pax2, is pivotal in the epigenetic specification of the definitive kidney (metanephros) from the caudal intermediate mesoderm. Responding to inductive signals, Pax2 recruits a histone H3K4 methyltransferase complex (PTIP-MLL complex) to nephric lineage genes. Pax2 associates with this complex by binding PTIP [31–33].
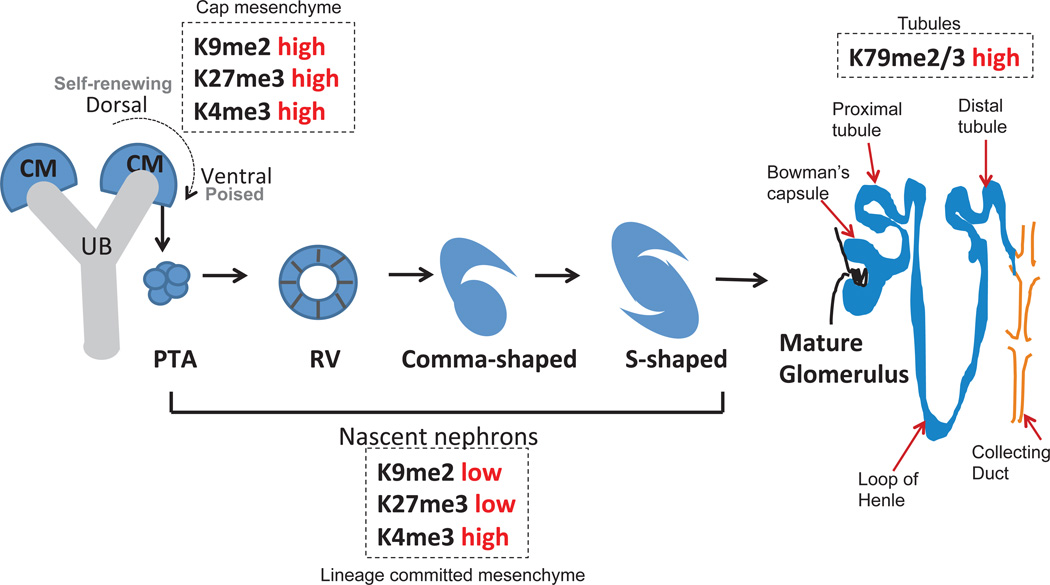
Two tissue lineages both arising from the caudal intermediate mesoderm form the metanephros. One is epithelial and forms the ureteric bud (UB) and ultimately collecting duct structures, while the other is a mesenchymal precursor to all the nephrons, stroma and parts of the vasculature (e.g., glomerular arterioles and juxtaglomerular cells). Both lineages undergo progressive fate restriction through reiterative, reciprocal signaling events. Subsets of the metanephric mesenchyme (MM) aggregate to form caps around each of the branching UB tips. The cap mesenchyme (CM) is a multipotent cell population representing progenitors of nephrons within the kidney [34, 35]. The CM cells have the ability to undergo self-renewal but remain “poised” to undergo differentiation in response to inductive Wnt signals from the UB [36]. The CM can be spatially demarcated into three sub-domains. A self-renewing population (Six2high/Cited1+/β-cateninlow) positioned immediately dorsal to the Ub tip; lateral to the Ub tip is the Six2high/Cited1−/BMPSMAD-responsive population which is primed for β-catenin-mediated differentiation and can be regarded as “poised”; further ventral to the Ub tip is Six2low/Cited1−/β-cateninhigh/Wnt4+ population which is committed to nephron formation (Figure 1).
Figure 1.
Bivalent chromatin code and its role in fate determination during nephrogenesis.
Initial forays into defining the chromatin signature in the nephron lineage were done in vitro. Whole genome Chip-seq and Chip-qPCR was used to compare two MM cell lines [37] which are broadly representative of the self-renewing Six2high/Wntlow cap cells (MK3 cell line) and nephron-fate committed Six2low/Wnthigh cells (MK4 cell line). With the onset of differentiation the MK3 cell line shows a loss of repressive histone marks, H3K9me2 and H3K27me3, and the retention of the activation mark, H3K4me3 on the promoters of nephrogenic lineage genes (e.g., Pax2, Pax8, Lhx1, Jag1 and Lef1). This conforms to observations in pluripotent cells wherein genes essential for lineage commitment carry permissive histone modifications and appear poised for differentiation. Accordingly, these genes typically possess a bivalent chromatin code: broad H3K27me3 enriched domains together with several peaks of H3K4me3 positioned around the transcriptional start site. Wnt signaling is the major trigger driving the commitment and differentiation events during nephrogenesis [35]. Wnt3a could substitute for Wnt9b in inducing nephrogenic gene expression in MK3 cells. In these assays Wnt-responsive genes gain β-catenin/H3K4me3 binding and lose Ezh2/H3K27me3 at the TCF/LEF binding sites [37, 38]. A departure from the H3K4/K27 bivalent code is seen in the pancreatic lineage gene Pdx1 in hepatoblasts [38]. Pdx1 silencing is accomplished by H3Kac/H3K27me3 instead. By contrast, the promoters of renewal genes, Six2, Osr1 and Eya1 show a gain of repressive marks and the loss of the activation histone marks.
