Abstract
Background: Protein-energy undernutrition during early development confers a lifelong increased risk of obesity-related metabolic disease. Mechanisms by which metabolic abnormalities persist despite catch-up growth are poorly understood.
Objective: We sought to determine whether abnormal metabolomic and intestinal microbiota profiles from undernourished neonatal mice remain altered during catch-up growth.
Methods: Male and female CD1 mouse pups were undernourished by timed separation from lactating dams for 4 h at 5 d of age, 8 h at 6 d of age, and 12 h/d from 7 to 15 d of age, then resumed ad libitum nursing, whereas controls fed uninterrupted. Both groups were weaned simultaneously to a standard unpurified diet. At 3 time points (0, 1, and 3 wk after ending feed deprivation), metabolites in urine, plasma, and stool were identified with the use of mass spectrometry, and fecal microbes were identified with the use of 16S metagenomic sequencing.
Results: Undernourished mice completely recovered deficits of 36% weight and 9% length by 3 wk of refeeding, at which time they had 1.4-fold higher plasma phenyllactate and 2.0-fold higher urinary p-cresol sulfate concentrations than did controls. Plasma serotonin concentrations in undernourished mice were 25% lower at 0 wk but 1.5-fold higher than in controls at 3 wk. Whereas most urine and plasma metabolites normalized with refeeding, 117 fecal metabolites remained altered at 3 wk, including multiple N-linked glycans. Microbiota profiles from undernourished mice also remained distinct, with lower mean proportions of Bacteroidetes (67% compared with 83%) and higher proportions of Firmicutes (26% compared with 16%). Abundances of the mucolytic organisms Akkermansia muciniphila and Mucispirillum schaedleri were altered at 0 and 1 wk. Whereas microbiota from undernourished mice at 0 wk contained 11% less community diversity (P = 0.015), refed mice at 3 wk harbored 1.2-fold greater diversity (P = 0.0006) than did controls.
Conclusion: Microbial-derived metabolites and intestinal microbiota remain altered during catch-up growth in undernourished neonatal mice.
Keywords: metagenomics, metabolomics, mucolytic bacteria, N-linked glycans, neonatal mice, protein-energy undernutrition, one-carbon metabolism, p-cresol sulfate, phenyllactate, serotonin
Introduction
Diarrheal diseases and protein-energy undernutrition (PEU)7 are ruthless coconspirators. They contribute to one-half of all deaths in children <5 y of age, and when present during critical periods of early development, they confer upon survivors lifelong sequelae of stunting, lower intelligence quotient, impaired vaccine efficacy, and predisposition to obesity, diabetes, and heart disease (1–3).
Recent studies implicate intestinal microbes in the pathogenesis of persistent PEU. First, in a randomized controlled trial, Malawian children with PEU given outpatient therapeutic feeding demonstrated improved weight gain when oral antibiotics were given (4), a result that is thought to be due to the restructuring of pathologic intestinal microbial communities (5). Second, Bangladeshi children with PEU had an immature fecal microbiota that correlated with a lack of sustained benefit from therapeutic refeeding (6). Third, gnotobiotic mice transplanted with fecal microbes derived from undernourished Malawian children demonstrated more severe weight loss on a regional diet than did mice receiving microbiota from a healthy twin (7). Together, these studies suggest that a pathologic microbiome may be present in PEU.
A neonatal mouse model of PEU was used recently to define the metabolome (all small molecules detected in multiple body compartments), revealing products of muscle and fat breakdown and evidence of oxidative stress, inflammation, liver injury, and early insulin resistance (8). Likewise, the intestinal microbiome of undernourished mice (U) is less diverse and contains fewer microbial genes that extract energy from nondigestible dietary components (9). The aim of the present study was to refeed U to determine whether these altered metabolite and intestinal microbial profiles resolve during catch-up growth. We hypothesized that a subset of metabolites and microbes harvested during refeeding and after deficits in weight and length had been recovered would identify mice that had been undernourished early in life.
Methods
Mouse model.
Newborn male and female outbred CD1 mice (Charles River Laboratories) were randomly assigned and reassigned to litters of 10/dam. Treatment groups were maintained in separate cages to avoid coprophagic contamination. The same 5 U/litter underwent feed deprivation (FD) by separation from mothers for 4 h at 5 d of age, 8 h at 6 d of age, and 12 h/d from 7 to 15 d of age, with nightly ad libitum nursing (8). Control mice (C) were maintained in litters of 10/dam without FD. All mice were weaned to standard unpurified rodent feed at 22 d of age. Irradiated Teklad Global Soy Protein-Free Extruded Rodent Diet (calories from protein, 24%; fat, 16%; and carbohydrate, 60%) was used for all dams and weaned pups. All protocols were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Samples were collected at 15 d of age, the final day of FD before resumption of ad libitum feeding (0 wk), and at 1 and 3 wk after ending FD (22 and 36 d of age). The final time point occurred 2 wk postweaning. Abdominal massage produced urine and stool, then blood was collected by exsanguination. Individual pups were too small to provide sufficient sample volumes for metabolomic analyses; thus, pooling was necessary. At 0 wk, samples from 4 pups were pooled to create each metabolomic data point; at 1 wk, samples from 2 pups were pooled to create each metabolomic data point. By 3 wk after ending FD, mice were large enough that pooling was not needed, and each 3 wk data point represents urine, stool, or blood from an individual mouse. In contrast, 16S sequencing requires minimal stool; thus, each sample represents microbial DNA from a single mouse. All samples were sealed in sterile freezer tubes and stored at −80°C until analysis.
Metabolomic profiling.
Samples of equivalent mass volumes or osmolality (urine) were divided into aliquots and analyzed on both LC-tandem MS and gas chromatography–mass spectrometry platforms as recently described (8). Raw spectrometry data were extracted, quality control measures were imposed, and biochemicals were identified with the use of a library of >4500 purified standards. Raw data were log-transformed and median-scaled, with minimal values imputed with the use of Array Studio (OmicSoft Corporation), and fold-changes reported as U/C at each time point.
