Abstract
Bloom (BLM) and Werner (WRN) syndrome proteins are members of the RecQ family of SF2 DNA helicases. In this paper, we show that restricting the rotational DNA backbone flexibility, by introducing vinylphosphonate internucleotide linkages in the translocating DNA strand, inhibits efficient duplex unwinding by these enzymes. The human single-stranded DNA binding protein replication protein A (RPA) fully restores the unwinding activity of BLM and WRN on vinylphosphonate-containing substrates while the heterologous single-stranded DNA binding protein from Escherichia coli (SSB) restores the activity only partially. Both RPA and SSB fail to restore the unwinding activity of the SF1 PcrA helicase on modified substrates, implying specific interactions of RPA with the BLM and WRN helicases. Our data highlight subtle differences between SF1 and SF2 helicases and suggest that although RecQ helicases belong to the SF2 family, they are mechanistically more similar to the SF1 PcrA helicase than to other SF2 helicases that are not affected by vinylphosphonate modifications.
INTRODUCTION
Helicases are motor proteins that use free energy from nucleotide triphosphate hydrolysis for unwinding of nucleic acid duplexes (1–4). Helicases can be classified as one of two types: those that translocate along a nucleic acid strand in a 3′ to 5′ direction and those that operate with the opposite polarity. Based on amino acid sequence homology, helicases have been grouped into five superfamilies (SF1–5) (5). Most of the known 3′–5′ DNA helicases are members of SF1 or SF2. These two superfamilies have similar sets of conserved motifs that are responsible for coupling of ATP hydrolysis to DNA translocation and unwinding. The SF1 and SF2 proteins are mostly monomeric, but the oligomeric state of the functional form of these enzymes remains to be clarified, since both monomeric and oligomeric models have been proposed (6–11). Out of various models proposed for DNA unwinding by helicases, the so-called inchworm model (12) seems to be the most plausible for the SF1 and SF2 enzymes (6,13). This model is reminiscent of a ‘snowplow’ which is pushed or pulled along the nucleic acid duplex mechanically forcing the duplex to dissociate into the constituent strands. Extensive structural and biochemical studies on the SF1 helicase PcrA suggested the mechanism of how the free energy of ATP hydrolysis is converted to the mechanical force required for the motion. The ATP binding site of the enzyme is located in the cleft between two RecA-like domains, with residues from seven helicase signature motifs mediating protein–nucleotide interactions (6). DNA is bound along a groove on the top of the RecA-like domain and the cleft closes and opens in response to ATP binding and hydrolysis, suggesting a mechanism for DNA translocation (6). A similar situation exists also in the other SF1 and SF2 helicases which have been analyzed to atomic resolution (14–16). However, it appears that there is a mechanistic distinction between the SF1 and SF2 helicases in the nature of contacts between the translocating motor and the DNA substrate (4). Structural studies of the SF1 helicases PcrA and Rep revealed that they bind to the substrate via hydrophobic interactions with the bases and thus translocate along single-stranded DNA (ssDNA). In contrast, structural data available for SF2 helicases suggest that these enzymes bind nucleic acids rather via contacts with the phosphodiester backbone, which would allow the helicase motor to translocate on both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) (4). In agreement with the structural data, ATP hydrolysis by SF1 helicases is stimulated only by ssDNA, whereas the ATPase activity of SF2 helicases is stimulated by both ssDNA and dsDNA (4).
The proposed base-flipping mechanism of ssDNA translocation by PcrA helicase has been examined using DNA substrates containing vinylphosphonate internucleotide linkages, which confer reduced rotational flexibility to the DNA backbone with minimal steric hindrance (17). In agreement with the model derived from structural studies, vinylphosphonate linkages have been found to completely inhibit the helicase activity of PcrA as long as they are situated in the translocating strand either at the single-strand/double-strand junction or inside the duplex region (18). The precise mechanism of this inhibitory effect is not known, but such data are consistent with unidirectional translocation of the enzyme along ssDNA mediated via base-stacking interactions between bases and aromatic residues of the enzyme, which is likely to depend on rotational flexibility of various bonds in the DNA backbone.
