Abstract
The inner surface of the retina contains a complex mixture of neurons, glia, and vasculature, including retinal ganglion cells (RGCs), the final output neurons of the retina and primary neurons that are damaged in several blinding diseases. The goal of the current work was two-fold: to assess the feasibility of using computer-assisted detection of nuclei and random forest classification to automate the quantification of RGCs in hematoxylin/eosin (H&E)-stained retinal whole-mounts; and if possible, to use the approach to examine how nuclear size influences disease susceptibility among RGC populations. To achieve this, data from RetFM-J, a semi-automated ImageJ-based module that detects, counts, and collects quantitative data on nuclei of H&E-stained whole-mounted retinas, were used in conjunction with a manually curated set of images to train a random forest classifier. To test performance, computer-derived outputs were compared to previously published features of several well-characterized mouse models of ophthalmic disease and their controls: normal C57BL/6J mice; Jun-sufficient and Jun-deficient mice subjected to controlled optic nerve crush (CONC); and DBA/2J mice with naturally occurring glaucoma. The result of these efforts was development of RetFM-Class, a command-line-based tool that uses data output from RetFM-J to perform random forest classification of cell type. Comparative testing revealed that manual and automated classifications by RetFM-Class correlated well, with 83.2% classification accuracy for RGCs. Automated characterization of C57BL/6J retinas predicted 54,642 RGCs per normal retina, and identified a 48.3% Jun-dependent loss of cells at 35 days post CONC and a 71.2% loss of RGCs among 16-month-old DBA/2J mice with glaucoma. Output from automated analyses was used to compare nuclear area among large numbers of RGCs from DBA/2J mice (n=127,361). In aged DBA/2J mice with glaucoma, RetFM-Class detected a decrease in median and mean nucleus size of cells classified into the RGC category, as did an independent confirmation study using manual measurements of nuclear area demarcated by BRN3A-immunoreactivity. In conclusion, we have demonstrated that histology-based random forest classification is feasible and can be utilized to study RGCs in a high-throughput fashion. Despite having some limitations, this approach demonstrated a significant association between the size of the RGC nucleus and the DBA/2J form of glaucoma.
Keywords: Retinal ganglion cells, image analysis, random forest-based classification, quantitative analysis, DBA/2J, glaucoma, Jun, nucleus size
1. Introduction
Retinal ganglion cells (RGCs) are the final output neuron of the retina; their axons transmit action potentials to the brain that encode all of the light and feature detection signals that our retinas can sense. Thus, RGCs are essential to normal vision and over a century of work has been done to better understand their basic biology. Because they are post-mitotic, loss of RGCs in disease leads to irreversible vision loss. RGCs can be damaged by a wide range of insults (Levin and Gordon 2002), several of which are very common, including glaucoma (Weinreb, Aung et al. 2014), Alzheimer’s disease (Dehabadi, Davis et al. 2014), diabetes (van Dijk, Verbraak et al. 2012), and forms of traumatic brain injury (Mohan, Kecova et al. 2013). The importance of RGCs in health and disease, coupled with their relative ease of accessibility, will continue to drive their study for many years to come.
Studies of RGCs, even in mouse models where extraneous variables can be minimized, face several challenges. It is currently estimated that there are likely up to 30 different sub-types of RGCs (Sanes and Masland 2015). There are several advantages to being able to distinguish and categorize these sub-types, and great progress has recently been made in utilizing computer-assisted approaches for using dendritic field morphologies to accomplish this on individual neurons (Sumbul, Song et al. 2014). Other challenges remain in the study of large populations of RGCs, two of which merit particular mention. First, in both health and disease, RGC density varies substantially. RGC density naturally varies ∼4-fold by anatomic position (Drager and Olsen 1981), and ∼2-fold by genetic background (Williams, Strom et al. 1996). Thus, most measurements of cell density reflect a substantial amount of pre-existing variation. Moreover, even within genetically identical strains of mice, there are relatively high degrees of individual-to-individual variation in RGC density (Williams, Strom et al. 1996), further exacerbating this issue. A second major challenge is that, in mice, RGCs are distributed across a heterogeneous environment in which displaced amacrine cells are approximately equal in number to RGCs, and a variety of other cell types are also present (Drager and Olsen 1981, Jeon, Strettoi et al. 1998, Schlamp, Montgomery et al. 2013).
Here, we set out to test the feasibility of a novel approach, based on computer-assisted detection of nuclei and classification by the random forest method, for studying RGC populations. The term “random forest” refers to an ensemble learning method that operates by building a multitude of decision trees, each built from random subsets of data describing the objects being classified, and ultimately assigning classes based upon the consensus of “votes” from all of the trees (Pedregosa, Varoquaux et al. 2011, Touw, Bayjanov et al. 2013); we have often found the random forest to be superior to other classification schemes in retinal image analysis (Niemeijer, Abramoff et al. 2011). In the inner retina, the nuclei of RGCs are morphologically distinct from those of other cells (Drager and Olsen 1981); as such, a random forest-based approach was expected to be able to make use of multiple histologically discernable features for inclusion in tree building to distinguish the RGCs from other cell types. To explore this possibility, we made use of RetFM-J, a semi-automated software module for the publicly available image processing program ImageJ (Abràmoff, Magalhães et al. 2004), that detects, counts, and collects quantitative data on 41 features of nuclei in the inner retina of H&E-stained whole-mounted retinas. Using these features, we developed a command-based software tool, RetFM-Class, which uses data from RetFM-J to perform random forest classification of cell type. Among initial tests of performance, we applied random forest classification to retinas of normal C57BL/6J mice, the most widely studied inbred strain of mice, as well as mice with controlled optic nerve crush (CONC) injury. We also studied retinas from DBA/2J mice with a naturally occurring form of glaucoma, and discovered that the mean nuclear size of surviving RGCs decreased in glaucoma. Modeling suggests that these glaucomatous changes occurred through a mechanism involving a subtle enlargement of all nuclei and preferential death of cells with the largest nuclei.
2. Materials and methods
2.1 Animal Husbandry
Inbred C57BL/6J and DBA/2J mouse strains (The Jackson Laboratory, Bar Harbor, ME) were housed at the University of Iowa Research Animal Facility, where they were maintained on a 4% fat NIH 31 diet provided ad libitum, housed in cages containing dry bedding (Cellu-dri; Shepherd Specialty Papers, Kalamazoo, MI), and kept in a 21°C environment with a 12-h light: 12-h dark cycle. Mice with conditional deletion of Jun in the retina, B6129;Jun(Six3-Cre) (abbreviated throughout as Jun−/− and Jun-deficient), were on a mixed C57BL/6J - 129 background and were generated by crossing mice with a floxed allele of Jun (Behrens, Sibilia et al. 2002) with mice expressing Cre recombinase under the control of the Six3 early retinal promoter (Furuta, Lagutin et al. 2000). Control littermate mice for experiments involving Jun contained the Six3-cre transgene and at least 1 copy of the wild-type Jun allele (abbreviated throughout as Jun+/? and Jun-sufficient). This colony was housed at the University of Rochester, where they were maintained on a 4% fat NIH 31 diet provided ad libitum, housed in cages containing dry bedding (Cellu-dri; Shepherd Specialty Papers, Kalamazoo, MI), and kept in a 21°C environment with a 12-h light: 12-h dark cycle. All experiments included mice of both genders. All mice were treated in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. All experimental protocols were approved by the Animal Care and Use Committee of the University of Iowa or the University of Rochester.
2.2 Microscopy and imaging of retinal whole-mounts for measurement of total retinal area
Retinas were collected for whole-mount preparations as previously described (Hedberg-Buenz, Christopher et al. 2015), with attention to keeping times for staining, mounting, and documentation as uniform as possible, and keeping experimental and control samples evenly dispersed between histologic batches. Whole-mounted retinas stained with H&E were scanned at 40x magnification with a digital whole-slide scanner (ScanScope CS, Aperio, Vista, CA). The total area of each retina was measured by outlining the sample, determining the stained area using the “Color Deconvolution v.9” algorithm, and then subtracting the area of the optic nerve. The areas of the central, mid-peripheral, and peripheral zones of eccentricity were calculated using circular regions of interest that divided each petal into approximately thirds. The length of each of the 4 petals (outer edge of optic nerve to edge of retina) was measured with the ruler tool and used to calculate average petal length. The central zone was defined as the area from the outer edge of the optic nerve to 1/3 of the average petal length, the mid-peripheral zone as the area from the edge of the central zone to 2/3 of the average petal length, and the peripheral zone as the area from the edge of the mid-peripheral zone to the edge of the tissue. The peripheral zone was calculated by subtracting the areas of central and mid-peripheral zones from the total retinal area.
2.3 Classification of nuclei with RetFM-Class
Stained whole-mount retinal preparations were imaged by light microscopy and images were subjected to analysis using RetFM-J as previously described (Hedberg-Buenz, Christopher et al. 2015). Quantitative measurements of 41 features relating to morphology and stain appearance (Supplemental Table 1) were computed for each extracted nucleus, so that further processing and classification could be performed. Included features were not specific to H&E, but rather included all relevant image metrics readily discerned by ImageJ for any region of interest. Feature information was applied to the development of a tool, called RetFM-Class, a command-line-based software tool (written in Python v. 2.7) that uses a random forest machine learning approach (Pedregosa, Varoquaux et al. 2011) to perform cell type classification. For training and leave-one-image-out evaluation of the random forest classifier, a set of retinal images was first manually classified, with every detected nucleus being assigned to 1 of 4 categories (RGC, DAC+, VEC, or ND) based on visual features previously described in the literature. RGC: Retinal ganglion cell. The nuclei of RGCs are lightly stained and have a prominent nucleolus (Drager and Olsen 1981, Levkovitch-Verbin, Quigley et al. 2001, Mavlyutov, Nickells et al. 2011); range broadly in size (∼30–160+ µm2) (Janssen, Mac Nair et al. 2013); and are distributed at a mean density of approximately 2,944 ± 262 to 3,817 ±160 cells/mm2 (Galindo-Romero, Aviles-Trigueros et al. 2011, Pang, Paul et al. 2013). DAC+: Displaced amacrine cells plus other cells with nuclei of similar appearance. These cells have small, dense, uniformly stained round nuclei and are present in large numbers (Drager and Olsen 1981). Other cell types detected in this population are astrocytes (also present in large numbers) (Bucher, Stahl et al. 2013), and pericytes (present in smaller numbers) (Cuthbertson and Mandel 1986, Chou, Rollins et al. 2014). The sizes of nuclei in displaced amacrine cells have been reported to range from 20–49 µm2 (Drager and Olsen 1981, Janssen, Mac Nair et al. 2013), and these cells constitute approximately 50% of neurons in the ganglion cell layer (Schlamp, Montgomery et al. 2013). Astrocytes and microglia are found in proximity to vessels (Jiang, Bezhadian et al. 1995, Checchin, Sennlaub et al. 2006), have small somas, and have mean densities of ∼800 cells/mm2 (Bucher, Stahl et al. 2013) and ∼178–500 cells/mm2 (Bosco, Steele et al. 2011, Liu, Li et al. 2012), respectively. Pericytes have densely stained ovoid nuclei ≤ 7 µm in diameter (Schallek, Geng et al. 2013), are inherent to the retinal vasculature, and are ∼2–5 times less common than endothelial cells (Cuthbertson and Mandel 1986, Chou, Rollins et al. 2014) with densities of ∼515 ± 94 cells/mm2 (Schallek, Geng et al. 2013). VEC: Vascular endothelial cells are particularly distinct, having densely stained, elongated nuclei (Cuthbertson and Mandel 1986, Schallek, Geng et al. 2013). ND: Nuclei with non-distinct features, not conclusively fitting into another category, were assigned to this category.