Metagenomic sequencing and alignment.
Sequencing methods have been reported in detail (9). Briefly, the 16S V4 region was sequenced on the MiSeq platform (Illumina), generating a median 35,889 mapped reads/sample. Reads were trimmed, quality-filtered, clustered into operational taxonomic units (OTUs) with a 97% similarity cutoff with the use of the UPARSE algorithm (10), and aligned to the SILVA database (11).
Statistical methods.
Weights and lengths were evaluated with 2-tailed t tests. Relative concentrations of metabolites were evaluated with Welch’s t-test, with α = 0.05 and Q values estimating the false-positive rate from multiple comparison testing. Each pool was treated as a single data point for statistical analyses. Heat maps were generated based on relative fold-change and significance. The randomForest package of R (12) was used to determine the most important metabolites distinguishing 2 groups. Principal coordinate analysis plots were built with the use of the Monte Carlo permutation test (13) to estimate P values, which were adjusted for multiple comparisons with Benjamini-Hochberg’s false discovery rate algorithm (14). Shannon’s diversity and richness indexes were calculated with the use of phyloseq in R (15) and evaluated with Mann-Whitney tests.
Results
Catch-up growth after undernutrition and global metabolomic trends.
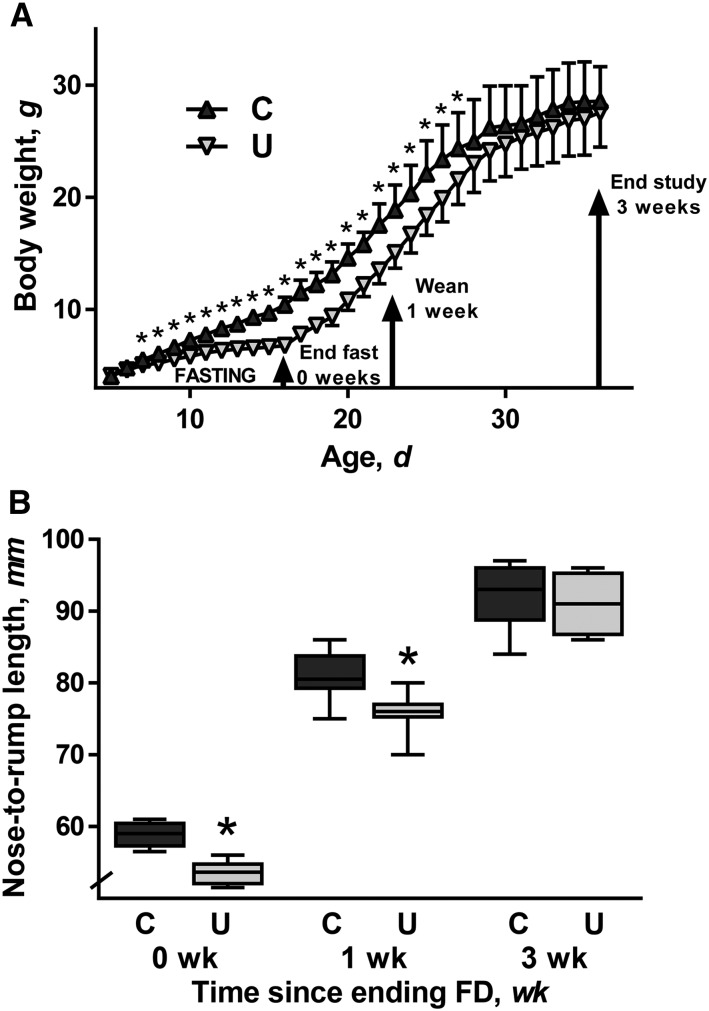
After 11 d of FD (15 d of age), mean body weight was 36% lower in U than in C (P < 0.001) and mean length was 9% lower (P < 0.001) (Figure 1), confirming previous results (8, 9). After initiation of ad libitum feeding, U had significantly lower body weight than did C for another 13 d (27 d of age). However, by 3 wk of refeeding (37 d of age), the mean weight in both groups equaled 28 g. Stunting persisted in U after 1 wk of refeeding (P = 0.005), but the 2 groups had equal mean lengths after 3 wk. Thus, U achieved catch-up growth through ad libitum feeding and were indistinguishable from C in terms of body weight and length at 3 wk.
FIGURE 1.
Body weight (A) and nose-to-rump length (B) of mouse pups from the C and U groups from 5 to 36 d of age. Triangles and error bars represent mean weights and SDs. The tops and bottoms of the boxes represent IQRs, the solid line represents median length, and the bars (whiskers) represent the range of data points; n = 10 mice/group. *Different from C, P < 0.05. C, control mice; FD, feed deprivation; U, mice undernourished from 5 to 15 d of age.
In all, 321 biochemicals of known identity were found in urine, 333 in plasma, and 423 in stool. Of these, 621 metabolite pairs differed in relative concentrations (P < 0.05) between U and C. The urine and plasma metabolomes of U became more similar to C during refeeding, with just 20 urine and 15 plasma metabolites different between the groups at 3 wk (Table 1). However, the stool metabolome of U did not normalize. Of the 116 altered fecal metabolite pairs at 0 wk, 91% were found in significantly lower concentrations in U than in C. Of the 117 altered metabolite pairs at 3 wk, 91% were found in significantly higher concentrations in U than in C.
TABLE 1.
Number of altered metabolite pairs found in the urine, plasma, and stool of mouse pups from the C and U groups at 15, 22, and 36 d of age1
| 0 wk |
1 wk |
3 wk |
|||||||
| Compartment | Total | U > C | U < C | Total | U > C | U < C | Total | U > C | U < C |
| Urine | 98 | 11 | 87 | 71 | 8 | 63 | 20 | 15 | 5 |
| Plasma | 73 | 19 | 54 | 32 | 7 | 25 | 15 | 1 | 14 |
| Stool | 116 | 105 | 11 | 79 | 65 | 14 | 117 | 11 | 106 |
n = 8 pools (4 mice/pool) at 0 wk; n = 8 pools (2 mice/pool) at 1 wk; and n = 8 mice at 3 wk after ending FD. C, control mice; FD, feed deprivation; U, mice undernourished from 5 to 15 d of age.