Here we have examined the effect of vinylphosphonate internucleotide linkages on DNA unwinding activity of two human SF2 helicases BLM and WRN. BLM and WRN belong to the highly-conserved RecQ family of 3′–5′ helicases that are essential for maintenance of genomic stability (19). Inherited mutations in both alleles of the BLM and WRN genes are associated with the cancer-prone disorders Bloom and Werner syndromes, respectively (19). BLM and WRN are DNA-structure-specific helicases that preferentially unwind G-quadruplex DNA and various branched DNA structures such as four-way junctions, D-loops and forked duplexes as compared to 3′-ssDNA-tailed duplexes (20). They can also promote branch migration of Holliday junctions along long stretches of dsDNA (21,22).
We present evidence that vinylphosphonate internucleotide linkages inhibit the helicase activity of both BLM and WRN as long as they are placed on the translocating strand. The observed inhibitory effect is not as drastic as in the case of PcrA but nevertheless it is significant. Moreover we show that the human ssDNA-binding factor replication protein A (RPA) relieves the inhibitory effect of vinylphosphonate modifications on BLM- and WRN-mediated unwinding, whereas it does not affect the PcrA-catalyzed reaction. We conclude that the rotational backbone flexibility of the DNA substrate is important for the action of the BLM and WRN helicases. The mechanism of DNA translocation by these enzymes may involve some base flipping but it is not based entirely on base-flipping as is the case of the SF1 PcrA helicase.
MATERIALS AND METHODS
DNA substrates
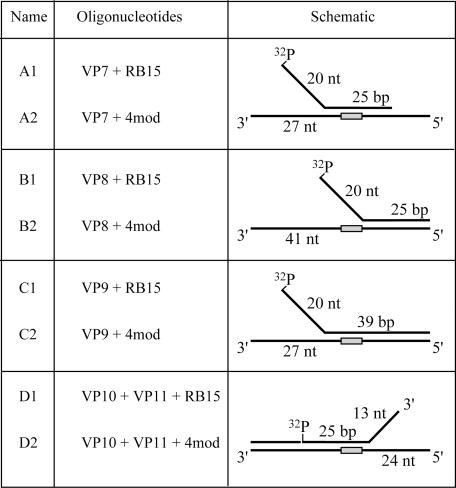
Unmodified oligonucleotides were purchased from Microsynth (Switzerland) and purified by PAGE. The sequences of the oligonucleotides used were as follows (5′ to 3′): VP7 (45mer), TCAAAGTCACGACCTAGACAATCCCCAAAAAAAAAGCTCGAATTC; VP8 (45mer), TCAAAGTCACGACCTAGACAAGCTCGAATTCGTAATCATGGTCAT; VP9 (59mer), TCAAAGTCACGACCTAGACAATCCCCAAAAAAAAAGCTCGAATTCGTAATCATGGTCAT; VP10(38mer), TCTAGAGGATCCCCAAAAAAAAACGAGCTTAAGCATTA; VP11 (19mer), TGCATGCCTGCAGGTCGAC; RB15 (66mer), ATGACCATGATTACGAATTCGAGCTTTTTTTTTGGGGATCCTCTAGAGTCGACCTGCAGGCATGCA. A derivative of RB15, named 4mod, containing a tandem of four vinylphosphonate internucleotide linkages (ATGACCATGATTACGAATTCGAGCTT*TT*TT*TT*TGGGGATCCTCTAGAGTCGACCTGCAGGCATGCA;positions of modifications are indicated by asterisks) was synthesized using 5′-DMTO-T*T-3′CEP phosphoramidite building blocks as described previously (17). It was purified using ABI OPC cartridges according to the manufacturer's instructions. To prepare the splayed-arm DNA substrates shown in Table 1, the VP7, VP8, VP9 and VP10 oligonucleotides were radioactively labeled at the 5′ end using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Amersham Pharmacia Biotech), and annealed to either RB15 (unmodified substrates) or 4mod (modified substrates) under previously described conditions (23).
Table 1. DNA substrates used in this study.