In an attempt to generate a training set with broadly generalizable performance, the training set consisted of 10 images (n=1,041 nuclei), taken from the following animals: 2 from glaucomatous (16 month) DBA/2J mice; 2 from pre-diseased (4 month) DBA/2J mice; 2 from young (2 month) unmanipulated C57BL/6J mice; 3 from young (4–7 month) unmanipulated Jun+/? mice; and 1 CONC- injured (35 days post-injury) C57BL/6J mouse. In 7 of these images, RetFM-J was utilized to identify all nuclei. The number of endothelial cells in the training set was boosted by analyzing 3 images by RetFM-J with an altered set of parameters such that only nuclei of vascular endothelial cells were identified. The 1,041 nuclei of the manually-classified training set included 339 RGC, 298 DAC+, 188 VEC, and 216 ND. The random forest classifier was evaluated using a leave-one-image-out strategy whereby, for each of the 10 images, the nuclei from the remaining 9 images were used to train the classifier (“learning” the relationship between the 41 features and the most likely cell type) and the trained classifier was used to automatically classify each nucleus in the “left out” image based on its features. Repeating this process 10 times provided the predicted cell type using the classifier for all the nuclei within the 10 images. Given the predicted cell types obtained from the classifier, the manual classifications and automated classifications were compared.
After the leave-one-image-out evaluation was performed, all 10 manually classified images were used as the training set so that the random forest classifier could be used to classify previously uncharacterized nuclei in images based only on the output of RetFM-J. The same training set was utilized in each subsequent analysis of unknown data. For inclusion in analyses of cell type classification, samples were required to pass an additional inclusion criterion. Specifically, for each retina assessed by RetFM-Class, the percentage of nuclei assigned to the ND category had to be within two standard deviations (SD) from the average for that particular strain (in the current studies, upper limits for inclusion ranged from 30.7 – 36.1%).
2.4 Tests of computational variability
To test the variability of RetFM-Class, data output from RetFM-J on 3 images from a C57BL/6J retina (1 central, 1 mid-peripheral, and 1 peripheral; all with manual exclusion of artifacts already performed) were classified repeatedly in 10 successive trials, with each trial also involving retraining of the classifier. To test content of the ND category, each instance of an ND classification in 4 auto-classified images from C57BL/6J mice was re-examined by 2 people and manually categorized using consensus.
2.5 Immunohistochemistry
Posterior eye cups were fixed in 4% paraformaldehyde, washed with PBS, and permeabilized with 0.3% Triton-X 100 in PBS (permeabilization buffer) overnight at 37°C.
For immunohistochemical labeling of BRN3A, posterior cups were further permeabilized with 0.3% Triton-X 100 in PBS at −80°C for 15 min. and then at room temperature for 30 min. All steps were carried out at room temperature, unless otherwise noted. Retinas were dissected and blocked with 2% normal donkey serum in permeabilization buffer for 3 h or overnight at 4°C. Retinas were incubated with an anti-BRN3A antibody (C-20, 1:200; Santa Cruz Biotechnology, Dallas, TX) in PBS with 2% normal donkey serum, 1% Triton-X 100, and 1% DMSO. Retinas were rinsed in permeabilization buffer, incubated with a donkey anti-goat Alexa488-conjugated secondary antibody (A11055, 1:200; Life Technologies, Madison, WI) in PBS with 5% normal donkey serum, 1% Triton-X 100, and 1% DMSO, rinsed in permeabilization buffer with DAPI, and then mounted using Aqua-Mount (Lerner, Pittsburgh, PA). Immunolabeled retinas from 4- and 22-month-old DBA/2J mice were imaged using fluorescence microscopy (BX-52 upright microscope, Olympus, Tokyo, Japan, with X-Cite 120 fluorescence illumination system, Exfo; Quebec, Canada). For each sample (n = 2 retinas per age, 1 retina from each of 2 mice), images (1360×1024 px, 0.07 mm2 image area) at 400X total magnification were collected from non-overlapping fields at each zone of eccentricity of the inner retina (n = 8 images total; 3 central, 3 mid-peripheral, 2 peripheral). Images were subsequently analyzed using the freehand drawing tool in ImageJ to manually trace and measure the area of all BRN3A–positive nuclei contained in the image set for each group (4-month-old, n = 2,369 nuclei; 22 month-old, n = 1,449 nuclei). For both groups, mean area of nuclei was expressed as a percentage relative to the mean area for nuclei from retinas of the 4-month-old mice.
For all other immunohistochemical labeling experiments, retinas were dissected and blocked with 1% BSA and 5% normal donkey serum in permeabilization buffer for 3 h or overnight at 4°C. Retinas were incubated with anti-GFAP antibody (SAB2500462, 1:100; Sigma-Aldrich Corp., St. Louis, MO), anti-COL4 antibody (AB756P, 1:200; Chemicon, Temecula, CA), anti-IBA1 antibody (#019–19741, 1:300; Wako, Richmond, VA), anti-AP-2α antibody (3B5, 1:100, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) or anti-RBPMS antibody (GTX118619, 1:200; GeneTex, Inc., Irvine, CA) in permeabilization buffer with 1% donkey serum and 1% BSA for 2–3 days at 4°C. Retinas were washed in PBS and then incubated with a 1:500 dilution of secondary antibody (biotin-conjugated donkey anti-rabbit and Alex568-conjugated donkey anti-goat). Retinas were washed in PBS and then incubated with Alexa488- or Alexa568-conjugated streptavidin (1:500). In the final wash, retinas were incubated in a 1:100 dilution of To-Pro-3 in PBS, and mounted with Aqua-Mount. Retinas not stained with To-Pro-3 were mounted using Vectashield medium containing DAPI (Vector Laboratories, Burlingame, CA). Retinas were imaged in standard confocal mode on a Leica SP8 STED Super Resolution Microscope (Leica Microsystems Inc., Buffalo Grove, IL). Individual retinas that were studied by both immunolabeling and H&E staining were labeled and imaged as described above, followed by the soaking off of coverslips by submersion of the slide in distilled water, H&E staining, and finally re-photographing the same retinal fields (using the vasculature to help guide registration) and conducting auto-classification with RetFM-Class. Correlations between auto-classifications and marker-based classifications were performed by: (1) examining 100 consecutive cells auto-classified as RGC and 100 consecutive cells auto-classified as DAC+ for the absence or presence of RBPMS labeling, (2) examining 100 consecutive cells auto-classified as DAC+ and 100 cells auto-classified as RGC for the absence or presence of AP-2α, and (3) examining 10 clearly delineated nuclei of each other type for similar correlations with GFAP, IBA1, or COL4 labeling.
2.6 Histology of optic nerves
In the analysis of glaucoma in DBA/2J mice, optic nerves were fixed and embedded in resin, sectioned, mounted, and stained with para-phenylenediamine (PPD), as previously described (Mao, Hedberg-Buenz et al. 2011).
2.7 Controlled optic nerve crush
Two groups of age-matched mice with conditional knock-out (Jun−/−) or at least one copy of the wild-type Jun allele (Jun+/?) were each subdivided into two treatment groups: those undergoing CONC, and naïve controls. For each mouse, both nerves underwent crush in the CONC group, whereas both nerves were un-manipulated in the naïve control group. CONC was performed as previously described (Libby, Li et al. 2005, Harder and Libby 2011), with optic nerves exposed and crushed using self-closing forceps (RS-5027; Roboz Surgical Instruments Co., Gaithersburg, MD) for 5 s, at a location 0.5 mm from the globe. Retinas were harvested 35 days after the procedure.
2.8 Modeling of glaucomatous changes to nuclear size
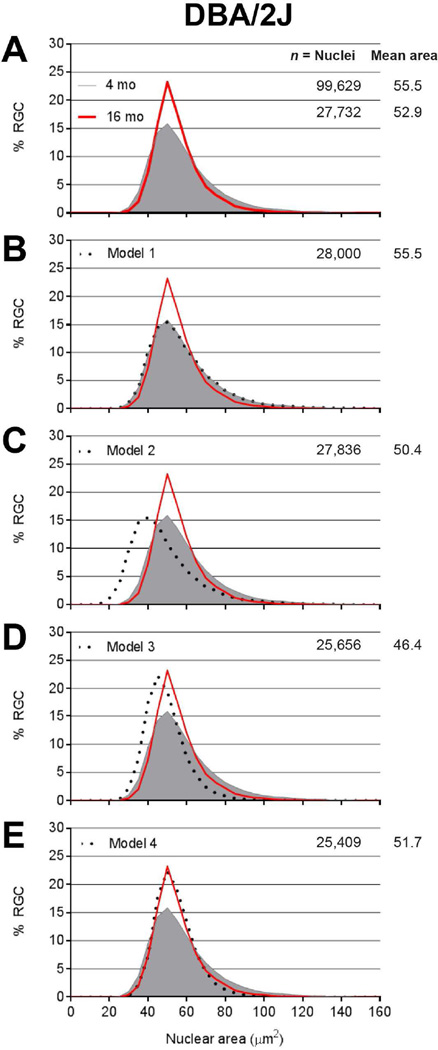
To model how various factors influence the relative frequency distribution of nuclear size in cells auto-classified in the RGC category, a spreadsheet of nuclear area from 4-month-old DBA/2J mice (n=99,629 nuclei) was manipulated to attempt matching of the observed changes in 16-month-old DBA/2J mice (n=27,732 nuclei), with particular attention to the reduction in cell number, mean size of nuclei in surviving cells, and shape of the curve. In the model shown in Figure 6E, 5 µm2 were added to the area of every nucleus, after which nuclei were randomly eliminated. In the models shown in Figure 6D and 6E, cells were randomly eliminated with half of the risk fixed at 50% and half of the risk continuously varying according to size rank ((rank/total) X 50). Thus, the smallest nuclei had a 50.001% chance of elimination and the largest had a 100.000% chance.
Figure 6. Subtle enlargement of all retinal ganglion cell nuclei and preferential death of cells with the largest nuclei in DBA/2J mice with glaucoma.
Frequency distribution plots of nuclear size for pre-disease 4-month-old DBA/2J (solid gray), glaucomatous 16-month old DBA/2J (red), and various models (dotted black) tested for whether a manipulation of the 4-month-old dataset could cause it to resemble the 16-month-old dataset. (A) Observed data for DBA/2J glaucoma showing reductions in cell number and nuclear area, with contractions of the distribution indicating a preferential loss of cells with largest and smallest nuclei. Asterisk, p=1.2E−24, Mann-Whitney Test. (B) Model 1 showing application of a random 72% loss of all nuclei. Note that the frequency distribution plot of the model completely overlaps that of pre-disease mice, indicating poor performance of the model. (C) Model 2 showing application of a uniform 5 µm2 atrophy to all nuclei followed by a random 72% loss of all nuclei. Note that that the frequency distribution plot of the model does not resemble that of the glaucomatous mice and that the mean nucleus size is too small, indicating poor performance of the model. (D) Model 3 showing a size-dependent loss of nuclei, with half of the risk fixed at 50% and the other half continuously increasing according to size rank amongst all retinal ganglion cell nuclei in the population. Note that the frequency distribution plot of the model does not resemble that of the glaucomatous mice and that the mean nucleus size is too small, indicating poor performance of the model. (E) Model 4 showing the same size-dependent loss of nuclei as Model 3, but with an accompanying and uniform 5 µm2 enlargement of all remaining nuclei. Note that the frequency distribution plot of the model closely resembles that of the glaucomatous mice, indicating good performance of the model.
2.9 Modeling influence of sampling coverage on statistical power
One set of 24 images at 200X total magnification was collected and analyzed in the standard positions described above (2 images in each of 3 zones of eccentricity in 4 petals) in 10 retinas of C57BL/6J mice. To model how the proportion of retinal area sampled (sampling coverage) relates to statistical power for detecting differences in RGC survival, these images were subsequently cropped to produce sampling fields matching the size of various combinations of coverage (equivalent to 24 images collected with 200X magnification, 12 images at 200X, 12 images at 1000X, and 4 images at 1000X), dropping 1 image per zone per petal for the analysis with 12 images, and using only data from the mid-peripheral region in the analysis with 4 images. To avoid introducing artificial border effects, RGCs in each retina were calculated as a percentage (RetFM-Class defined RGCs divided by RetFM-J defined total cells), and the mean (± SD) percentage of RGCs calculated for the 10 retinas with each method. Thus, the analysis utilized successively smaller regions of an identical image set from purely inbred and unmanipulated mice, ignoring the additional variation that would presumably be introduced in the context of a disease and/or treatment. Power calculations solving for sample size were performed using the Piface Java applet (version 1.76) developed by Russell Lenth, analyzing a two-sample t-test with sigmas derived from the SD of each method (assumptions of equal sigmas), alpha of 0.05, and power of 0.9; calculated for a true difference of the means corresponding to either 10% or 20% difference in RGCs.