Altered aromatic amino acid metabolism, inflammation, and protein glycosylation.
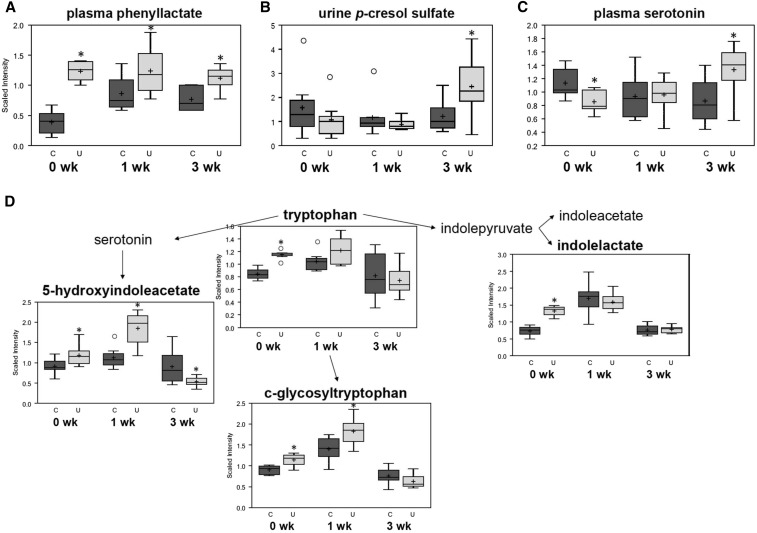
The most perturbed group of metabolites during refeeding were aromatic amino acid derivatives, including Phe and its catabolite phenyllactate. Plasma quantities of phenyllactate were consistently higher in U than in C, by 3.2-fold at 0 wk (P = 5 × 10−4), 1.4-fold at 1 wk (P = 0.040), and 1.5-fold at 3 wk (P = 0.004) after ending FD (Figure 2A). Altered quantities of phenyllactate and Phe were found in urine from U, and numerous aromatic amino acid metabolites were found in abnormal concentrations in stool (Supplemental Figure 1). Concentrations of another Phe derivative, p-cresol sulfate, were 2.0-fold higher in the urine from U than in that from C at 3 wk (P = 0.047; Figure 2B). Furthermore, plasma concentrations of tryptophan derivative serotonin were 25% lower in U than in C at 0 wk (P = 0.011), unchanged after 1 wk, and 1.5-fold higher in U than in C at 3 wk (P = 0.033; Figure 2C). Urine from U also contained altered concentrations of multiple tryptophan metabolites (Figure 2D). These data suggest global abnormalities in aromatic amino acid metabolism during refeeding.
FIGURE 2.
Select aromatic amino acid metabolites differing between mouse pups from the C and U groups at 15, 22, and 36 d of age. Phenyllactate in plasma (A); p-cresol sulfate in urine (B); serotonin in plasma (C); and catabolites of tryptophan and serotonin in urine (D). The top and the bottom of the boxes represent IQRs, the solid line represents the median, the + sign represents the mean, and the bars (whiskers) represent the range of data points, excluding extreme values denoted by open circles. The y axes depict relative scaled intensity; the x axes depict time since ending FD; n = 8 pools (4 mice/pool) at 0 wk; n = 8 pools (2 mice/pool) at 1 wk; and n = 8 mice at 3 wk after ending FD. *Different from C, P < 0.05. C, control mice; FD, feed deprivation; U, mice undernourished from 5 to 15 d of age.
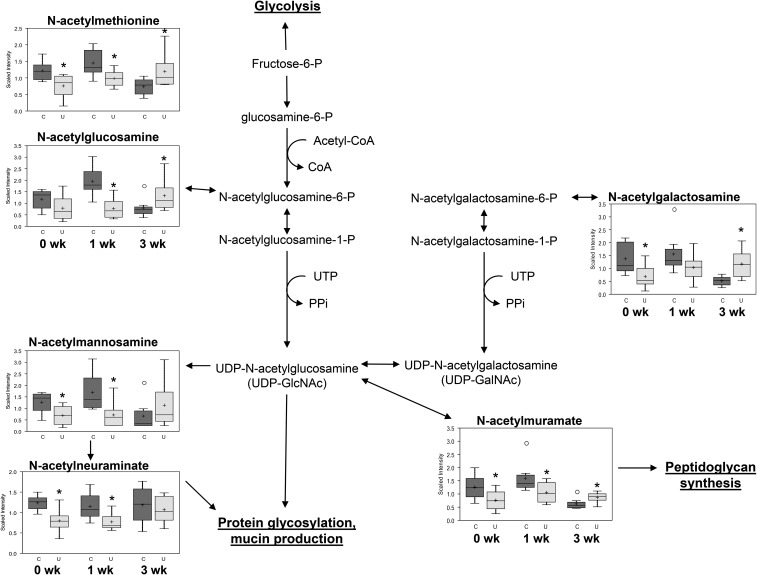
Relative concentrations of fecal N-linked glycans differed at each time point (Figure 3), with quantities being lower in U than in C at 0 wk and higher in U than in C at 3 wk after ending FD. These include the peptidoglycan cell wall components N-acetylglucosamine and N-acetylmuramate; the mucus layer component N-acetylneuraminate; and the amino sugars N-acetylmannosamine, N-acetylgalactosamine, and N-acetylmethionine. Multiple markers of inflammation were present in U at 0 wk, and many of these remained altered during refeeding (Supplemental Table 1). Together, these data suggest abnormal protein glycosylation and inflammatory responses, despite normalization of growth markers.
FIGURE 3.