Sequences of the constituent oligonucleotides are described in Materials and Methods. A1, B1, C1 and D1 are unmodified substrates. A2, B2, C2 and D2 are modified substrates. Positions of the four tandem vinylphosphonate modifications in the schemes of individual substrates are indicated as light gray boxes. Note that in substrates D1 and D2, a 19mer oligonucleotide, VP11, was annealed to the 3′ end of RB15 and 4mod, respectively, to ensure that the enzymes enters the duplex only from the splayed-arm side and thereby translocates only along unmodified strand.
Protein purifications
BLM642-1290, WRN and PcrA helicases were prepared as previously described (23–26). Escherichia coli single-stranded DNA binding (SSB) protein was purchased from Promega. Human RPA was prepared essentially as described in (27). Concentrations of proteins are expressed in moles of monomer except for RPA, which is expressed in moles of heterotrimeric complex.
Oligonucleotide-displacement assays
Reactions were carried out at 37°C in a buffer containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 2 mM ATP, 1 mM DTT and 50 μg/ml BSA. Reaction mixtures contained 1 nM 5′ 32P-labeled DNA substrate and 20 nM BLM642–1290 or 5 nM WRN. Control reactions with 100 nM PcrA and 0.5 nM DNA substrate were carried out in a buffer containing 20 mM Tris (pH 7.5), 10 mM MgCl2, 50 mM NaCl, 4 mM DTT, 2.5 mM ATP. Where required, RPA was added to a concentration of 24 nM and SSB to a concentration of 120 nM. Reactions were started with enzyme. Aliquots of 10 μl were removed at indicated time points and mixed with 5 μl of stop solution containing 150 mM EDTA, 2% (w/v) SDS, 30% (v/v) glycerol and 0.1% (w/v) bromophenol blue. The reaction mixtures were resolved on 10% (w/v) polyacrylamide gels (acrylamide to bis-acrylamide ratio 19:1) run in TBE buffer (90 mM Tris–borate, pH 8.3, and 2 mM EDTA) at 7 V/cm and room temperature. Radiolabeled DNA species were visualized by autoradiography and quantified using a Molecular Dynamics Typhoon 9400 scanner with associated IMAGEQUANT software. The relative concentration of displaced products was expressed as a percentage of total DNA.
RESULTS
Inhibition of the DNA helicase activity of BLM by vinylphosphonate internucleotide linkages in the translocating strand
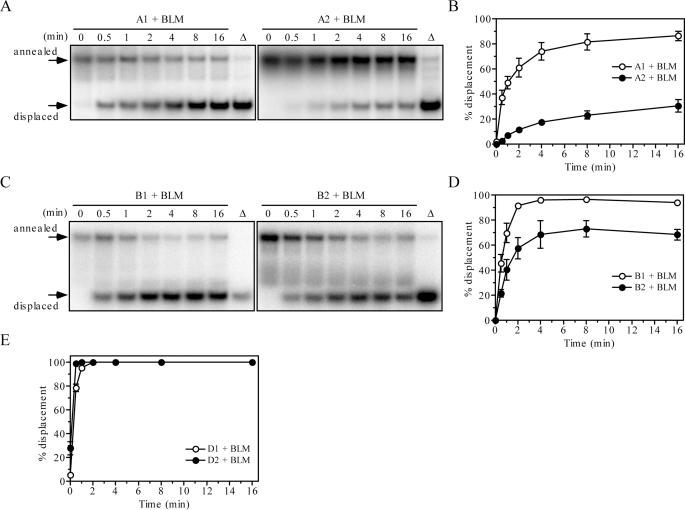
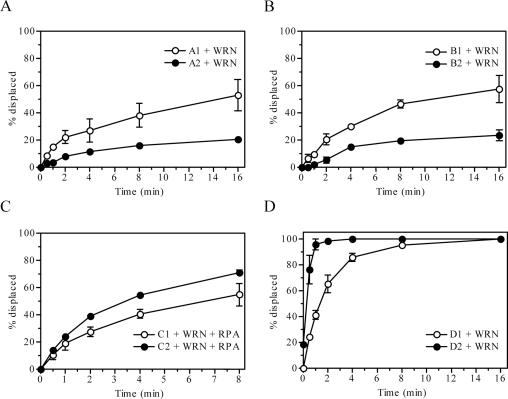
To investigate the effect of vinylphosphonate internucleotide linkages on the helicase activity of BLM, we prepared a series of splayed-arm DNA substrates containing four consecutive modifications either in the translocating or displaced strands at varying positions relative to the single-strand/double-strand junction (Table 1). A fragment of BLM comprising the helicase catalytic core (residues 642–1290, BLM642–1290) was used in these experiments. This variant was found to exhibit a similar specific activity as the full-size BLM protein on a number of DNA structures including splayed arm (23). Using a simple gel-based, oligonucleotide displacement assay, we observed that the BLM642–1290-mediated unwinding of a 25 bp duplex was inhibited when the modifications were located inside the duplex