2.10 Software accessibility
RetFM-J is available for download from the ImageJ website. The RetFM-Class tool is available upon request. The training set utilized here is also available upon request, though RetFM-Class can equally be utilized with any user provided training set consisting of images and a spreadsheet with RetFM-J assigned cell identification numbers, RetFM-J determined features, and manual cell type classifications.
3. Results
3.1 Coupling of RetFM-J with RetFM-Class enables quantification and random forest classification
We coupled the functionalities of RetFM-J and RetFM-Class to perform histology-based random forest classification (Figure 1). In mouse, the inner retinal surface consists primarily of neurons, glia, and a robust vasculature. As in other mammals (Stone and Fukuda 1974, Hughes 1975, Wassle, Levick et al. 1975, Tiao and Blakemore 1976, Peichl 1992, Gellrich and Gellrich 1996, Gramlich, Joachim et al. 2011), these populations have anatomic and histologic characteristics that help distinguish them (see Methods) (Drager and Olsen 1981). Light microscopy of H&E-stained retinal whole-mounts distinguishes among three categories of cells (Figure 1A, B): RGCs, which contain nuclei of diverse sizes that are consistently lightly-stained and have prominent nucleoli (categorized RGC), vascular endothelial cells, which have elongated nuclei that delineate vessels (categorized VEC), and a large number of cells containing small, round, and densely-stained nuclei, which represent mostly displaced amacrine cells, but likely also some astrocytes and microglia (hence categorized DAC+). There are also a fraction of apparent nuclei with non-distinct features that are manually difficult to assign to another cellular category (categorized ND). We designed RetFM-Class to utilize RetFM-J-generated feature information from a training set of images that had been classified manually based on these features, thereby generating a random forest trained classifier.
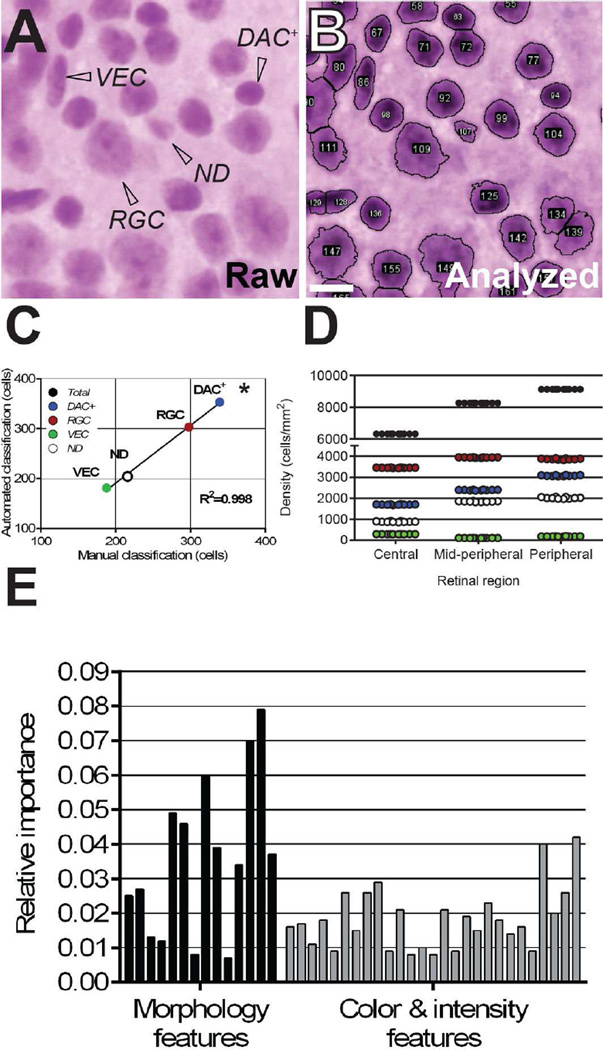
Figure 1. RetFM-Class performs automated cell type classification with an accuracy and precision similar to those of manual classification.
(A) Representative image from an H&E-stained retinal whole-mount containing three categories of distinguishable cells (RGC, retinal ganglion cells; VEC, vascular endothelial cells; DAC+, displaced amacrine cells plus other cells with similar-appearing nuclei), as well as a fraction of what appear to be nuclei but are difficult to assign to another cellular category manually because their features are not distinct (ND). (B) Automated nucleus recognition and counting of the same image shown in panel A by RetFM-J. (C) Comparison of manual- and auto-classification (n=10 images; 1,041 nuclei). (D) Comparison of 10 repeat classifications of cells with RetFM-Class from an individual image. Cell type is color coded as in panel C for RGC (red), DAC+(blue) VEC (black), and ND (open). Note that the Y-axis is discontinuous and not equally scaled in the upper and lower portions. (E) Auto-classifications by RetFM-Class were multifactorial, using multiple image features related to morphology and color and intensity to varying extents. In consideration of space, labels for individual bars may be found in Supplemental Table 1, with the order of the features in the bar graph (left to right) matching the presentation in the Table (top to bottom). Scale bar = 10 µm.
Based on a leave-one-image-out evaluation of the random forest classifier, the overall accuracy between manual- and auto-classifications was 79.5% (828/1041), representing 83.2% for RGC, 85.6%, for DAC+, 84.6% for VEC, and 61.1% for ND. Manual and auto-classifications disagreed in 20.5% (213/1041) of cases, with the following mismatches occurring: nuclei manually identified as RGC nuclei mismatched as DAC+, VEC, and ND in 28, 1, and 28 from a total of 339 cases, respectively; nuclei manually identified as DAC+ nuclei mismatched as RGC, VEC, and ND in 25, 1, and 17 from a total of 298 cases, respectively; nuclei manually identified as VEC mismatched as RGC and ND in 2 and 27 from a total of 188 cases, respectively; and nuclei manually identified as ND mismatched as RGC, DAC+, and VEC in 44, 20, and 20 from a total of 216 cases, respectively. Thus, the misclassifications were approximately symmetrical between categories, and the overall numbers of each nuclear type counted manually correlated well with the number counted by automated analysis (Figure 1C, R2= 0.998, p<0.001). In testing the influence that various technical factors may have on RetFM-Class output, we found that with repeat training of the classifier, subsequent classification of cells from an individual image using RetFM-Class resulted in less than 1.1% coefficient of variation (Figure 1D), and that auto-classifications by RetFM-Class were based on multiple features (Figure 1E).
To better understand the nature of the ND category, we manually re-assessed the H&E-stained samples of 61 cases of nuclei that had been auto-classified to this category. Of these 61 cases, the majority (n=41) involved clumped nuclei that were individually identifiable but overlapping; most often a vascular endothelial cell was involved (7/41 RGC-VEC, 16/41 DAC+-VEC, 12/41 VEC-VEC, 6/41 non-VEC associated clumps). The ND category also contained several cells (n=20) in which RetFM-J had recognized the nuclear border atypically, for example mistaking the nucleolus for the nucleus, in a cell that could have been classified manually (11/20 RGC, 4/20 DAC+, 1/20 VEC). Finally, the ND category contained a few objects that were recognized as nuclei by RetFM-J but were not compatible with any of the defined criteria for manual classification (n=4). Of relevance for quantification for RGCs, 29.5% of objects in the ND category included RGCs, with 11.5% of these involving an RGC in a clump.
As an additional test of performance of this auto-classification system, we utilized immunohistochemistry to assess the correlation of these random forest-based classifications with the absence or presence of markers for the most common cell types in the inner retina (Figure 2). Because of incompatibility between H&E staining and immunofluorescence-based labeling, this experiment was conducted by first performing antibody labeling and capturing images, and then soaking off the cover slips, performing H&E staining, re-photographing the same cell fields, and conducting auto-classification. Although this procedure was not well-suited for large-scale comparisons, instances of well-registered images allowed assessments. In areas of the inner retina that were devoid of vasculature, where the vast majority of cells are RGCs or displaced amacrine cells, 73% (73/100) of cells auto-classified into the RGC category were indeed positive for the RGC-marker RBPMS (Rodriguez, de Sevilla Muller et al. 2014) and 95% (95/100) of cells auto-classified into the DAC+ category were RBPMS-negative (Figure 2A–C). Of cells auto-classified into the DAC+ category, 86% (86/100) were positive for the amacrine-marker AP-2α (Bassett, Pontoriero et al. 2007) and 77% (77/100) of cells auto-classified into the RGC category were AP-2α-negative (Figure 2M–O). Within blood vessels, as delineated by COL4 labeling, elongated nuclei characteristic of vascular endothelial cells were mostly classified into the VEC category (9/10 VEC, 1/10 ND), as were most oval nuclei within vessels characteristic of pericytes (5/10 VEC, 4/10 ND, 1/10 DAC+). In contrast, nearby cells positive for the astrocyte marker GFAP were mostly auto-classified into the ND category (5/10 ND, 3/10 DAC+, 1/10 RGC, 1 VEC; Figure 2D–F). Cells positive for the microglial marker IBA1 were auto-classified into several categories, regardless of activation state (activated small ovoid morphology, 1/2 RGC, 1/2 DAC+; resting ramified morphology, 4/8 RGC, 2/8 DAC+, 1/8 VEC, 1/8 ND, Figure 2G–L).
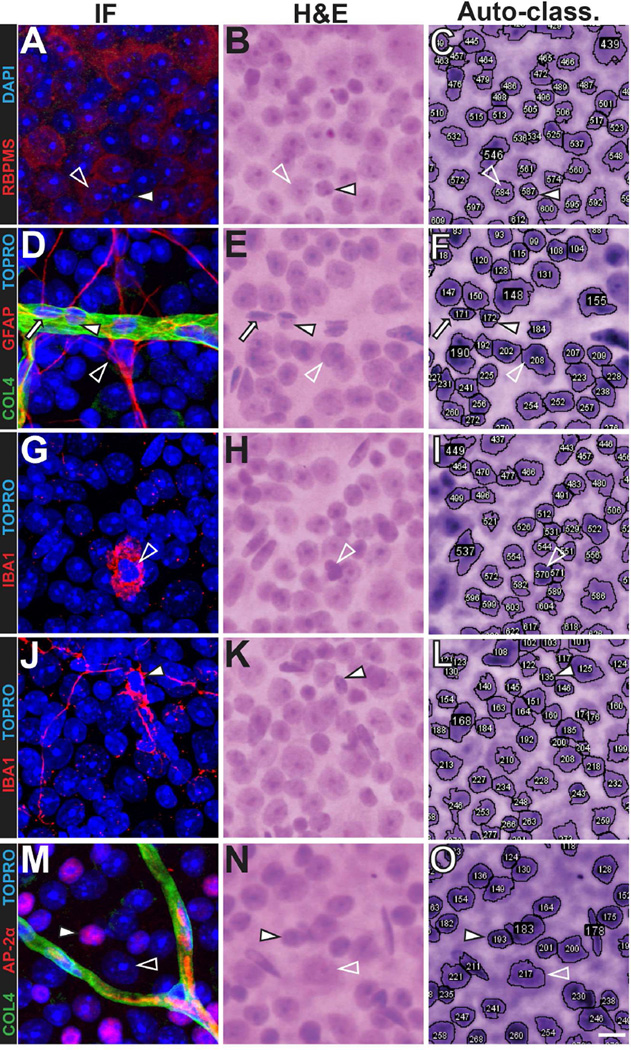
Figure 2. Immunohistochemical markers for the most common cell types in the inner retina correspond to random forest classifications.