Fecal metabolites involved in amino sugar metabolism differing between mouse pups from the C and U groups at 15, 22, and 36 d of age. The top and the bottom of the boxes represent IQRs, the solid line represents the median, the + sign represents the mean, and the bars (whiskers) represent the range of data points, excluding extreme values denoted by open circles. The y axes depict relative scaled intensity; the x axes depict time since ending FD; n = 8 pools (4 mice/pool) at 0 wk; n = 8 pools (2 mice/pool) at 1 wk; n = 8 mice at 3 wk after ending FD. *Different from C, P < 0.05. C, control mice; FD, feed deprivation; P, phosphate; PPi, inorganic pyrophosphate; U, mice undernourished from 5 to 15 d of age.
The fecal metabolome during refeeding.
A global heat map was constructed to illustrate the direction and significance of change of biochemicals in stool from U after ending FD (Supplemental Figure 2A). Quantities of most altered metabolites were lower in U than in C at 0 wk and higher in U than in C at 3 wk, including BCAA catabolites (Supplemental Figure 2B) and methionine and N-acetylmethionine, which mediate one-carbon metabolism (Supplemental Figure 2C). Plasma and urine methionine was also higher in U than in C at 3 wk (Supplemental Figure 2D). Altered γ-glutamyltransferase activity, evidenced by abnormal γ-glutamyl amino acid concentrations in the stool of U at 0 and 1 wk, was not observed at 3 wk (Supplemental Figure 2E). On the other hand, quantities of dipeptide protein digestion products that were not altered during FD were higher in the stool of U than in that of C at 3 wk; among several examples were the leucine-conjugated dipeptides (Supplemental Figure 2F). Finally, concentrations of glucose and other mediators of carbohydrate energy metabolism were lower in the stool of U than in that of C at 0 wk and higher in the stool of U than in that of C at 3 wk (Supplemental Figure 2G). The urine from U demonstrated glucosuria at 1 wk (Supplemental Figure 2H), along with altered quantities of mediators of pentose metabolism (Supplemental Figure 3A–G) and of fructose, mannose, and galactose metabolism (Supplemental Figure 3H–K). Altogether, these data suggest abnormal macronutrient and energy metabolism during catch-up growth.
To determine which metabolites most strongly distinguished the fecal profiles of U compared with C 3 wk after ending FD, we constructed a random forest model, which correctly classified the 16 stool samples as derived from U or C with 87.5% accuracy (Supplemental Figure 4). Fifteen of the 30 biochemicals that were most important to this classification were dipeptides found in greater quantities in U than in C, including the 5 highest-ranked metabolites, serylisoleucine, asparagylisoleucine, asparagylleucine, phenylalanylglutamate, and alanylglutamate. Another 5 of the top 30—mannose, glucose-6-phosphate, fructose, ribulose, and galactose—were carbohydrate mediators of energy metabolism and were present in greater quantities in U than in C. Random forest models for plasma and urine at 3 wk were less effective predictors than stool models, achieving just 62% and 69% predictive accuracy. Thus, hallmark metabolites identifying mice that previously were feed deprived were macronutrient digestion products found in greater quantities in the stool of U than in that of C.
Microbes present in stool during catch-up growth.
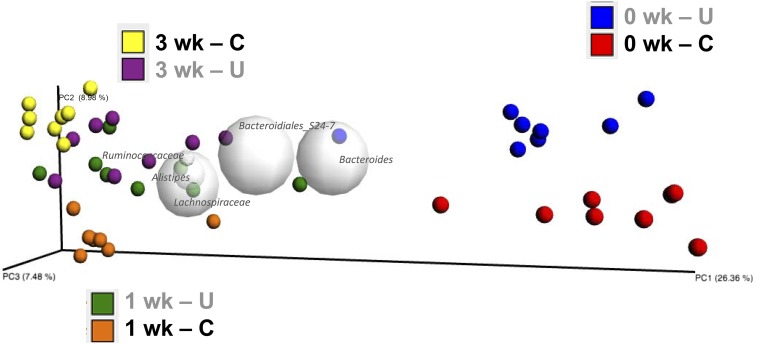
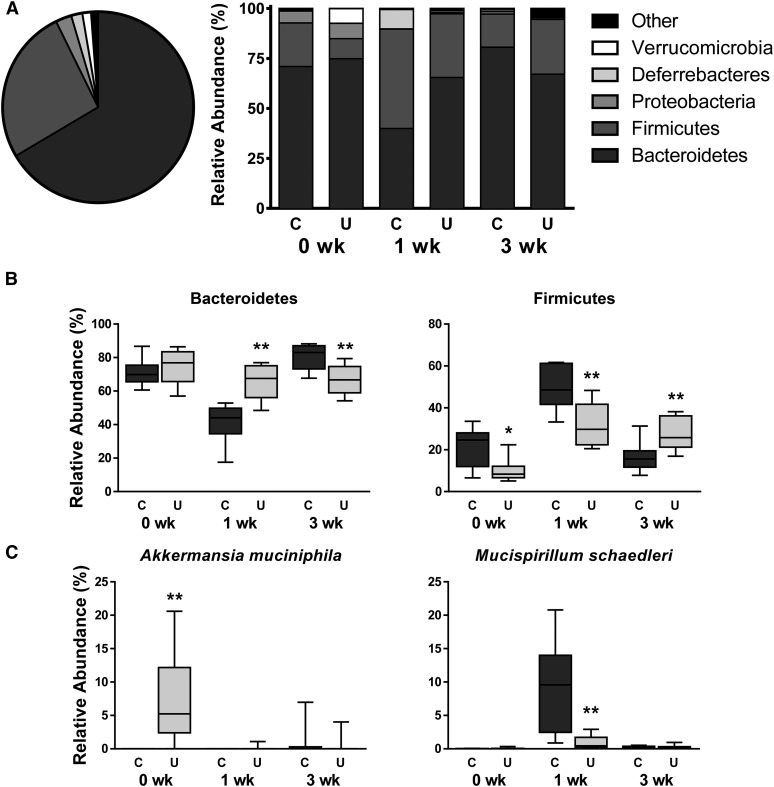
We sought to determine whether the altered fecal metabolites during refeeding corresponded with altered gut microbiota. On a principal coordinate analysis plot, profiles from U compared with C separated at each time point (Figure 4), indicating that microbial community composition differed throughout catch-up growth. Altogether, 93% of bacteria belonged to 2 phyla, Bacteroidetes and Firmicutes (Figure 5A), both of which were altered during refeeding (Figure 5B), whereas 63% of OTUs belonged to just 3 genera: an unclassified genus in the order Bacteroidales, Bacteroides, and Blautia (Supplemental Figure 5). Two phylum-level changes were driven by 2 organisms residing in the mucus layer (Figure 5C). The greater abundance of Verrucomicrobia in the stool of U than in that of C at 0 wk was due to a 2000-fold bloom of Akkermansia muciniphila (P = 0.001), consistent with previous findings (9), and the lower abundance of Deferribacteres in the stool of U than in that of C at 1 wk was due to an order-of-magnitude loss of Mucispirillum schaedleri (P = 0.004). Together, these data suggest that differences in fecal microbial community composition due to PEU do not resolve with refeeding.