region on the translocating strand (Figure 1A and B). Modifications incorporated just prior to the duplex on the translocating strand also reduced the helicase activity of BLM642–1290 though to a much lesser extent than the modifications inside the duplex (Figure 1C and D). No inhibitory effect was apparent when the modifications were present on the displaced strand inside the duplex (Figure 1E). Overall, these data indicate that restricting the rotational DNA backbone flexibility in the translocating DNA strand impairs BLM-mediated DNA unwinding.
Figure 1.
Effect of vinylphosphonate internucleotide linkages on helicase activity of BLM642–1290. (A) Kinetics of unwinding of 1 nM A1 (left panel) and 1 nM A2 (right panel) by 20 nM BLM642–1290. (B) Quantification of the reactions shown in (A). Data points represent mean values from three independent experiments. (C) Kinetics of unwinding of 1 nM B1 (left panel) and 1 nM B2 (right panel) by 20 nM BLM642–1290. (D) Quantification of the reactions shown in (C). Data points represent mean values from three independent experiments. (E) Kinetics of unwinding of 1 nM D1 (open circles) and 1 nM D2 (filled circles) by 20 nM BLM642–1290. The graph shows mean values from three independent experiments. All reactions were carried out at 37°C and analyzed as described in Materials and Methods. The relative concentration of the strand-displacement products is expressed as a percentage of total DNA. Lanes labeled with triangles contain heat-denatured substrate. DNA substrates are shown in Table 1.
RPA relieves the inhibitory effect of vinylphosphonate internucleotide linkages on BLM-mediated DNA unwinding
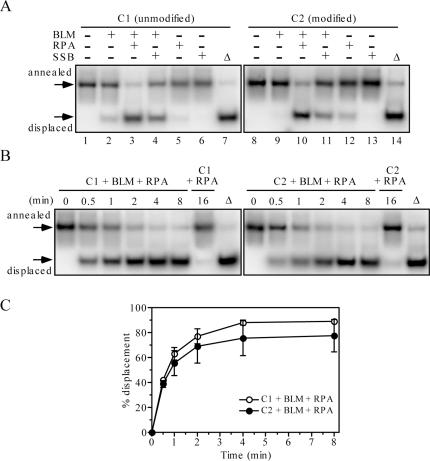
The human RPA has been shown to specifically promote unwinding of long DNA duplexes by BLM and WRN. This stimulatory effect is mediated by physical interaction between RPA and these helicases (28,29). We set up to determine whether RPA can alleviate the inhibitory effect of vinylphoshonate internucleotide linkages on the helicase activity of BLM. In these experiments, we used 39 bp forked duplex substrates with or without four consecutive modifications on the translocating strand located inside the duplex (Table 1; substrates C1 and C2). Under the conditions used, BLM642–1290 alone could only partially unwind the unmodified substrate and showed no detectable helicase activity on the modified substrate (Figure 2A; compare lanes 2 and 9). In contrast, in the presence of RPA, both unmodified and modified substrates were unwound by BLM642–1290 efficiently (Figure 2A; lanes 3 and 10). Substitution of RPA with the E.coli SSB protein also resulted in a stimulation of BLM-catalyzed unwinding of both modified and unmodified substrates, but to a lesser extent than in the case of the RPA-promoted reactions (Figure 2A; lanes 4 and 11). Time-course experiments revealed that the RPA-promoted unwinding reaction on the modified substrate displayed similar kinetics as that with unmodified substrate (Figure 2B and C). Thus, these data indicate that RPA completely relieves the inhibitory effect of vinylphosphonate internucleotide linkages on BLM-catalyzed DNA unwinding.