Our laboratory has also examined the spatio-temporal expression of the histone methylation marks in nephron progenitors and their progeny during mid-organogenesis [39]. Notably, the self-renewing Six2+ nephron progenitors have higher levels of H3K9me2 and H3K27me3, both of which are associated with transcriptional repression of genes (Figure 1). The corresponding histone modifiers, G9a and Ezh2 show a similar distribution. Commitment of these cells to the nascent nephron fate coincides with the loss of these repressive marks. By contrast, the distribution of the activation mark, H3K4me3 does not show any spatial bias between nephron progenitors and nascent nephrons. Elevated expression of H3K9me2 was previously reported in mitotically active cells of the developing neural tube in mice [40]. This would suggest that repressive histone marks may be a signature of actively proliferating cells. Terminal epithelial differentiation of maturing nephrons is accompanied by an increase in H3K79me2/3 and its histone modifier, Dot1L. Therefore, the spatially-defined epigenetic modifications and their modifiers guide differential gene regulation during nephrogenesis under normal physiological states [41].
Biological processes such as cell proliferation, differentiation and survival in the developing kidney can also be regulated by chromatin modifiers affecting the acetylation status of specific gene loci. This is accomplished by two opposing classes of enzymes: (1) histone acetyl transferases or HATs which add acetyl groups to histone and nonhistone proteins, and (2) histone deacetylases (HDACs; Classes I-IV) that remove these modifications. In zebrafish, HDACs act to prevent renal progenitor expansion [42]. Using pharmacological inhibitors of HDACs we identified key signaling pathways being modulated during renal development in the mouse [43, 44]. These included the Wnt/β-catenin, TGFβ/Smad and PI3K-AKT pathway components. The renal genes under HDAC modulation included Pax2/Pax8, Sfrp1 and Gdnf. Widespread cell cycle arrest and apoptosis also impairs ureteric bud branching upon inhibition of HDAC function. Congruent with this data, conditional elimination of HDAC1 and HDAC2 from the ureteric bud lineage causes renal hypoplasia owing to hyper acetylation of the tumor suppressor p53 and impaired Wnt signaling [45]. Modulation of chromatin by HDACs is also known to affect cardiovascular, oligodendrocyte, epidermal and chondrocyte development.
Clinical relevance of epigenetics
Our expanding knowledge of epigenetics can serve diagnostic and therapeutic purposes. Epigenetic characterization of a pediatric tumor, Wilm’s tumor, which originates in the metanephric blastema, has been performed [46]. Using ChIP-Seq, this group mapped the promoter chromatin states of genes in this tumor relative to adult and fetal kidney. They delineated four gene sets: set 1 includes kidney-specific renewal genes (GDNF, SIX2, EYA1, OSR1) with broad domains of H3K4me3; set 2 comprising genes shared between Wilm’s tumor and fetal kidney. These include epigenetic modulators like JMJD2B, HDACs, JARID2, and components of the Polycomb repressive complex. These genes are believed to be relevant to lineage commitment and show H3K4me3 peaks at their promoters. Set 3 represents poised genes which are common to Wilm’s tumor and fetal kidney and harbor the bivalent code H3K4me3/H3K27me3. Set 4 enlists genes that have enrichment of H3K27me3, such as repressed genes. It is surmised that the persistence of the bivalent chromatin code on the promoters of genes essential for kidney development subverts normal development in the Wilm’s tumor.
Inhibitors of DNMTs (5’-azacitidine drug inhibits fibrosis in mice kidneys) or HDACs (treatment of experimentally-induced glomerulosclerosis and acute kidney injury) could have clinical relevance [47, 48]. However, owing to the pleiotropic effects of the available drugs, efforts to improve specificity of action is essential before any kind of human trials.
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health grants P50 DK096373-03 and P30GM103337.
Footnotes
Conflict of Interest
The authors of this article do not have any conflict of interest.