FIGURE 4.
Principal coordinate analysis plot of microbes derived from stool of mouse pups from the C and U groups at 15, 22, and 36 d of age; n = 8 mice/group. C, control mice; PC, principal coordinate; U, mice undernourished from 5 to 15 d of age.
FIGURE 5.
Relative abundance of fecal bacteria in mouse pups from the C and U groups at 15, 22, and 36 d of age. (A) Phylum-level relative abundance plots depicting all groups combined on one pie chart and individual treatment groups in stacked bars. (B) Changes over time in most abundant phyla, Bacteroidetes and Firmicutes. (C) Changes over time in mucolytic species A. muciniphila and M. schaedleri; see also Supplemental Figure 5. The x axes depict time since ending FD; n = 8 mice/group. *,**Different from C: *P < 0.05, **P < 0.01. C, control mice; FD, feed deprivation; U, mice undernourished from 5 to 15 d of age.
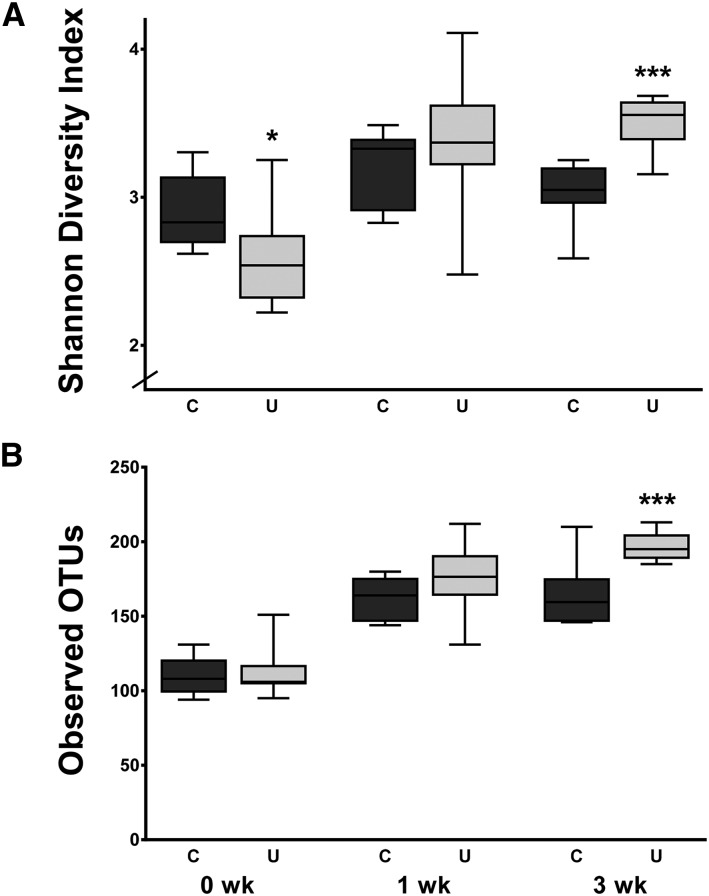
Also confirming previous findings (9), microbial communities in the stool of U were less diverse than they were in C during FD (P = 0.015; Figure 6A), although this difference dissipated after 1 wk of refeeding. Unexpectedly, after 3 wk of refeeding, the stool of U harbored 1.2-fold greater microbiota diversity than did that of C (P = 0.0006). This greater diversity at 3 wk was driven by greater richness of the fecal microbiota of U; a median 35.5 additional unique OTUs were detected in the stool of U than in that of C at 3 wk (P = 0.0064; Figure 6B). These data suggest that a PEU-induced loss of fecal microbial diversity is reparable in neonatal mice that are given sufficient dietary intake.
FIGURE 6.
Microbial community diversity in the feces of mouse pups from the C and U groups at 15, 22, and 36 d of age. Shannon diversity index (A) and richness defined as the number of unique OTUs per sample (B); the x axes depict time since ending FD; n = 8 mice/group. *,***Different from C: *P < 0.05, ***P < 0.001. C, control mice; FD, feed deprivation; OTU, operational taxonomic unit; U, mice undernourished from 5 to 15 d of age.
Discussion
Timed separation of suckling mice from dams reproduces pathologies seen in the vicious cycle of childhood diarrheal diseases and PEU (1, 2). This model replicates underweight and stunting, worsens Cryptosporidium parvum and enteroaggregative Escherichia coli infections to perpetuate weight loss (16–18), and blunts probiotic enhancement of antibodies, enterocyte proliferation, and villus repopulation in rotavirus-infected mice (19). PEU models have not yet revealed why long-term metabolic and cognitive comorbidities develop after recovery from PEU (1–3), although recent data indicate that timed separation drastically alters the metabolome (8) and intestinal microbiome (9) in pups from the U group. The aim of the present study was to determine whether the altered metabolomic and intestinal microbial profiles would normalize with refeeding.