Figure 2.
Single-stranded DNA binding proteins promote unwinding of vinylphosphonate-containing DNA substrates by BLM642–1290. (A) Effect of 24 nM human RPA and 120 nM E.coli SSB on unwinding of 1 nM C1 and 1 nM C2 by 20 nM BLM642–1290. Reactions were incubated for 20 min. (B) Kinetics of unwinding of 1 nM C1 (left panel) and 1 nM C2 (right panel) by 20 nM BLM642–1290 in the presence of 24 nM RPA. In control reactions, 1 nM DNA was incubated with 24 nM RPA for 16 min. (C) Quantification of the reactions shown in (B). Data points represent mean values from three independent experiments. The relative concentration of the strand-displacement products is expressed as a percentage of total DNA. All reactions were carried out at 37°C and analyzed as described in Materials and Methods. Lanes labelled with triangles contain heat-denatured substrate. DNA substrates are shown in Table 1.
Single-stranded DNA binding proteins do not stimulate the helicase activity of PcrA on DNA substrates containing vinylphosphonate linkages
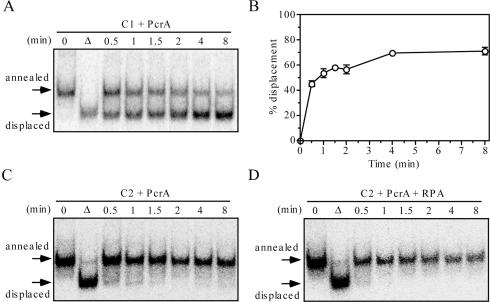
As mentioned above the helicase activity of PcrA has been shown to be inhibited by vinylphosphonate internucleotidelinkages located in the translocating DNA strand (18). In agreement with these findings, the 39 bp splayed arm substrate containing modifications inside the duplex region was not unwound by PcrA, although unmodified substrate was unwound efficiently (Figure 3A, B and C). In contrast to BLM, RPA did not alleviate the inhibitory effect of vinylphosphonate modifications on PcrA-mediated DNA unwinding reaction (Figure 3D). In addition, PcrA helicase activity on modified substrate was not stimulated by E.coli SSB protein (data not shown). These data provide further evidence that the RPA-promoted bypass of regions containing vinyphosphonate internucleotide linkages by BLM is a specific reaction.
Figure 3.
RPA does not alleviate inhibitory effect of vinylphosphonate internucleotide linkages on PcrA-catalyzed DNA unwinding. (A) Kinetics of unwinding of 0.5 nM C1 by 100 nM PcrA. (B) Quantification of the reaction shown in (A). Data points represent mean values from two independent experiments. (C) Time course of the reaction of 100 nM PcrA with 0.5 nM C2. (D) Time course of the reaction of 100 nM PcrA with 0.5 nM C2 in the presence of 24 nM RPA. Reactions were carried out at 37°C and analyzed as described in Materials and Methods. The relative concentration of the strand-displacement products is expressed as a percentage of total DNA. Lanes labelled with triangles contain heat-denatured substrate. DNA substrates are shown in Table 1.
Effect of vinylphosphonate internucleotide linkages on the helicase activity of WRN
Next we sought to examine whether other RecQ DNA helicases display the same characteristics as BLM on DNA substrates containing vinylphosphonate internucleotide linkages. To this end, we tested the WRN helicase for the ability to unwind these substrates in the absence and presence of RPA. Similar to BLM, we found that vinylphosphonate modifications significantly reduced the helicase activity of WRN if located in the translocating strand (Figure 4A and B), but did not produce an inhibitory effect when located on the displaced strand (Figure 4D). In fact, the substrate with modifications in the displaced strand was unwound by WRN more efficiently than the corresponding unmodified substrate. This could be explained partly by an assumption that the vinylphosphonate modifications decrease somewhat the stability of the DNA duplex, since DNA melting studies have revealed that such modifications decrease the melting temperature of DNA duplexes by ∼3.5°C per modification (data not shown). In the case of BLM, such striking differences was not apparent as the reactions were nearly completed at the first time point analyzed (Figure 1E). It should also be emphasized that these modifications do not affect the overall structure of normal B-DNA as revealed by NMR studies (18).