Identical fields of cells are shown in each row, with immunofluorescence (IF), H&E staining (H&E), and RetFM-J-recognized cells (Auto-class.) shown in successive columns. (A–C) The retinal ganglion cell marker RBPMS is present in a nucleus auto-classified as RGC (open arrowhead) and absent in a nucleus that has features characteristic of a displaced amacrine cell and was auto-classified as DAC+ (solid arrowhead). (D–F) The astrocyte marker GFAP is present in: a nucleus auto-classified as ND (open arrowhead); an elongated nucleus within a vessel marked by COL4, that has features characteristic of a vascular endothelial cell and was auto-classified as a VEC (solid arrow); and an oval nucleus within a vessel marked by COL4, that has features characteristic of a pericyte and was auto-classified as DAC+ (solid arrowhead). (G–L) The microglial marker IBA1 is present in activated (G–I, open arrowhead) and resting (J-L, solid arrowhead) microglia, both of which were auto-classified as DAC+(M–O) The amacrine marker AP-2α is present in a nucleus auto-classified as DAC+ (closed arrowhead) and absent in a nucleus that has features characteristic of a retinal ganglion cell and was auto-classified as RGC (open arrowhead). C57BL/6J mice at 9–12 weeks of age; scale bar = 10 µm.
3.2 Tests of random forest classification meet predictive expectations for C57BL/6J mice
We next extended random forest classification to previously uncharacterized images of C57BL/6J mice (Figure 3). Qualitatively, comparing images before and after analysis indicated that the vast majority of cells were correctly recognized, with cell clumps typically being associated with instances of less certainty (Figure 3A, B). After classification, ∼70% of the cells were classified into the RGC, DAC+, or VEC categories and the remaining ∼30% were grouped into the ND category (Figure 3C).
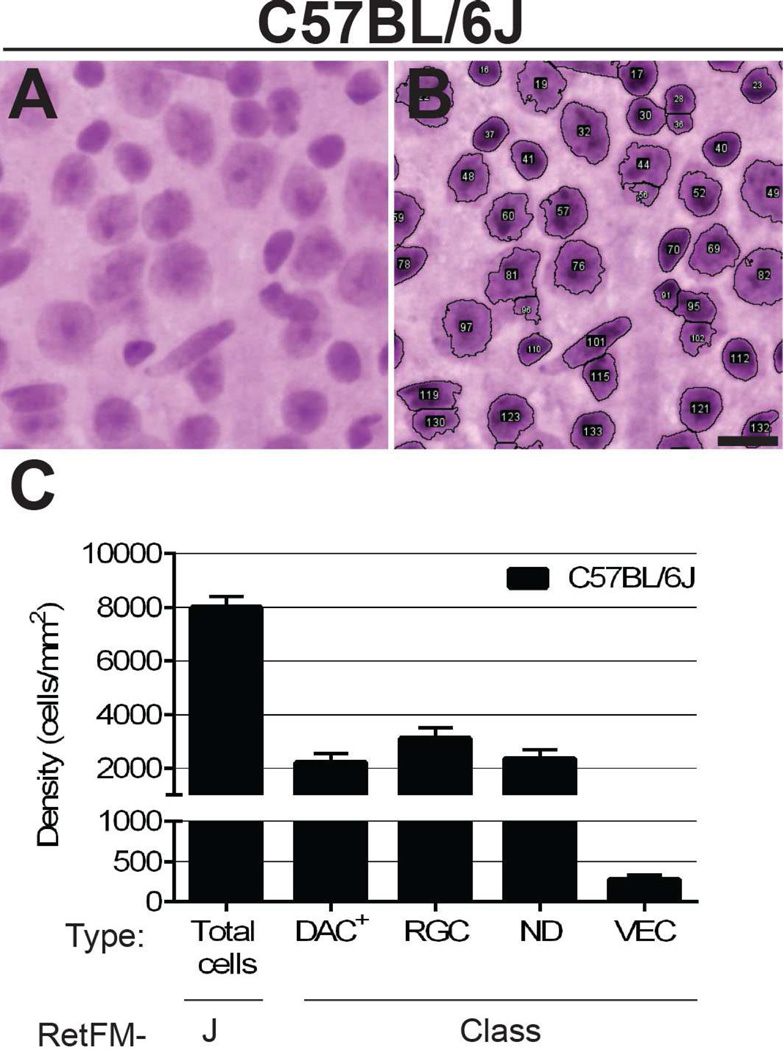
Figure 3. RetFM-J and RetFM-Class performance matches expectations in C57BL/6J mice.
(A) Raw and (B) analyzed images captured from the mid-peripheral zone. Most nuclei are clearly recognized and separated by RetFM-J, but there is less certainty in areas of cell clustering (for example, involving objects 91, 95, and 102 at the lower right). Scale bar = 10µm. (C) Bar graph of cell density versus cell category. Note that the Y-axis is discontinuous and not equally scaled in the upper and lower portions. C57BL/6J mice (n = 10 retinas, 254,135 nuclei); 8–9 wks of age; mean ± 1 SD.
As a test of performance, we compared the values obtained using our random forest approach to previously published estimates of cellularity (total number of inner retinal cells) in whole-mounts of C57BL/6J mice; these have ranged from 94,229 (based on counts of DAPI-labeled nuclei but not the elongated nuclei of endothelial cells) (Quigley, Cone et al. 2011) to 117,000 (based on counts of Nissl-stained cells but not the elongated endothelial cells) (Drager and Olsen 1981). To obtain analogous estimates from our data, we characterized retinas of C57BL/6J mice (n=10, 8–9 weeks of age) with RetFM-J and RetFM-Class, then subtracted the VEC category from total nuclei to obtain the density of all non-endothelial nuclei in each zone of eccentricity (8,140 ± 627 central, 8,038 ± 533 mid-peripheral, 6,974 ± 491 peripheral; 7,717 ± 550.4 cells/mm2 overall average). In order to convert densities to total numbers of cells, we measured the total area of the same whole-mounts (14.4 ± 0.7 mm2) and each zone of eccentricity (2.4 ± 0.1 central, 5.3 ± 0.3 mid-peripheral, and 6.7 ± 0.4 mm2 peripheral). Finally, we multiplied the average density for each zone by its average area and summed the values for all zones, arriving at a final estimate of 108,863 non-endothelial cells per retina.
Similarly, we compared the values obtained using random forest classification to previously published estimates of RGCs in C57BL/6J mice. Estimates of RGC number in C57BL/6J mice have varied, with those based on quantification of optic nerve axons typically predicting from 44,860 ± 3,125 (Jeon, Strettoi et al. 1998) to 54,416 ± 4,896 RGCs (Williams, Strom et al. 1996), and those based on labeling with retrograde tracers typically identifying from 42,658 ± 1,540 (Salinas-Navarro, Jimenez-Lopez et al. 2009) to 49,823 ± 1,792 (Pang and Wu 2011). To estimate the total number of RGCs predicted by our random forest approach, we performed a computation analogous to the one described above, multiplying the density of RGCs calculated in each zone (2,767 ± 325 central, 3,450 ± 611 mid-peripheral, 3,106 ± 491 peripheral; 3,107 ± 476 cells/mm2 overall average) of eccentricity by its area and summing the values for each zone to arrive at a total number of RGCs at 45,736. If extended to also include 29.5% of cells in the ND category of each zone (see above, the percentage of objects in the ND category that are RGCs; 3,467 ± 554 central, 2,264 ± 284 mid-peripheral, 1,473 ± 545 peripheral; 2,401 ± 461 cells/mm2 overall average), the total number calculated is 54,642.
3.3 Changes in RGCs detected by random forest-based analysis in mice subjected to CONC
We next tested the performance of this random forest classification approach by examining retinas 35 days after CONC (Figure 4). Because this insult is specific to RGCs, the expectation was that only RGCs would be lost. As an additional test, the experiment was carried out on Jun-deficient (Jun−/−) mice, in which significantly more RGCs survive CONC-induced apoptosis at 35 days post injury (Fernandes, Harder et al. 2012); this would allow assessment of how pre-apoptotic changes, such as nuclear atrophy (Janssen, Mac Nair et al. 2013), influence performance of the classifier. Thus, the overall experimental design was to compare mice of the same cross, genetic background, and experimentally-induced injury, but with differing susceptibilities to apoptosis.
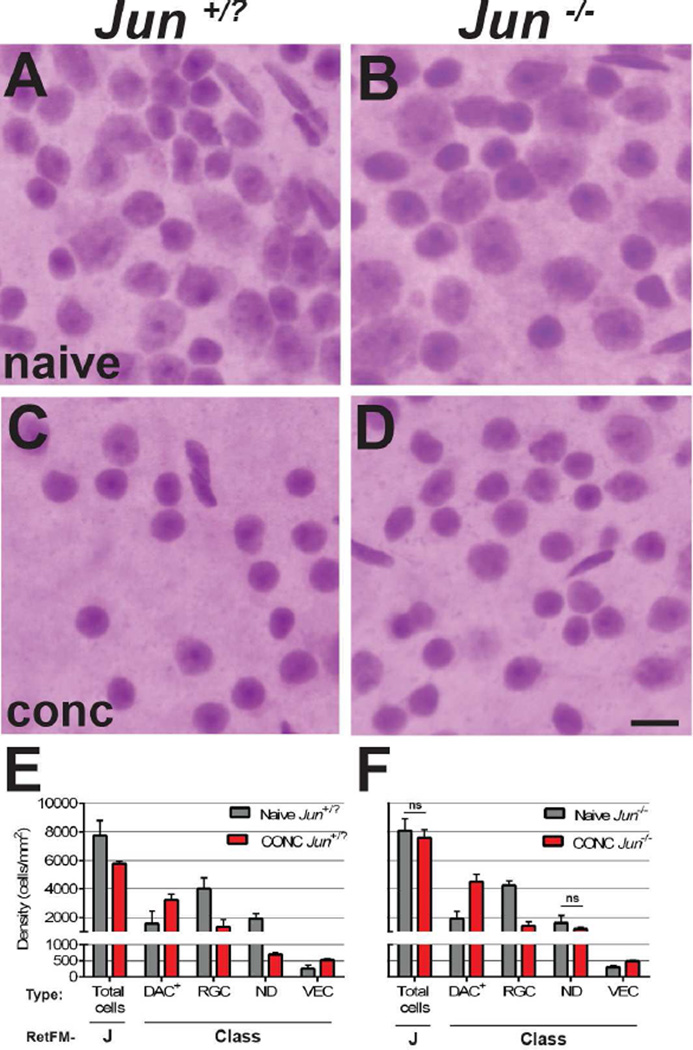
Figure 4. RetFM-J, but not RetFM-Class, performance matches expectations in acute damage induced by CONC.
Mice of the same cross and genetic background. Jun+/? mice (left column) undergo apoptosis in response to CONC. Jun−/− mice (right column) have resistance to CONC-induced apoptosis but are capable of pre-apoptotic changes such as nuclear atrophy. In the naïve control groups, retinas from (A) Jun+/? and (B) Jun−/− retinas both exhibit dense populations of cells of varying type in the inner retina. In the CONC-injured groups assessed 35 days after induction, (C) Jun+/? retinas have decreased cell density and the majority of remaining cells have small, densely stained nuclei. (D) Jun−/− retinas have nearly normal cellular density and the majority of remaining cells again appear to have small, densely stained nuclei. Note that the diversity in appearance of nuclei is greater in panel D than C. Scale bar = 10µm. – Bar graph of cell density versus cell category. RetFM-J detected a loss of total cells in Jun+/− and not Jun−/− retinas, matching expectations, but RetFM-Class predicted statistically significant differences between CONC and naïve mice in nearly all cellular categories of both genotypes, indicating that classification performance was negatively influenced by the induced synchronous insult of CONC (p<6.1E−3, Bonferroni-corrected 2-tailed Student’s t-test for all comparisons except those noted with ns). Note that the Y-axis is discontinuous and not equally scaled in the upper and lower portions. Jun+/? naïve (n = 8 retinas, 269,743 nuclei), CONC (n = 4 retinas, 75,589 nuclei); Jun−/− naïve (n=6 retinas, 157,735 nuclei), CONC (n=4 retinas, 100,410 nuclei); mean ± 1 SD.