Whereas the urine and plasma metabolomes of U improved with refeeding, the fecal metabolome and microbiota of U remained markedly perturbed despite complete recovery of weight and length deficits. This finding suggests that refeeding after early PEU affects intestinal and systemic compartments differently. Given that biochemicals in urine and plasma may reflect metabolism inside the body more accurately than biochemicals in stool, the lack of persistence of abnormalities in these compartments could imply that most metabolic derangements are correctable with diet, despite an altered intestinal environment. However, of the abnormal urine and plasma markers 3 wk after ending FD (Supplemental Tables 2 and 3), a surprising number are microbial-derived molecules, suggesting that gut bacteria might have a greater than expected influence on host metabolism during refeeding.
The biomarker that most consistently separated the 2 groups over time was phenyllactate. In the absence of inborn errors of metabolism, phenyllactate is derived almost entirely by gut microbes from the Phe derivative phenylpyruvate. The mammalian commensal genera Bifidobacterium, Lactobacillus, and Lactococcus have phenyllactate synthetic machinery (20–23), and analogous genes are found in diverse phyla, suggesting an evolutionary advantage to deriving fuel from Phe and other alternative sources (24–26). Enriching for phenyllactate-synthesizing microbes could affect host health. In vitro, phenyllactate decreases production of reactive oxygen species by mitochondria (20, 27) and inhibits growth of fungal (28, 29) and bacterial (30, 31) pathogens. Another microbial-derived product of aromatic amino acid metabolism, p-cresol, is linked to neuropsychiatric sequelae (32–35). At 3 wk, p-cresol sulfate was significantly elevated in the urine of U, with a trend toward elevation in plasma (1.8-fold; P = 0.070); this metabolite warrants exploration as a biomarker or mediator of the persistent neurocognitive morbidities in children with early-life PEU (3).
Our studies highlight serotonin as another potential link between PEU and neurogastroenterologic effects. The majority (95%) of the body’s serotonin, which does not cross the blood–brain barrier (36), is synthesized by duodenal enterochromaffin cells; serotonin regulates transit and peristalsis, mucosal and enteric nervous system development, bone growth, and innate inflammatory responses (37, 38). Thus, altered serotonin metabolism could contribute to the inflammatory markers observed in U during refeeding (Supplemental Table 1). Intriguingly, concentrations of plasma serotonin may be regulated by gut microbes. Candida albicans (39, 40) and certain bacteria (41) synthesize serotonin, and germ-free mice have significantly lower concentrations of plasma serotonin than do C (42, 43). Recent reports identified microbial-derived small molecules that stimulate serotonin release from cell cultures (44, 45), 4 of which were altered in the stool of U: α-tocopherol, cholate, and deoxycholate were found in lower concentrations in U than in C at 0 wk (when serotonin was decreased), whereas tyramine was higher in the stool of U than in C at 3 wk (when plasma serotonin was increased). Whether the dysbiosis in PEU is responsible for altering serotonin, and whether serotonin metabolism in PEU affects intestinal function and growth, are under exploration.
How PEU leads to dysbiosis is unknown. Although effects from stress because of maternal separation cannot be excluded, mucosal inflammatory or architectural changes are not observed (19). One potential mechanism was suggested by metabolomics: the differential influx of glycans. In the gut, most glycans are nondigestible and derived from diet (N-linked) or intestinal mucus (O-linked). Glycans serve as fermentation substrates for specific bacteria, shaping microbiota composition and function (46). Decreased breast-milk intake in PEU could yield decreased fecal N-linked glycans and could select for microbes, including A. muciniphila (47–49), that prefer O-linked glycans of the mucus layer. Given that the intestinal microbiome during PEU lacks N-linked glycan-metabolizing genes (9), the higher concentrations of N-linked glycans in stool from U at 3 wk could alternatively reflect unused substrate from persistent loss of metagenomic function; whole metagenome sequencing during refeeding could test this hypothesis. Mechanisms underlying the persistence of intestinal changes are not known, although our finding of altered mediators of one-carbon metabolism suggest a potential epigenetic basis (50, 51), a possibility strengthened by evidence that DNA methylation is sensitive to nutrient availability (52, 53).
Altered abundances of mucolytic organisms may reflect host systemic changes. A. muciniphila blooms with inflammation due to cold temperature-induced stress (54), IL-10 deficiency (55), and dextran sodium sulfate–induced colitis (56); in the latter instance, the bacterium appears to be protective (57). Similarly, populations of M. schaedleri (58) expand during mucosal disruption, including murine dextran sodium sulfate–induced (56) and infectious colitis (59), porcine Trichuris suis infection (60), and colonization of germ-free mice with bacteria (61). Given that low-grade inflammation transiently accompanies weaning in healthy mice (62, 63), rats (64), and piglets (65, 66), the absence of M. schaedleri in U 1 wk after ending FD could indicate delayed intestinal maturation. Our results also confirm others’ findings that the abundance of A. muciniphila is inversely proportional to body weight in mice (9, 67–69) and humans (70–72) by a yet-uncharacterized mechanism. Whether these altered mucosal microbes influence the mucous barrier, immune system, or nutrient assimilation in PEU warrants further study.
Our finding of less fecal microbial diversity in U than in C at 0 wk (during FD) was consistent with previous studies (9); however, we were surprised to find that U had greater diversity than did C 3 wk after ending FD. Microbiome diversity is thought to benefit the host. For example, in the elderly, decreased diversity is linked to frailty (73). Microbiota of children with PEU are delayed in maturation to diverse, adult-like microbial communities; this finding is associated with anthropomorphic deficits refractory to refeeding (6). Our data reveal evidence of a regenerative capacity for microbiota diversity in response to diet alone. Whether these changes persist, and whether they may enhance energy extraction from the diet, responsiveness to live oral vaccines, or resistance to enteric infections is unknown. It is also possible that greater microbial diversity helps an acutely undernourished host rapidly regain weight, but also predisposes the host to obesity and its comorbidities. Of note, fewer proportions of Bacteroidetes and greater proportions of Firmicutes were found in the stool of U 3 wk after ending FD; these have been reported in genetically obese mice (74) and obese humans (75). Future studies will explore whether U have body composition differences after ending FD despite normal weight and length, and whether this greater microbial diversity is a transient rebound effect or a more lasting change beyond the 3 wk period of this study.