Figure 4.
Effect of vinylphosphonate internucleotide linkages on helicase activity of WRN in the absence and the presence of RPA. (A) Kinetics of unwinding of 1 nM A1 (open circles) and 1 nM A2 (filled circles) by 5 nM WRN. (B) Kinetics of unwinding of 1 nM B1 (open circles) and 1 nM B2 (filled circles) by 5 nM WRN. (C) Kinetics of unwinding of 1 nM C1 (open circles) and 1 nM C2 (filled circles) by 5 nM WRN in the presence of 24 nM RPA. (D) Kinetics of unwinding of 1 nM D1 (open circles) and 1 nM D2 (filled circles) by 5 nM WRN. Reactions were carried out at 37°C and analyzed as described in Materials and Methods. The relative concentration of the strand-displacement products is expressed as a percentage of total DNA. Data points represent mean values from three independent experiments. DNA substrates are shown in Table 1.
The inhibitory effect of vinylphosphonate modifications on WRN-catalyzed unwinding was lost upon addition of RPA (Figure 4C), again highlighting differences between RecQ helicases and PcrA in the mode of DNA translocation.
DISCUSSION
Based on amino acid sequence homology, the RecQ proteins belong to the SF2 superfamily of helicases (5). Several crystal structures of SF2 helicases, including E.coli RecQ (16), HCV NS3 (15,30), UvrB (31), RecG (13) and eIF4a (32) have been solved and revealed structural similarities, but also intriguing differences from the SF1 helicases PcrA (6,33) and Rep (14). While the SF1 helicases interact with ssDNA using mainly hydrophobic interactions with the DNA bases, the SF2 helicases seem to interact with the DNA via non-specific electrostatic interactions with the phosphodiester backbone (4,34). These differences may reflect mechanistic differences, with SF1 helicases apparently able to translocate along ssDNA and the SF2 helicases along both ssDNA and dsDNA.
The translocation mechanism of helicases has been a contentious issue (35), but in the case of PcrA a base-flipping unidirectional mode of translocation with one ATP molecule utilized per base translocated has been suggested (6,36). The base-flipping mechanism requires the DNA base to flip in and out of certain pockets on the surface of the protein as it moves along the ssDNA, which is enabled by rotational flexibility of the DNA backbone. Consistent with this model, restricting backbone rotational freedom by introducing vinylphosphonate internucleotide linkages has a detrimental effect on the PcrA helicase activity (18). Vinylphosphonate backbone modifications do not affect regular bending and ‘stiffness’ of DNA, unless there are sharp kinks induced by the translocating helicase that also require rotational backbone freedom. There is no structural or functional evidence that such sharp kinks are part of the translocation mechanism of any helicase.