In Jun-sufficient mice (Jun+/?; Figure 4A,C,E), RetFM-J detected a CONC-induced 25.4% decrease in total cell number compared to naïve controls (naïve 7,743 ± 1,049 cells/mm2; CONC 5,772 ± 173 cells/mm2; p=4.7E−3, Bonferroni-corrected 2-tailed Student’s t-Test). Because RGCs constitute 52.6% of the total cell population in naïve mice from this particular cross and genetic background, a 25.4% decrease in total cells corresponds to a 48.3% decrease in RGCs, reasonably matching expectations for the extent of cell loss resulting from CONC at this time point based on previous reports (Fernandes, Harder et al. 2012). In Jun-deficient mice (Jun−/−; Figure 4B, D, F), RetFM-J detected only a modest, statistically insignificant, reduction in total cell density. Thus, RetFM-J performance matched expectations for these mice. In both Jun+/? and Jun−/− mice, RetFM-Class detected a similar pattern, characterized by symmetrical increases in density in the DAC+ and VEC categories, and decreases in density in the RGC and ND categories. The observed differences in multiple categories indicate that RetFM-Class performance did not match expectations for these mice.
3.4 Cell type specific changes detected in random forest-based analysis of DBA/2J mice with glaucoma
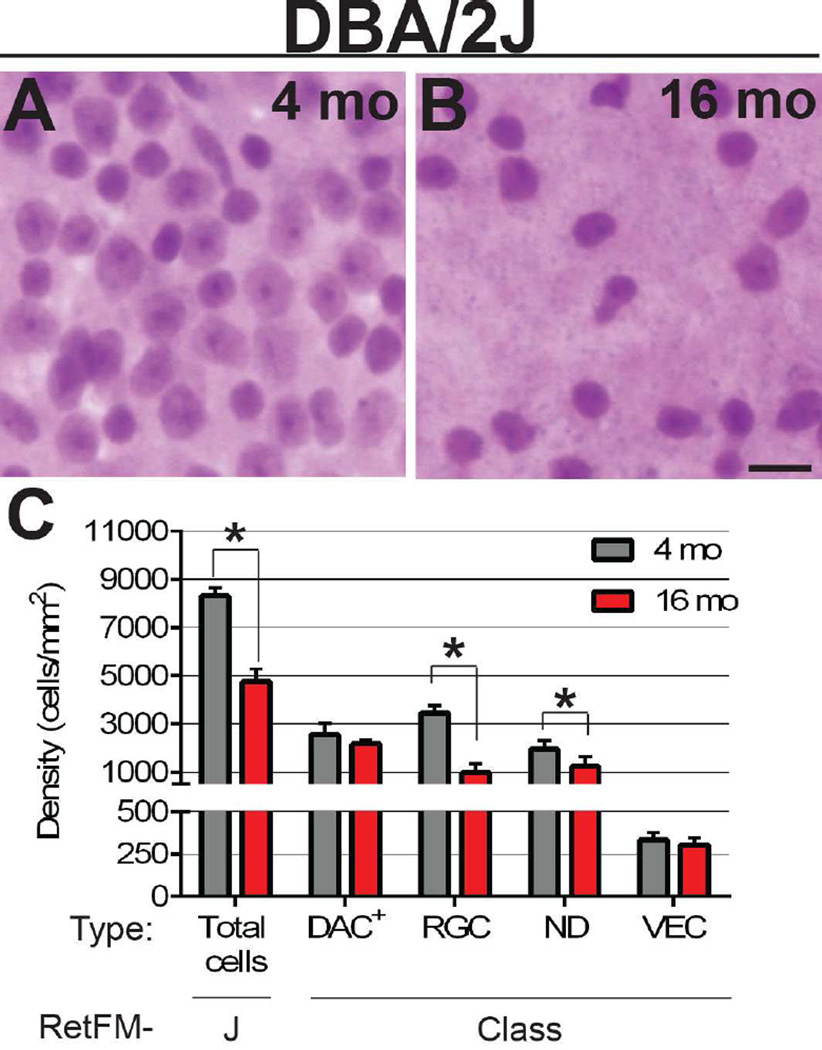
Having identified caveats for using the random forest approach to study RGCs in the acute damage setting of CONC, we next tested performance in the context of chronic damage, examining DBA/2J mice with glaucoma (Figure 5). In pre-diseased, DBA/2J mice (4-month-old, n = 9 retinas), retinas had slightly greater RGC densities than in those of their C57BL/6J (2–5-month-old, n = 13 retinas; p=1.1E−2, 2-tailed Student’s t-test) counterparts, consistent with the outcomes of previous studies of these strains (Williams, Strom et al. 1996, Strom and Williams 1998). At 16 months, an age at which 80% of DBA/2J mice have late-stage glaucoma, total cell density decreased by 42.6% (4-month-old 8,297 ± 347 cells/mm2, 16-month-old 4,761 ± 515 cells/mm2; p=6.5E−9, Bonferroni-corrected 2-tailed Student’s t-test). This change was attributable primarily to a 71.2% reduction in RGC density (4-month-old 3,458 ± 309 cells/mm2, 16-month-old 995 ± 362 cells/mm2; p=2.8E−9, Bonferroni-corrected 2-tailed Student’s t-test), closely matching expectations (Libby, Anderson et al. 2005, Howell, Libby et al. 2007). Using a previously described three-point optic nerve damage grading scheme (Howell, Libby et al. 2007), the optic nerves from 16-month-old mice included two nerves with "moderate" damage and six nerves with "severe" damage. Consistent with observations in related DBA mouse strains (Danias, Lee et al. 2003), the total area of the whole-mounts increased by ∼10% (15.3 ± 1.0 mm2 at 4 months, n=9 retinas; 17.0 ± 1.1 mm2 at 16 months, n=8 retinas).
Figure 5. RetFM-J and RetFM-Class performance matches expectations in chronic glaucoma of DBA/2J mice.
Representative images from the mid-peripheral zone. (A) In retinas of 4-month-old mice the distribution of cells of the inner retina whose nuclei are characteristic of retinal ganglion cells , displaced amacrine cells, and vascular endothelial cells is dense. (B) In retinas of 16-month-old mice the number of nuclei in the inner retina is dramatically reduced, reflective of a reduction in cell density. Of the remaining nuclei, most resemble those of displaced amacrine cells; very few nuclei resemble those of retinal ganglion cells. (C) Bar graph of cell density versus cell category. RetFM-J and RetFM-Class detected parallel losses of total cells and retinal ganglion cells, respectively. (*) p < 7.0E−3, Bonferroni-corrected 2-tailed Student’s t-test. Note that the Y-axis is discontinuous and not equally scaled in the upper and lower portions. DBA/2J 4 month (n=9 retinas, 99,629 nuclei); DBA/2J 16 month (n=8 retinas, 27,732 nuclei); mean ± 1 SD.
In the DBA/2J mice, the size of nuclei of cells in the RGC category changed between the 4- and 16-month time points (Figure 6). Median size of these nuclei decreased with glaucoma (4-month-old 51.8 µm2; 16-month-old 50.6 µm2; p=1.2E−24, Mann-Whitney test), as did the mean size (4-month-old 55.5 ± 17.2 µm2, n=99,629; 16-month-old 52.9 ± 12.8 µm2, n=27,732). Examination of relative frequency distribution plots of nuclear size revealed a complex pattern characterized by an overall contraction in the distribution plot (Figure 6A). In considering various models (Figure 6B–D) capable of causing such rates of RGC death, changes in nuclear size, and changes in frequency distribution profiles, a model evoking subtle enlargement of all nuclei (causing a statistical "loss" of nuclei as they are shifted from smaller to larger bins) and preferential death of cells with the largest nuclei fits the data well qualitatively (Figure 6E).
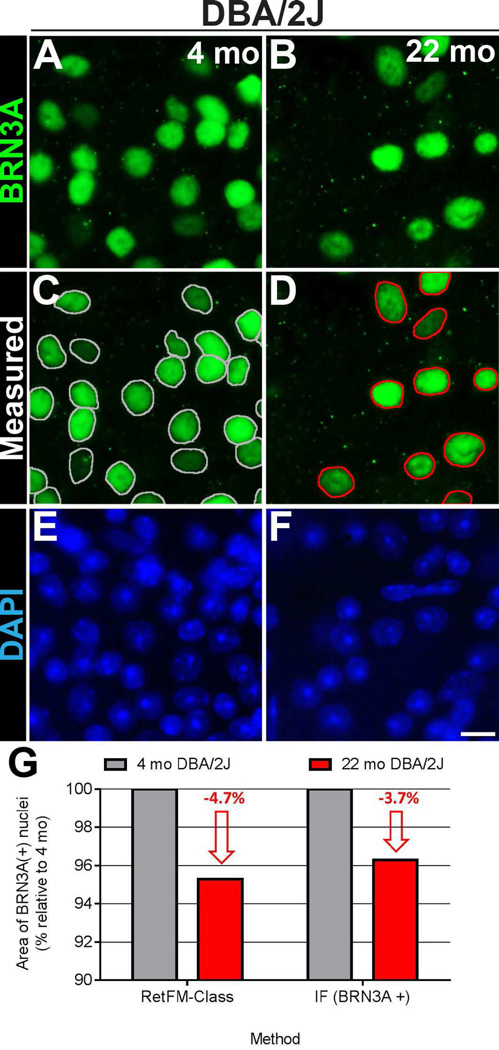
As an independent test for assessing the relationship between nuclear size and glaucoma, we performed a similar analysis on independent DBA/2J mice using standard immunofluorescent microscopy and an antibody recognizing the RGC-specific nuclear transcription factor BRN3A (Xiang, Zhou et al. 1995). In this experiment, median size of surviving nuclei in glaucoma again decreased (4-month-old 67.3 µm2; 22-month-old 64.9 µm2; p=2.37E−7, Mann-Whitney test), as did the mean size (4-month-old 68.6 ± 16 µm2, n = 2,369; 22-month-old 66.1 ± 15.8 µm2, n = 1,449; Figure 7).
Figure 7. Confirmation that surviving retinal ganglion cells in DBA/2J mice with glaucoma have smaller nuclei.
Immunofluorescent images from the inner retina of whole-mount preparations showing labeling of BRN3A–positive nuclei of presumptive retinal ganglion cells in (A) pre-diseased DBA/2J mice at 4 months of age (left column) and (B) glaucomatous DBA/2J mice at 22 months of age (right column). Note that there are decreased numbers of BRN3A–positive nuclei in retinas from the older DBA/2J mice with glaucoma with age and onset of glaucoma. The same fields are also shown below with (C, D) nuclear outlines indicating the areas that were measured and with (E, F) DAPI-labeling. (G) Graph comparing the change in mean area of DBA/2J nuclei (as a percentage relative to that at 4 months) in glaucoma as measured by RetFM-class (left; n = 99,629 nuclei at 4 months of age; 27,732 nuclei at 16 months of age) and immunofluorescence (right; n = 2,369 nuclei at 4 months of age; 1,449 nuclei at 22 months of age). Note that both methods reveal a similar 3.7–4.7% reduction in the mean nuclear area of retinal ganglion cells. Scale bar = 10 µm.
4. Discussion
The inner retinal surface is a complex environment, consisting of: a robust vasculature with endothelial cells and pericytes, astrocytes, microglia, axons of the nerve fiber layer, a variety of phagocytic cells (Koeberle, Gauldie et al. 2004, Kezic, Chrysostomou et al. 2013, Wang, Chen et al. 2015), and the ganglion cell layer itself, which contains up to 30 different sub-types of RGCs (Badea and Nathans 2004, Kong, Fish et al. 2005, Coombs, van der List et al. 2006, Volgyi, Chheda et al. 2009, Kim, Zhang et al. 2010, Sanes and Masland 2015), and 10 different types of DAC (Perez De Sevilla Muller, Shelley et al. 2007). As is the case for many other CNS tissues, this cellular diversity can be a challenge in carrying out experiments. Here, we have combined the advantages of H&E histology (Chan 2014), random forest classification (Touw, Bayjanov et al. 2013), and computer automation (Gurcan, Boucheron et al. 2009) in a new methodology that was compared to multiple “truth” sets, including manually-classified cells (n=1,041), immunohistochemically-classified cells (n=240), and functionally-classified cells from mouse models exhibiting normal (C57BL/6J) or diseased retinas (CONC and DBA/2J). Results of these experiments have relevance to the methodology itself, and its application to the discovery of new aspects of RGC pathophysiology.