In summary, we identified an altered intestinal environment during refeeding after PEU that, unlike the plasma and urine metabolomes, remains significantly perturbed despite recovery of growth deficits. Further study of microbial-derived metabolites that remain altered in plasma and urine, including those that may persist indefinitely, will be needed to determine whether therapeutic manipulation of the intestinal microbiome during refeeding might have clinical impact in terms of healthy weight gain in childhood with minimal risk of long-term metabolic comorbidities later in life.
Acknowledgments
We thank Kirk Pappan from Metabolon for his assistance with generating Figures 2 and 3. GAP designed the research and had primary responsibility for the final content; GAP and BCB conducted the research; GAP, NJA, MCW, MEC, and JFP analyzed the data; and GAP, NJA, MEC, and JFP wrote the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: C, control mice; FD, feed deprivation; OTU, operational taxonomic unit; PEU, protein-energy undernutrition; U, undernourished mice.
References
- 1.Guerrant RL, Oria RB, Moore SR, Oria MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev 2008;66:487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology 2011;140:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrant RL, Deboer MD, Moore SR, Scharf RJ, Lima AA. The impoverished gut-a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 2013;10:220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trehan I, Goldbach HS, LaGrone LN, Meuli GJ, Wang RJ, Maleta KM, Manary MJ. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med 2013;368:425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gough EK, Moodie EE, Prendergast AJ, Johnson SM, Humphrey JH, Stoltzfus RJ, Walker AS, Trehan I, Gibb DM, Goto R, et al. The impact of antibiotics on growth in children in low and middle income countries: systematic review and meta-analysis of randomised controlled trials. BMJ 2014;348:g2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014;510:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013;339:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preidis GA, Keaton MA, Campeau PM, Bessard BC, Conner ME, Hotez PJ. The undernourished neonatal mouse metabolome reveals evidence of liver and biliary dysfunction, inflammation, and oxidative stress. J Nutr 2014;144:273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Composition and function of the undernourished neonatal mouse intestinal microbiome. J Nutr Biochem 2015;26:1050–7. [DOI] [PubMed] [Google Scholar]
- 10.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 11.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Core Team. A language and environment for statistical computing [Internet]. Vienna (Austria): R Foundation for Statistical Computing; 2014. [cited 2015 Nov 28]. Available from: http://www.R-project.org.
- 13.Dwass M. Modified randomization tests for nonparametric hypotheses. Ann Math Stat 1957;28:181–7. [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;Series B 57:289–300. [Google Scholar]
- 15.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coutinho BP, Oria RB, Vieira CM, Sevilleja JE, Warren CA, Maciel JG, Thompson MR, Pinkerton RC, Lima AA, Guerrant RL. Cryptosporidium infection causes undernutrition and, conversely, weanling undernutrition intensifies infection. J Parasitol 2008;94:1225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche JK, Cabel A, Sevilleja J, Nataro J, Guerrant RL. Enteroaggregative Escherichia coli (EAEC) impairs growth while malnutrition worsens EAEC infection: a novel murine model of the infection malnutrition cycle. J Infect Dis 2010;202:506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa LB, Noronha FJ, Roche JK, Sevilleja JE, Warren CA, Oria R, Lima A, Guerrant RL. Novel in vitro and in vivo models and potential new therapeutics to break the vicious cycle of Cryptosporidium infection and malnutrition. J Infect Dis 2012;205:1464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preidis GA, Saulnier DM, Blutt SE, Mistretta TA, Riehle KP, Major AM, Venable SF, Barrish JP, Finegold MJ, Petrosino JF, et al. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. J Pediatr Gastroenterol Nutr 2012;55:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beloborodova N, Bairamov I, Olenin A, Shubina V, Teplova V, Fedotcheva N. Effect of phenolic acids of microbial origin on production of reactive oxygen species in mitochondria and neutrophils. J Biomed Sci 2012;19:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pudlik AM, Lolkema JS. Rerouting citrate metabolism in Lactococcus lactis to citrate-driven transamination. Appl Environ Microbiol 2012;78:6665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spaapen LJ, Ketting D, Wadman SK, Bruinvis L, Duran M. Urinary D-4-hydroxyphenyllactate, D-phenyllactate and D-2-hydroxyisocaproate, abnormalities of bacterial origin. J Inherit Metab Dis 1987;10:383–90. [DOI] [PubMed] [Google Scholar]
- 23.Jia J, Mu W, Zhang T, Jiang B. Bioconversion of phenylpyruvate to phenyllactate: gene cloning, expression, and enzymatic characterization of D- and L1-lactate dehydrogenases from Lactobacillus plantarum SK002. Appl Biochem Biotechnol 2010;162:242–51. [DOI] [PubMed] [Google Scholar]
- 24.Hamid MA, Iwaku M, Hoshino E. The metabolism of phenylalanine and leucine by a cell suspension of Eubacterium brachy and the effects of metronidazole on metabolism. Arch Oral Biol 1994;39:967–72. [DOI] [PubMed] [Google Scholar]
- 25.Prasuna ML, Mujahid M, Sasikala C, Ramana Ch V. L-phenylalanine catabolism and L-phenyllactic acid production by a phototrophic bacterium, Rubrivivax benzoatilyticus JA2. Microbiol Res 2012;167:526–31. [DOI] [PubMed] [Google Scholar]
- 26.Parthasarathy A, Kahnt J, Chowdhury NP, Buckel W. Phenylalanine catabolism in Archaeoglobus fulgidus VC-16. Arch Microbiol 2013;195:781–97. [DOI] [PubMed] [Google Scholar]
- 27.Fedotcheva NI, Kazakov RE, Kondrashova MN, Beloborodova NV. Toxic effects of microbial phenolic acids on the functions of mitochondria. Toxicol Lett 2008;180:182–8. [DOI] [PubMed] [Google Scholar]