Helicases that interact with DNA mainly via the phosphodiester backbone, such as the SF2 HCV NS3 (Eckhard Jankowsky, personal communication) and RecG (Robert G. Lloyd, personal communication), are able to translocate over vinylphosphonate modifications without apparent problems. In contrast, here we show that two other SF2 helicases, BLM and WRN, are inhibited by vinylphosphonate modifications. As these modifications in the A2 and C2 substrates reside inside the duplex region, it is unlikely that they will inhibit the initial binding of BLM and WRN proteins to the ssDNA/dsDNA junction. Instead, the inhibition must be related to the actual translocation process. Although the inhibitory effect of vinylphosphonate internucleotide linkages on BLM- and WRN-mediated DNA unwinding is not as drastic as that observed for PcrA, it is significant, suggesting that rotational DNA backbone freedom is an essential requirement for efficient helicase activity in these helicases. Therefore, the translocation mechanism of BLM and WRN seems to involve some base-flipping aspect, as in PcrA, but it is not entirely dependent on base-flipping, unlike PcrA (partial inhibition versus complete inhibition). As mentioned above, other SF2 helicases such as HCV NS3 and RecG are unaffected by vinylphosphonate modifications, suggesting that their mechanism of translocation does not involve base-flipping and is instead based upon interaction with the phosphodiester backbone in agreement with structural studies. Moreover, the SF2 helicase NPH-II has been recently shown to tolerate abasic sites in the translocating strand, while it essentially requires physical continuity of the phosphodiester linkage, again suggesting a mechanism in which the protein makes contact with the sugar-phosphate backbone during translocation (37). The recent crystal structure of the E.coli RecQ revealed a patch of conserved aromatic and charged residues (residues 156–159, C-terminal to motif II) mapping to the same relative surface location as similar residues in PcrA that are used in DNA binding (residues 257–260, C-terminal to motif III) (16). This structural feature suggests that E.coli RecQ may utilize a combination of base-flipping and phosphodiester backbone interactions to translocate along ssDNA, although the structure of RecQ complexed with DNA is required to confirm these observations.
The stimulatory effect of RPA on BLM- and WRN-mediated unwinding of vinylphosphonate-containing DNA substrates and the failure of RPA to stimulate PcrA imply a specific interaction between RPA and both BLM and WRN helicases. This is further supported by the fact that heterologous SSB from E.coli stimulated BLM and WRN helicases on modified substrates only marginally. This effect is likely to reflect a simple non-specific trapping of displaced strands by SSB. BLM and WRN helicases exhibit a very low processivity (28,29). RPA has been shown to specifically stimulate BLM- and WRN-mediated unwinding through direct protein–protein interactions (28,29). It has been postulated that in addition to trapping displaced single-strands, RPA may tether the helicase to the DNA substrate at the single-strand/double-strand junction to facilitate the progression of the enzyme through relatively long DNA duplex tracts (28,29). Such a mechanism would explain the observed stimulatory effect of RPA on BLM- and WRN-mediated unwinding of vinylphosphonate-linked substrates. Providing that the modifications cause a pause in enzyme translocation followed by its dissociation from the DNA substrate, one can assume that tethering the enzyme to the translocating strand via interaction with ssDNA-bound RPA would allow it to cope with a more rotationally rigid region in the DNA template during the unwinding reaction.
Our study provides evidence of subtle differences and similarities between SF1 and SF2 helicases. Although RecQ helicases belong to SF2, they appear to be different from other SF2 helicases such as HCV NS3, RecG and NPH-II. They may translocate along ssDNA by using a combination of base-flipping (characteristic of the SF1 PcrA helicase) and non-specific phosphodiester–backbone interactions (characteristic of SF2 HCV NS3, RecG and NPH-II helicases). In that sense RecQ helicases may mechanistically belong to a sub-family of SF2 helicases.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Richard Bertram and John Keyte for help with the preparation of vinylphosphonate DNA substrates. This work was supported by a Swiss National Science Foundation grant to P.J. and a Wellcome Trust grant (064751/Z/01/Z) to P.S.