4.1 Applicability and caveats regarding methodology
Sufficiently powered studies of neuroprotection are costly and time-consuming, and typically require the measurement of multiple disease parameters in relatively large cohorts of animals. Accordingly, the quality and throughput capacity of methods for quantifying changes to the inner retina are technical factors of particular importance to studies of glaucoma, Alzheimer’s disease, and traumatic brain injury. As a methodology for the quantification of RGCs, the tools and approach presented here could be utilized in a wide variety of experiments.
Most approaches for RGC quantification utilize antibodies, transgenic animals, or retrograde labeling techniques, each of which has potential limitations. In the case of antibody-based techniques, these depend on the specificity of the antibody with respect to retinal cell type, RGC sub-type, disease state, and (too often) the precise manufacturer and lot of the antibody, all of which can confound analyses (Soto, Oglesby et al. 2008, Rodriguez, de Sevilla Muller et al. 2014, Baker 2015). In the case of transgenic approaches, these offer powerful opportunities for many types of studies, yet transgenes themselves are highly sensitive to position-dependent integration effects that can confound certain kinds of analyses. Furthermore, the husbandry required to cross such genes into other strains of mice while also controlling genetic background is typically too time-consuming to be practical, and therefore, often ignored. Finally, retrograde labeling has the advantage of being highly specific for RGCs, but it fails to differentiate among those that have died, lost their axons, or are defective for axonal transport (Kobbert, Apps et al. 2000, Soto, Oglesby et al. 2008). The advantages of histology-based random forest classification are that it simultaneously utilizes multiple features to classify cells, requires no special husbandry, is independent of axonal properties, yields simultaneous information on multiple cell types, is rapid and inexpensive, and is readily applicable to high-throughput analyses.
With respect to specificity, our study reveals that 83.2% of cells manually classified as RGC were also auto-classified as RGC, and 73% of cells auto-classified as RGC were positive for the RGC marker RBPMS. Our data show that one of the factors contributing to imperfect specificity is confusion between classification categories, which is an area that might be improved upon in future iterations. To help encourage this, we have designed our approach to utilize a user specified set of training images, which could be the set we have described herein, or new ones, thus, allowing continuing improvement of the approach without recoding software. Of relevance to specificity, it is also worth noting that the selection of a “gold-standard” independently defining an RGC is problematic; for example, RBPMS is a robust RGC marker but fails to label 15.8% of RGCs marked in Thy1-CFP transgenic mice (Rodriguez, de Sevilla Muller et al. 2014). To our knowledge, other approaches for labeling an RGC all share similar caveats preventing them from having perfect RGC specificity. Our current data do not support that the current iteration of histology-based random forest classification improves accuracy or precision over those of RBPMS-labeling or other approaches for identifying RGCs. It may in fact have less. However, it does not lag by much and overall is a promising complement to existing options.
One advantage of the histology-based random forest classification method pertains to uniformity. All manual classification methods are prone to higher variation than automated methodologies. Among the reports of computer-assisted approaches for quantitating cells in retinal whole-mounts (Danias, Lee et al. 2003, Salinas-Navarro, Jimenez-Lopez et al. 2009), ours is the first to use open-source software and widely available reagents, making it possible for two people to analyze the same retina and obtain the same data in a uniform fashion. Thus, the approach, and its continued evolution, has the potential to be a useful complement to the subjective grading schemes commonly used in many mouse studies of RGCs.
A second advantage pertains to sampling size. As a consequence of the naturally occurring variability in RGC density, quantitative approaches are limited by the size of the sampling region – typically with the trade-off that counting cells in larger areas yields higher statistical power but consumes corresponding amounts of labor time. The most time-consuming aspects of histology-based random forest classification are in the preparation of the whole-mounts and collection/preparation of images for analysis; the actual classification is automated and rapid. Therefore, the approach can readily be used to quantitate large retinal areas. We have empirically settled on using 24 images collected at 200X total magnification representing an average of 23.5% of total retinal area. This coverage provides a large sampling field, with magnification and resolution sufficient for distinguishing cell type while also avoiding overlap of fields. The consequences of this for statistical power are important (Table 1). Our study with C57BL/6J mice found an average RGC density of 3,119 ± 397 cells/mm2 (mean ± SD), yielding a coefficient of variation of 12.7%. This is similar to results from other studies assessing density across large areas of the retina – which have likewise given rise to coefficients of variation between 8.9% and 13.7% in various inbred mouse strains (Salinas-Navarro, Jimenez-Lopez et al. 2009, Galindo-Romero, Aviles-Trigueros et al. 2011, Germain, Istillarte et al. 2013, Himori, Maruyama et al. 2014). With this degree of natural variability present, even among completely inbred mice with retinas free of overt disease, quantifications using greater sampling coverage are a prudent measure to avoid spurious results. In the context of diseases such as glaucoma, greater sampling coverage will also increase the potential for detection and examination of specific patterns of RGC loss that others have described to occur in DBA/2J and related mouse strains (Danias, Lee et al. 2003, Schlamp, Li et al. 2006, Howell, Libby et al. 2007, Perez de Lara, Santano et al. 2014).
Table 1.
Modeling influence of sampling coverage on statistical power.
| Retinal sampling (fields x total mag.) |
Coverage (%) | SD |
n= retinas required with 90% power to detect: |
|
|---|---|---|---|---|
| 10% loss | 20% loss | |||
| 24 × 200x | 23.5 | 0.0446 | 28 | 8 |
| 12 × 200x | 11.8 | 0.0455 | 29 | 8 |
| 12 × 1000x | 2.4 | 0.0572 | 46 | 12 |
| 4 × 1000x | 0.8 | 0.0727 | 73 | 19 |
Using the current iteration of RetFM-Class, histology-based random forest classification also has limitations. First, some classification categories are currently not unique, with DAC+ and ND both containing substantial numbers of multiple cell types. Even the RGC category is not pure, with some displaced amacrine cells, astrocytes, and microglia likely contributing to it. Although multiple publications have commented on the apparent differences in histologic appearance of RGCs and displaced amacrine cells (Drager and Olsen 1981, Levkovitch-Verbin, Quigley et al. 2001), a subset of each may have overlapping features, and thus be currently indistinguishable by this method. Second, as we demonstrated with the use of Jun-deficient mice, situations dissociating nuclear atrophy from cell death can lead to misclassification. Our results hint that RGCs with condensing nuclei may be particularly prone to misclassification into the DAC+ category. Importantly, the ability of RetFM-J to count total nuclei is not similarly influenced; thus, the observation of decreases in the number of one cell type without a corresponding change in total cellularity can serve as an indication of potential misclassification. Third, we have ignored some comparatively rare cell types known or presumed to be present under various conditions (e.g., polymorphonuclear leukocytes (Neufeld, Kawai et al. 2002), monocytes (Koeberle, Gauldie et al. 2004), hyalocytes (Kezic, Chrysostomou et al. 2013), and displaced horizontal cells (Silveira, Yamada et al. 1989)); their presence or absence is currently undetectable. Finally, because the H&E staining protocol is incompatible with immunofluorescence (Fischer, Jacobson et al. 2008), histology-based random forest classification is cumbersome when used in conjunction with immunohistochemistry or the detection of retrograde tracers. Although we have obtained some data by performing antibody labeling with subsequent imaging first and then performing H&E staining and imaging of the same cells or areas second, this is a laborious process and cannot be easily adapted for routine comparisons.
Random forest classification is an approach that could equally be performed with tissue labeled with a wide variety of different histologic stains or immunohistochemical markers; however, the cell segmentation approach implemented in RetFM-J and exact features extracted may have to be adapted accordingly. We chose to utilize H&E for several reasons, including its complex dichromatic label (random forest classifiers are well-suited to accommodate multiple features); resilience to disease-state (unlike antibodies, which mark an individual epitope of a single protein that may have altered expression in disease, hematoxylin stains nucleic acids and eosin stains proteins, which in bulk are less likely to be influenced by disease state); ease of uniform imaging; and its low cost and familiarity to biologists. We opted against Nissl stain, a frequent stain used in other studies of retinal whole-mounts, because it is monochromatic, and in pilot studies, the labeling of cytoplasmic granules made detection of nuclei more problematic. Like H&E, Nissl also stains multiple cell types of the inner retina, including RGCs, displaced amacrine cells, and vascular endothelial cells. With new training image sets and adjustments to inclusion criteria, RetFM-Class could be utilized to classify tissues labeled with other histochemical stains; the adjustments to workflow for immunohistochemically-labeled tissues would likely be significant.
Our work has also revealed opportunities for improvement in this type of experimental approach, as well as raising some fundamental challenges that are unavoidable. Among opportunities for improvement, the most significant relate to problems related to cell clumping, a biological phenomenon that is particularly common in the central retina. In our comparisons of auto- versus manual-classification, mismatches involving cell clumps within the ND category were frequent. The development of algorithms with an improved ability to distinguish tightly packed cells would undoubtedly improve performance. Among challenges that may be unavoidable, one is that the nerve fiber layer can partially conceal imaging of stained whole-mounts. Using the proportion of cells in the ND category as an indication, cell classification improved as the nerve fiber layer thinned. This was true with respect to the disease state (better in diseased retinas with fewer cells than in wild-type eyes), as well as the anatomic features of particular regions (better in the peripheral vs. central retina).
For users considering utilization of the RetFM-J and RetFM-Class software tools to quantify RGCs in mouse studies of diseases such as glaucoma, we additionally offer the following suggestions. First, users should specify whether they are reporting mathematical or anatomically weighted averages of RGC densities, whether they are reporting only cells in the RGC category or RGC plus a fraction of ND, and whether corrections for expansion of the total retinal area were considered. Second, it should be kept in mind that because of clumping, and specifically the “de-clumping” of cells that will occur as disease leads to reduced cell density, the number of cells detected in non-RGC categories is expected to increase slightly in disease. Likewise, microgliosis could cause increases specific to disease stage in some categories (Bosco, Steele et al. 2011, Liu, Li et al. 2012). Third, although performance of RetFM-J is robust with regard to retinas stained in different batches (Hedberg-Buenz, Christopher et al. 2015), the same has not yet been stringently tested for RetFM-Class. In an attempt to minimize this potential confounder, the current training set for RetFM-Class purposefully included samples from multiple histologic batches. None-the-less, it is advisable to design experiments in such a way that glaucomatous and control retinas are included in each batch of tissues subjected to H&E staining. Fourth, if a condition being studied would typically be analyzed using more than one assay (such as two markers of RGCs, or analysis of retinas and optic nerves), the same will likely be true with experiments utilizing RetFM-Class. Finally, for studies of CONC and other forms of disease that are likely to involve rapid-onset, largely synchronous disease, assessment of total cellularity using RetFM-J, without RetFM-Class classification, is currently recommended.
4.2 Selective cell death in glaucoma
Our results identify a reduction in the mean and median sizes of the nuclei in RGCs surviving in DBA/2J mice that have developed glaucoma. As shown in Figure 6, it is clear that glaucoma does not cause a uniform loss of RGCs with nuclei of all sizes (Figure 6B), a population-wide nuclear atrophy (Figure 6C), or a preferential loss of RGCs with the largest nuclei without concurrent changes to nuclei of surviving cells (Figure 6D). Though speculative, a model evoking subtle enlargement of all RGC-nuclei (causing “loss” of the smallest nuclei as they shift into a larger size bin) and preferential death of the cells with the largest nuclei closely fits the data (Figure 6E). This scenario is also supported theoretically. Somal hypertrophy of RGCs (perhaps as a consequence of decreasing cell density creating space for surviving cells to expand over time) has been observed in various other mouse models, as described in detail by Urcola et. al (Urcola, Hernandez et al. 2006). As further discussed below, studies in other species have also indicated that large RGCs are particularly susceptible to glaucomatous damage. Thus, there are precedents for the changes that we observed.