References
- 1.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S. ENCODE and our very busy genome. Nat Genet. 2007;39:817–818. doi: 10.1038/ng0707-817. [DOI] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y, Clarke D, Eastman C, Euskirchen G, Frietze S, Fu Y, Gertz J, Grubert F, Harmanci A, Jain P, Kasowski M, Lacroute P, Leng J, Lian J, Monahan H, O'Geen H, Ouyang Z, Partridge EC, Patacsil D, Pauli F, Raha D, Ramirez L, Reddy TE, Reed B, Shi M, Slifer T, Wang J, Wu L, Yang X, Yip KY, Zilberman-Schapira G, Batzoglou S, Sidow A, Farnham PJ, Myers RM, Weissman SM, Snyder M. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem. 2010;285:15111–15118. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic Development following Somatic Cell Nuclear Transfer Impeded by Persisting Histone Methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-typespecific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dressler The specification and maintenance of renal cell type by epigenetic factors. Organogenesis. 2009;5:73–82. doi: 10.4161/org.5.2.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burney MJ, Johnston C, Wong KY, Teng SW, Beglopoulos V, Stanton LW, Williams BP, Bithell A, Buckley NJ. An epigenetic signature of developmental potential in neural stem cells and early neurons. Stem Cells. 2013;31:1868–1880. doi: 10.1002/stem.1431. [DOI] [PubMed] [Google Scholar]
- 29.Foret MR, Sandstrom RS, Rhodes CT, Wang Y, Berger MS, Lin CH. Molecular targets of chromatin repressive mark H3K9me3 in primate progenitor cells within adult neurogenic niches. Front Genet. 2014;5:1–11. doi: 10.3389/fgene.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large organized chromatin K9-modifications (LOCKs) distringuish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dressler GR, Patel SR. Epigenetics in kidney development and renal disease. Trans Res. 2015;165:166–176. doi: 10.1016/j.trsl.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel SR, Ranghini E, Dressler GR. Mechanisms of gene activation and repression by Pax proteins in the developing kidney. Pediatr Nephrol. 2014;29:589–595. doi: 10.1007/s00467-013-2603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JS, Ma W, O'Brien LL, Chung E, Guo JJ, Cheng JG, Valerius MT, McMahon JA, Wong WH, McMahon AP. Six2 and Wnt Regulate Self-Renewal and Commitment of Nephron Progenitors through Shared Gene Regulatory Networks. Dev Cell. 2012;23:637–651. doi: 10.1016/j.devcel.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 37.McLaughlin N, Yao X, Li Y, Saifudeen Z, El-Dahr SS. Histone signature of metanephric mesenchyme cell lines. Epigenetics. 2013;8:970–978. doi: 10.4161/epi.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin "prepattern" and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin N, Wang F, Saifudeen Z, El-Dahr SS. In situ histone landscape of nephrogenesis. Epigenetics. 2014;9:222–235. doi: 10.4161/epi.26793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biron VL, McManus KJ, Hu N, Hendzel MJ, Underhill DA. Distinct dynamics and distribution of histone methyl-lysine derivatives in mouse development. Dev Biol. 2004;276:337–351. doi: 10.1016/j.ydbio.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 41.Adli M, Parlak M, Li Y, El-Dahr SS. Epigenetic States of nephron progenitors and epithelial differentiation. J Cell Biochem. 2015;116:893–902. doi: 10.1002/jcb.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Groh ED, Swanhart LM, Cosentino CC, Jackson RL, Dai W, Kitchens CA, Day BW, Smithgall TE, Hukriede NA. Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol. 2010;21:794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, Bellew C, Yao X, Stefkova J, Dipp S, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone Deacetylase (HDAC) Activity Is Critical for Embryonic Kidney Gene Expression, Growth, and Differentiation. J Biol Chem. 2011;286:32775–32789. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenberg SL, Chen S, McLaughlin N, El-Dahr SS. Regulation of kidney development by histone deacetylases. Pediatr Nephrol. 2011;26:1445–1452. doi: 10.1007/s00467-011-1796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Yao X, Li Y, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone deacetylase 1 and 2 regulate Wnt and p53 pathways in the ureteric bud epithelium. Development. 2015;142:1180–1192. doi: 10.1242/dev.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aiden AP, Rivera MN, Rheinbay E, Ku M, Coffman EJ, Truong TT, Vargas SO, Lander ES, Haber DA, Bernstein BE. Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell. 2010;6:591–602. doi: 10.1016/j.stem.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bechtel W, McGoohan S, Zeisberg EM, Muller GA, Kalbacher H, Salant DJ, Muller CA, Kalluri R, Zeisberg M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Beneden K, Mannaerts I, Pauwels M, Van den Branden C, van Grunsven LA. HDAC inhibitors in experimental liver and kidney fibrosis. Fibrogenesis Tissue Repair. 2013;6:1. doi: 10.1186/1755-1536-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]