REFERENCES
- 1.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 2.Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. [DOI] [PubMed] [Google Scholar]
- 3.von Hippel P.H. and Delagoutte,E. (2001) A general model for nucleic acid helicases and their ‘coupling’ within macromolecular machines. Cell, 104, 177–190. [DOI] [PubMed] [Google Scholar]
- 4.Singleton M.R. and Wigley,D.B. (2002) Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol., 184, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 6.Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 7.Levin M.K. and Patel,S.S. (1999) The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem., 274, 31839–31846. [DOI] [PubMed] [Google Scholar]
- 8.Cheng W., Hsieh,J., Brendza,K.M. and Lohman,T.M. (2001) E.coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol., 310, 327–350. [DOI] [PubMed] [Google Scholar]
- 9.Ha T., Rasnik,I., Cheng,W., Babcock,H.P., Gauss,G.H., Lohman,T.M. and Chu,S. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature, 419, 638–641. [DOI] [PubMed] [Google Scholar]
- 10.Maluf N.K., Fischer,C.J. and Lohman,T.M. (2003) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol., 325, 913–935. [DOI] [PubMed] [Google Scholar]
- 11.Levin M.K., Wang,Y.H. and Patel,S.S. (2004) The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem., 279, 26005–26012. [DOI] [PubMed] [Google Scholar]
- 12.Yarranton G.T. and Gefter,M.L. (1979) Enzyme-catalyzed DNA unwinding: studies on Escherichia coli Rep protein. Proc. Natl Acad. Sci. USA, 76, 1658–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singleton M.R., Scaife,S. and Wigley,D.B. (2001) Structural analysis of DNA replication fork reversal by RecG. Cell, 107, 79–89. [DOI] [PubMed] [Google Scholar]
- 14.Korolev S., Hsieh,J., Gauss,G.H., Lohman,T.M. and Waksman,G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- 15.Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein D.A., Zittel,M.C. and Keck,J.L. (2003) High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J., 22, 4910–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbas S., Bertram,R.D. and Hayes,C.J. (2001) Commercially available 5′-DMT phosphoramidites as reagents for the synthesis of vinylphosphonate-linked oligonucleic acids. Org. Lett., 3, 3365–3367. [DOI] [PubMed] [Google Scholar]
- 18.Bertram R.D., Hayes,C.J. and Soultanas,P. (2002) Vinylphosphonate internucleotide linkages inhibit the activity of PcrA DNA helicase. Biochemistry, 41, 7725–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachrati C.Z. and Hickson,I.D. (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J., 374, 577–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohaghegh P., Karow,J.K., Brosh,R.M.Jr., Bohr,V.A. and Hickson,I.D. (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Constantinou A., Tarsounas,M., Karow,J.K., Brosh,R.M., Bohr,V.A., Hickson,I.D. and West,S.C. (2000) Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep., 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janscak P., Garcia,P.L., Hamburger,F., Makuta,Y., Shiraishi,K., Imai,Y., Ikeda,H. and Bickle,T.A. (2003) Characterization and mutational analysis of the RecQ core of the Bloom syndrome protein. J. Mol. Biol., 330, 29–42. [DOI] [PubMed] [Google Scholar]
- 24.Orren D.K., Brosh,R.M.,Jr, Nehlin,J.O., Machwe,A., Gray,M.D. and Bohr,V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nature Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- 26.Bird L.E., Brannigan,J.A., Subramanya,H.S. and Wigley,D.B. (1998) Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res., 26, 2686–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 28.Brosh R.M. Jr, Li,J.L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 29.Brosh R.M. Jr, Orren,D.K., Nehlin,J.O., Ravn,P.H., Kenny,M.K., Machwe,A. and Bohr,V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- 30.Yao N., Hesson,T., Cable,M., Hong,Z., Kwong,A.D., Le,H.V. and Weber,P.C. (1997) Structure of the hepatitis C virus RNA helicase domain. Nature Struct. Biol., 4, 463–467. [DOI] [PubMed] [Google Scholar]
- 31.Machius M., Henry,L., Palnitkar,M. and Deisenhofer,J. (1999) Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl Acad. Sci. USA, 96, 11717–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caruthers J.M., Johnson,E.R. and McKay,D.B. (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA, 97, 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 34.Korolev S., Yao,N., Lohman,T.M., Weber,P.C. and Waksman,G. (1998) Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci., 7, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soultanas P. and Wigley,D.B. (2001) Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci., 26, 47–54. [DOI] [PubMed] [Google Scholar]
- 36.Dillingham M.S., Wigley,D.B. and Webb,M.R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry, 39, 205–212. [DOI] [PubMed] [Google Scholar]
- 37.Kawaoka J., Jankowsky,E. and Pyle,A.M. (2004) Backbone tracking by the SF2 helicase NPH-II. Nature Struct. Mol. Biol., 11, 526–530. [DOI] [PubMed] [Google Scholar]