The question of whether or not specific RGC sub-types are preferentially susceptible to glaucomatous damage is of intense interest, but also controversial. Early work involving quantitative studies of Nissl-stained RGCs in a non-human primate model of glaucoma revealed that the larger cells that give rise to the magnocellular pathway are particularly vulnerable to apoptosis (Glovinsky, Quigley et al. 1991, Vickers, Schumer et al. 1995). Similar observations were subsequently made in some, but not all, follow-up studies with primate (Quigley 1999, Morgan, Uchida et al. 2000, Morgan 2002) and human tissue (Quigley, Dunkelberger et al. 1989, Chaturvedi, Hedley-Whyte et al. 1993, Quigley 1999). Because the size of the nucleus directly correlates with size of the soma in mouse RGCs (Janssen, Mac Nair et al. 2013), and the predominant effect we observed is that the mean and median areas of the RGC nuclei decreased, our current experiments support a model whereby large RGCs are preferentially lost in the context of glaucoma. Our result differs from those of studies of RGC somal size in inducible models of glaucoma in rats (Urcola, Hernandez et al. 2006), which did not find evidence for size-dependent cell loss. It is unclear whether these differences result from differences in the glaucoma models, including species used, differing responses of soma versus nuclei, or differences in the methodology used for quantification.
Our analysis of nuclear size also involves some caveats. First, because the numbers of mice and cells analyzed with BRN3A remains relatively small, we have not analyzed nuclear size distribution plots. We also note that the absolute value for nuclear size varied between the retinas analyzed with RetFM-Class versus BRN3A. We assume that these differences involve imperfect recognition of auto-segmented nuclear borders and methodologic differences in sample processing (retinas were dehydrated for the RetFM-Class, but not for BRN3A analyses), but it remains possible that one of the techniques has an unknown bias. Second, the literature also suggests a plausible alternative hypothesis. Instead of reflecting selective cell loss based on nuclear size, selective nuclear atrophy might also explain our RGC data. It is known that nuclear atrophy of RGCs can precede the BAX-dependent stage of apoptosis (Janssen, Mac Nair et al. 2013), and that there is an early stage of apoptotic volume decrease that can be induced independently of mitochondrial signaling or initiator caspase activation (Maeno, Tsubata et al. 2012). These phenomena have previously been studied in timeframes of hours to days, using potent pharmacologic or genetic manipulations, and it is unclear how they might relate to the physiologic context of a slow progressive disease in an otherwise healthy animal. In 16- or 22-month old DBA/2J mice, the number of RGCs engaged in active apoptosis is fleetingly small, <100/retina (Reichstein, Ren et al. 2007); thus, they are unlikely to explain the reduction in nuclear size we observed. However, it may be plausible that some RGCs insulted by glaucoma could undergo nuclear atrophy without full induction of apoptosis and provide an alternative explanation for our observation.
5. Conclusions
In conclusion, we have demonstrated that histology-based random forest classification is feasible and can be utilized to detect and quantitate features of RGCs in a high-throughput fashion. Taking advantage of the ability to quantitate nuclear area for large numbers of cells in the retinas of DBA/2J mice, we have shown for the first time, in a naturally occurring animal model of glaucoma, that surviving RGCs have a decreased mean nuclear area. Modeling suggests that the glaucomatous changes occurred through a mechanism involving a subtle enlargement of all RGC-nuclei and preferential death of cells with the largest nuclei.
Supplementary Material
Highlights.
H&E-stained retinal whole-mounts can be used with a random forest classifier to discern cell-type.
No loss of displaced amacrine cells in glaucomatous DBA/2J mice was detected.
Glaucomatous DBA/2J mice exhibit non-random changes in nuclear size of RGCs.
Acknowledgements
We thank John Fingert of the Department of Ophthalmology and Visual Sciences at the University of Iowa Hospitals and Clinics for helpful discussions. We thank Victoria Utter of the Department of Electrical and Computer Engineering at the University of Iowa for her help in development of the RetFM-Class tool. This work was supported by Merit Review Award #I01 RX001481 from the United States (U.S.) Department of Veterans Affairs Rehabilitation Research and Development Service, and National Institutes of Health, NEI (R01EY017673) grants to MGA. MC and TS are supported by National Institutes of Health, NEI grant (R01023187). The contents do not represent the views of the U.S. Department of Veterans Affairs or the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11(7):36–43. [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480(4):331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521(7552):274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- Bassett EA, Pontoriero GF, Feng W, Marquardt T, Fini ME, Williams T, West-Mays JA. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol. 2007;27(21):7497–7510. doi: 10.1128/MCB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Sibilia M, David JP, Mohle-Steinlein U, Tronche F, Schutz G, Wagner EF. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. Embo j. 2002;21(7):1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco A, Steele MR, Vetter ML. Early microglia activation in a mouse model of chronic glaucoma. J Comp Neurol. 2011;519(4):599–620. doi: 10.1002/cne.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher F, Stahl A, Agostini HT, Martin G. Hyperoxia causes reduced density of retinal astrocytes in the central avascular zone in the mouse model of oxygen-induced retinopathy. Mol Cell Neurosci. 2013;56:225–233. doi: 10.1016/j.mcn.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Chan JK. The wonderful colors of the hematoxylin-eosin stain in diagnostic surgical pathology. Int J Surg Pathol. 2014;22(1):12–32. doi: 10.1177/1066896913517939. [DOI] [PubMed] [Google Scholar]
- Chaturvedi N, Hedley-Whyte ET, Dreyer EB. Lateral geniculate nucleus in glaucoma. Am J Ophthalmol. 1993;116(2):182–188. doi: 10.1016/s0002-9394(14)71283-8. [DOI] [PubMed] [Google Scholar]
- Checchin D, Sennlaub F, Levavasseur E, Leduc M, Chemtob S. Potential role of microglia in retinal blood vessel formation. Invest Ophthalmol Vis Sci. 2006;47(8):3595–3602. doi: 10.1167/iovs.05-1522. [DOI] [PubMed] [Google Scholar]
- Chou JC, Rollins SD, Ye M, Batlle D, Fawzi AA. Endothelin receptor-A antagonist attenuates retinal vascular and neuroretinal pathology in diabetic mice. Invest Ophthalmol Vis Sci. 2014;55(4):2516–2525. doi: 10.1167/iovs.13-13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140(1):123–136. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- Cuthbertson RA, Mandel TE. Anatomy of the mouse retina. Endothelial cell-pericyte ratio and capillary distribution. Invest Ophthalmol Vis Sci. 1986;27(11):1659–1664. [PubMed] [Google Scholar]
- Danias J, Lee KC, Zamora MF, Chen B, Shen F, Filippopoulos T, Su Y, Goldblum D, Podos SM, Mittag T. Quantitative analysis of retinal ganglion cell (RGC) loss in aging DBA/2NNia glaucomatous mice: comparison with RGC loss in aging C57/BL6 mice. Invest Ophthalmol Vis Sci. 2003;44(12):5151–5162. doi: 10.1167/iovs.02-1101. [DOI] [PubMed] [Google Scholar]
- Dehabadi MH, Davis BM, Wong TK, Cordeiro MF. Retinal manifestations of Alzheimer's disease. Neurodegener Dis Manag. 2014;4(3):241–252. doi: 10.2217/nmt.14.19. [DOI] [PubMed] [Google Scholar]
- Drager UC, Olsen JF. Ganglion cell distribution in the retina of the mouse. Invest Ophthalmol Vis Sci. 1981;20(3):285–293. [PubMed] [Google Scholar]
- Fernandes KA, Harder JM, Fornarola LB, Freeman RS, Clark AF, Pang IH, John SW, Libby RT. JNK2 and JNK3 are major regulators of axonal injury-induced retinal ganglion cell death. Neurobiol Dis. 2012;46(2):393–401. doi: 10.1016/j.nbd.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4986. pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26(2):130–132. [PubMed] [Google Scholar]
- Galindo-Romero C, Aviles-Trigueros M, Jimenez-Lopez M, Valiente-Soriano FJ, Salinas-Navarro M, Nadal-Nicolas F, Villegas-Perez MP, Vidal-Sanz M, Agudo-Barriuso M. Axotomy-induced retinal ganglion cell death in adult mice: quantitative and topographic time course analyses. Exp Eye Res. 2011;92(5):377–387. doi: 10.1016/j.exer.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Gellrich MM, Gellrich NC. Quantitative relations in the retinal ganglion cell layer of the rat: neurons, glia and capillaries before and after optic nerve section. Graefes Arch Clin Exp Ophthalmol. 1996;234(5):315–323. doi: 10.1007/BF00220707. [DOI] [PubMed] [Google Scholar]
- Germain F, Istillarte M, Gomez-Vicente V, Perez-Rico C, de la Villa P. Electroretinographical and histological study of mouse retina after optic nerve section: a comparison between wild-type and retinal degeneration 1 mice. Clin Experiment Ophthalmol. 2013;41(6):593–602. doi: 10.1111/ceo.12046. [DOI] [PubMed] [Google Scholar]
- Glovinsky Y, Quigley HA, Dunkelberger GR. Retinal ganglion cell loss is size dependent in experimental glaucoma. Invest Ophthalmol Vis Sci. 1991;32(3):484–491. [PubMed] [Google Scholar]
- Gramlich OW, Joachim SC, Gottschling PF, Laspas P, Cuny CS, Pfeiffer N, Grus FH. Ophthalmopathology in rats with MBP-induced experimental autoimmune encephalomyelitis. Graefes Arch Clin Exp Ophthalmol. 2011;249(7):1009–1020. doi: 10.1007/s00417-011-1633-9. [DOI] [PubMed] [Google Scholar]
- Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder JM, Libby RT. BBC3 (PUMA) regulates developmental apoptosis but not axonal injury induced death in the retina. Mol Neurodegener. 2011;6:50. doi: 10.1186/1750-1326-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg-Buenz A, Christopher MA, Lewis CJ, Meyer KJ, Rudd DS, Dutca LM, Wang K, Scheetz TE, Abramoff MD, Harper MM, Anderson MG. RetFM-J, an ImageJ-based module for automated counting and quantifying features of nuclei in retinal whole-mounts. Exp Eye Res. 2015 doi: 10.1016/j.exer.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himori N, Maruyama K, Yamamoto K, Yasuda M, Ryu M, Omodaka K, Shiga Y, Tanaka Y, Nakazawa T. Critical neuroprotective roles of heme oxygenase-1 induction against axonal injury-induced retinal ganglion cell death. J Neurosci Res. 2014;92(9):1134–1142. doi: 10.1002/jnr.23398. [DOI] [PubMed] [Google Scholar]
- Howell GR, Libby RT, Jakobs TC, Smith RS, Phalan FC, Barter JW, Barbay JM, Marchant JK, Mahesh N, Porciatti V, Whitmore AV, Masland RH, John SW. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol. 2007;179(7):1523–1537. doi: 10.1083/jcb.200706181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. A quantitative analysis of the cat retinal ganglion cell topography. J Comp Neurol. 1975;163(1):107–128. doi: 10.1002/cne.901630107. [DOI] [PubMed] [Google Scholar]
- Janssen KT, Mac Nair CE, Dietz JA, Schlamp CL, Nickells RW. Nuclear atrophy of retinal ganglion cells precedes the bax-dependent stage of apoptosis. Invest Ophthalmol Vis Sci. 2013;54(3):1805–1815. doi: 10.1167/iovs.11-9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18(21):8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Bezhadian MA, Caldwell RB. Astrocytes modulate retinal vasculogenesis: effects on endothelial cell differentiation. Glia. 1995;15(1):1–10. doi: 10.1002/glia.440150102. [DOI] [PubMed] [Google Scholar]
- Kezic JM, Chrysostomou V, Trounce IA, McMenamin PG, Crowston JG. Effect of anterior chamber cannulation and acute IOP elevation on retinal macrophages in the adult mouse. Invest Ophthalmol Vis Sci. 2013;54(4):3028–3036. doi: 10.1167/iovs.13-11865. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30(4):1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbert C, Apps R, Bechmann I, Lanciego JL, Mey J, Thanos S. Current concepts in neuroanatomical tracing. Prog Neurobiol. 2000;62(4):327–351. doi: 10.1016/s0301-0082(00)00019-8. [DOI] [PubMed] [Google Scholar]
- Koeberle PD, Gauldie J, Ball AK. Effects of adenoviral-mediated gene transfer of interleukin-10, interleukin-4, and transforming growth factor-beta on the survival of axotomized retinal ganglion cells. Neuroscience. 2004;125(4):903–920. doi: 10.1016/S0306-4522(03)00398-1. [DOI] [PubMed] [Google Scholar]
- Kong JH, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. J Comp Neurol. 2005;489(3):293–310. doi: 10.1002/cne.20631. [DOI] [PubMed] [Google Scholar]
- Levin LA, Gordon LK. Retinal ganglion cell disorders: types and treatments. Prog Retin Eye Res. 2002;21(5):465–484. doi: 10.1016/s1350-9462(02)00012-5. [DOI] [PubMed] [Google Scholar]
- Levkovitch-Verbin H, Quigley HA, Kerrigan-Baumrind LA, D'Anna SA, Kerrigan D, Pease ME. Optic nerve transection in monkeys may result in secondary degeneration of retinal ganglion cells. Invest Ophthalmol Vis Sci. 2001;42(5):975–982. [PubMed] [Google Scholar]
- Libby RT, Anderson MG, Pang IH, Robinson ZH, Savinova OV, Cosma IM, Snow A, Wilson LA, Smith RS, Clark AF, John SW. Inherited glaucoma in DBA/2J mice: pertinent disease features for studying the neurodegeneration. Vis Neurosci. 2005;22(5):637–648. doi: 10.1017/S0952523805225130. [DOI] [PubMed] [Google Scholar]
- Libby RT, Li Y, Savinova OV, Barter J, Smith RS, Nickells RW, John SW. Susceptibility to neurodegeneration in a glaucoma is modified by Bax gene dosage. PLoS Genet. 2005;1(1):17–26. doi: 10.1371/journal.pgen.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li ZW, Weinreb RN, Xu G, Lindsey JD, Ye C, Yung WH, Pang CP, Lam DS, Leung CK. Tracking retinal microgliosis in models of retinal ganglion cell damage. Invest Ophthalmol Vis Sci. 2012;53(10):6254–6262. doi: 10.1167/iovs.12-9450. [DOI] [PubMed] [Google Scholar]
- Maeno E, Tsubata T, Okada Y. Apoptotic Volume Decrease (AVD) Is Independent of Mitochondrial Dysfunction and Initiator Caspase Activation. Cells. 2012;1(4):1156–1167. doi: 10.3390/cells1041156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M, Hedberg-Buenz A, Koehn D, John SW, Anderson MG. Anterior segment dysgenesis and early-onset glaucoma in nee mice with mutation of Sh3pxd2b. Invest Ophthalmol Vis Sci. 2011;52(5):2679–2688. doi: 10.1167/iovs.10-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavlyutov TA, Nickells RW, Guo LW. Accelerated retinal ganglion cell death in mice deficient in the Sigma-1 receptor. Mol Vis. 2011;17:1034–1043. [PMC free article] [PubMed] [Google Scholar]
- Mohan K, Kecova H, Hernandez-Merino E, Kardon RH, Harper MM. Retinal ganglion cell damage in an experimental rodent model of blast-mediated traumatic brain injury. Invest Ophthalmol Vis Sci. 2013;54(5):3440–3450. doi: 10.1167/iovs.12-11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JE. Retinal ganglion cell shrinkage in glaucoma. J Glaucoma. 2002;11(4):365–370. doi: 10.1097/00061198-200208000-00015. [DOI] [PubMed] [Google Scholar]
- Morgan JE, Uchida H, Caprioli J. Retinal ganglion cell death in experimental glaucoma. Br J Ophthalmol. 2000;84(3):303–310. doi: 10.1136/bjo.84.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld AH, Kawai S, Das S, Vora S, Gachie E, Connor JR, Manning PT. Loss of retinal ganglion cells following retinal ischemia: the role of inducible nitric oxide synthase. Exp Eye Res. 2002;75(5):521–528. doi: 10.1006/exer.2002.2042. [DOI] [PubMed] [Google Scholar]
- Niemeijer M, Abramoff MD, Joshi N, Brady M. Comparison of classifier performance for information fusion in automated diabetic retinopathy screening. Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on, IEEE. 2011 [Google Scholar]
- Pang JJ, Paul DL, Wu SM. Survey on amacrine cells coupling to retrograde-identified ganglion cells in the mouse retina. Invest Ophthalmol Vis Sci. 2013;54(8):5151–5162. doi: 10.1167/iovs.13-11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JJ, Wu SM. Morphology and immunoreactivity of retrogradely double-labeled ganglion cells in the mouse retina. Invest Ophthalmol Vis Sci. 2011;52(7):4886–4896. doi: 10.1167/iovs.10-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. Scikit-learn: Machine learning in Python. The Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011 Oct;12:2825–2830. [Google Scholar]
- Peichl L. Topography of ganglion cells in the dog and wolf retina. J Comp Neurol. 1992;324(4):603–620. doi: 10.1002/cne.903240412. [DOI] [PubMed] [Google Scholar]
- Perez de Lara MJ, Santano C, Guzman-Aranguez A, Valiente-Soriano FJ, Aviles-Trigueros M, Vidal-Sanz M, de la Villa P, Pintor J. Assessment of inner retina dysfunction and progressive ganglion cell loss in a mouse model of glaucoma. Exp Eye Res. 2014;122:40–49. doi: 10.1016/j.exer.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Perez De Sevilla Muller L, Shelley J, Weiler R. Displaced amacrine cells of the mouse retina. J Comp Neurol. 2007;505(2):177–189. doi: 10.1002/cne.21487. [DOI] [PubMed] [Google Scholar]
- Quigley HA. Neuronal death in glaucoma. Prog Retin Eye Res. 1999;18(1):39–57. doi: 10.1016/s1350-9462(98)00014-7. [DOI] [PubMed] [Google Scholar]
- Quigley HA, Cone FE, Gelman SE, Yang Z, Son JL, Oglesby EN, Pease ME, Zack DJ. Lack of neuroprotection against experimental glaucoma in c-Jun N-terminal kinase 3 knockout mice. Experimental eye research. 2011;92(4):299–305. doi: 10.1016/j.exer.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Dunkelberger GR, Green WR. Retinal ganglion cell atrophy correlated with automated perimetry in human eyes with glaucoma. Am J Ophthalmol. 1989;107(5):453–464. doi: 10.1016/0002-9394(89)90488-1. [DOI] [PubMed] [Google Scholar]
- Reichstein D, Ren L, Filippopoulos T, Mittag T, Danias J. Apoptotic retinal ganglion cell death in the DBA/2 mouse model of glaucoma. Exp Eye Res. 2007;84(1):13–21. doi: 10.1016/j.exer.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Rodriguez AR, de Sevilla Muller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol. 2014;522(6):1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Navarro M, Jimenez-Lopez M, Valiente-Soriano FJ, Alarcon-Martinez L, Aviles-Trigueros M, Mayor S, Holmes T, Lund RD, Villegas-Perez MP, Vidal-Sanz M. Retinal ganglion cell population in adult albino and pigmented mice: a computerized analysis of the entire population and its spatial distribution. Vision research. 2009;49(6):637–647. doi: 10.1016/j.visres.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu Rev Neurosci. 2015 doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Schallek J, Geng Y, Nguyen H, Williams DR. Morphology and topography of retinal pericytes in the living mouse retina using in vivo adaptive optics imaging and ex vivo characterization. Invest Ophthalmol Vis Sci. 2013;54(13):8237–8250. doi: 10.1167/iovs.13-12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlamp CL, Li Y, Dietz JA, Janssen KT, Nickells RW. Progressive ganglion cell loss and optic nerve degeneration in DBA/2J mice is variable and asymmetric. BMC Neurosci. 2006;7:66. doi: 10.1186/1471-2202-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlamp CL, Montgomery AD, Mac Nair CE, Schuart C, Willmer DJ, Nickells RW. Evaluation of the percentage of ganglion cells in the ganglion cell layer of the rodent retina. Molecular vision. 2013;19:1387–1396. [PMC free article] [PubMed] [Google Scholar]
- Silveira LC, Yamada ES, Picanco-Diniz CW. Displaced horizontal cells and biplexiform horizontal cells in the mammalian retina. Vis Neurosci. 1989;3(5):483–488. doi: 10.1017/s0952523800005988. [DOI] [PubMed] [Google Scholar]
- Soto I, Oglesby E, Buckingham BP, Son JL, Roberson ED, Steele MR, Inman DM, Vetter ML, Horner PJ, Marsh-Armstrong N. Retinal ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci. 2008;28(2):548–561. doi: 10.1523/JNEUROSCI.3714-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, Fukuda Y. The naso-temporal division of the cat's retina re-examined in terms of Y-, X- and W-cells. J Comp Neurol. 1974;155(4):377–394. doi: 10.1002/cne.901550402. [DOI] [PubMed] [Google Scholar]
- Strom RC, Williams RW. Cell production and cell death in the generation of variation in neuron number. J Neurosci. 1998;18(23):9948–9953. doi: 10.1523/JNEUROSCI.18-23-09948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbul U, Song S, McCulloch K, Becker M, Lin B, Sanes JR, Masland RH, Seung HS. A genetic and computational approach to structurally classify neuronal types. Nat Commun. 2014;5:3512. doi: 10.1038/ncomms4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiao YC, Blakemore C. Regional specialization in the golden hamster's retina. J Comp Neurol. 1976;168(4):439–457. doi: 10.1002/cne.901680402. [DOI] [PubMed] [Google Scholar]
- Touw WG, Bayjanov JR, Overmars L, Backus L, Boekhorst J, Wels M, van Hijum SA. Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform. 2013;14(3):315–326. doi: 10.1093/bib/bbs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcola JH, Hernandez M, Vecino E. Three experimental glaucoma models in rats: comparison of the effects of intraocular pressure elevation on retinal ganglion cell size and death. Exp Eye Res. 2006;83(2):429–437. doi: 10.1016/j.exer.2006.01.025. [DOI] [PubMed] [Google Scholar]
- van Dijk HW, Verbraak FD, Kok PH, Stehouwer M, Garvin MK, Sonka M, DeVries JH, Schlingemann RO, Abramoff MD. Early neurodegeneration in the retina of type 2 diabetic patients. Invest Ophthalmol Vis Sci. 2012;53(6):2715–2719. doi: 10.1167/iovs.11-8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers JC, Schumer RA, Podos SM, Wang RF, Riederer BM, Morrison JH. Differential vulnerability of neurochemically identified subpopulations of retinal neurons in a monkey model of glaucoma. Brain Res. 1995;680(1–2):23–35. doi: 10.1016/0006-8993(95)00211-8. [DOI] [PubMed] [Google Scholar]
- Volgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol. 2009;512(5):664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Chen SD, Zhang XL, Jonas JB. Retinal Microglia in Glaucoma. J Glaucoma. 2015 doi: 10.1097/IJG.0000000000000200. [DOI] [PubMed] [Google Scholar]
- Wassle H, Levick WR, Cleland BG. The distribution of the alpha type of ganglion cells in the cat's retina. J Comp Neurol. 1975;159(3):419–438. doi: 10.1002/cne.901590308. [DOI] [PubMed] [Google Scholar]
- Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. Jama. 2014;311(18):1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Strom RC, Rice DS, Goldowitz D. Genetic and environmental control of variation in retinal ganglion cell number in mice. J Neurosci. 1996;16(22):7193–7205. doi: 10.1523/JNEUROSCI.16-22-07193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15(7 Pt 1):4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.