- 28.Svanström Å, Boveri S, Bostrom E, Melin P. The lactic acid bacteria metabolite phenyllactic acid inhibits both radial growth and sporulation of filamentous fungi. BMC Res Notes 2013;6:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, Gobbetti M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl Environ Microbiol 2000;66:4084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieuleveux V, Gueguen M. Antimicrobial effects of D-3-phenyllactic acid on Listeria monocytogenes in TSB-YE medium, milk, and cheese. J Food Prot 1998;61:1281–5. [DOI] [PubMed] [Google Scholar]
- 31.Dieuleveux V, Lemarinier S, Gueguen M. Antimicrobial spectrum and target site of D-3-phenyllactic acid. Int J Food Microbiol 1998;40:177–83. [DOI] [PubMed] [Google Scholar]
- 32.Altieri L, Neri C, Sacco R, Curatolo P, Benvenuto A, Muratori F, Santocchi E, Bravaccio C, Lenti C, Saccani M, et al. Urinary p-cresol is elevated in small children with severe autism spectrum disorder. Biomarkers 2011;16:252–60. [DOI] [PubMed] [Google Scholar]
- 33.Clayton TA. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS Lett 2012;586:956–61. [DOI] [PubMed] [Google Scholar]
- 34.Persico AM, Napolioni V. Urinary p-cresol in autism spectrum disorder. Neurotoxicol Teratol 2013;36:82–90. [DOI] [PubMed] [Google Scholar]
- 35.Gabriele S, Sacco R, Cerullo S, Neri C, Urbani A, Tripi G, Malvy J, Barthelemy C, Bonnet-Brihault F, Persico AM. Urinary p-cresol is elevated in young French children with autism spectrum disorder: a replication study. Biomarkers 2014;19:463–70. [DOI] [PubMed] [Google Scholar]
- 36.Mann JJ, McBride PA, Brown RP, Linnoila M, Leon AC, DeMeo M, Mieczkowski T, Myers JE, Stanley M. Relationship between central and peripheral serotonin indexes in depressed and suicidal psychiatric inpatients. Arch Gen Psychiatry 1992;49:442–6. [DOI] [PubMed] [Google Scholar]
- 37.Gershon MD. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr Opin Endocrinol Diabetes Obes 2013;20:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li N, Ghia JE, Wang H, McClemens J, Cote F, Suehiro Y, Mallet J, Khan WI. Serotonin activates dendritic cell function in the context of gut inflammation. Am J Pathol 2011;178:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokoloff B, Saelhof CC, Yoshino A. Candida albicans as a producer of serotonin. Growth 1967;31:297–300. [PubMed] [Google Scholar]
- 40.Hurley R, Leask BG, Ruthven CR, Sandler M, Southgate J. Investigation of 5-hydroxytryptamine production by Candida albicans in vitro and in vivo. Microbios 1971;4:133–43. [PubMed] [Google Scholar]
- 41.Hsu SC, Johansson KR, Donahue MJ. The bacterial flora of the intestine of Ascaris suum and 5-hydroxytryptamine production. J Parasitol 1986;72:545–9. [PubMed] [Google Scholar]
- 42.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 2009;106:3698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjögren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, Backhed F, Ohlsson C. The gut microbiota regulates bone mass in mice. J Bone Miner Res 2012;27:1357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015;161:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reigstad CS, Salmonson CE, Rainey JF 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J 2015;29:1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 2012;10:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- 48.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 2007;73:7767–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol 2008;74:1646–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001;293:1089–93. [DOI] [PubMed] [Google Scholar]
- 51.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 2002;3:662–73. [DOI] [PubMed] [Google Scholar]
- 52.Canani RB, Costanzo MD, Leone L, Bedogni G, Brambilla P, Cianfarani S, Nobili V, Pietrobelli A, Agostoni C. Epigenetic mechanisms elicited by nutrition in early life. Nutr Res Rev 2011;24:198–205. [DOI] [PubMed] [Google Scholar]
- 53.de Moura EG, Passos MC. Neonatal programming of body weight regulation and energetic metabolism. Biosci Rep 2005;25:251–69. [DOI] [PubMed] [Google Scholar]
- 54.Chevalier C, Stojanovic O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanovic A, Hagemann S, et al. Gut microbiota orchestrates energy homeostasis during cold. Cell 2015;163:1360–74. [DOI] [PubMed] [Google Scholar]
- 55.Bassett SA, Young W, Barnett MP, Cookson AL, McNabb WC, Roy NC. Changes in composition of caecal microbiota associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species. Nutrients 2015;7:1798–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, et al. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J 2012;6:2091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One 2013;8:e76520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 2005;55:1199–204. [DOI] [PubMed] [Google Scholar]
- 59.Belzer C, Gerber GK, Roeselers G, Delaney M, DuBois A, Liu Q, Belavusava V, Yeliseyev V, Houseman A, Onderdonk A, et al. Dynamics of the microbiota in response to host infection. PLoS One 2014;9:e95534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li RW, Wu S, Li W, Navarro K, Couch RD, Hill D, Urban JF Jr. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect Immun 2012;80:2150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El Aidy S, Derrien M, Aardema R, Hooiveld G, Richards SE, Dane A, Dekker J, Vreeken R, Levenez F, Dore J, et al. Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef Microbes 2014;5:67–77. [DOI] [PubMed] [Google Scholar]
- 62.Vázquez E, Gil A, Garcia-Olivares E, Rueda R. Weaning induces an increase in the number of specific cytokine-secreting intestinal lymphocytes in mice. Cytokine 2000;12:1267–70. [DOI] [PubMed] [Google Scholar]
- 63.Manzano M, Abadia-Molina AC, Garcia-Olivares E, Gil A, Rueda R. Absolute counts and distribution of lymphocyte subsets in small intestine of BALB/c mice change during weaning. J Nutr 2002;132:2757–62. [DOI] [PubMed] [Google Scholar]
- 64.Mengheri E, Ciapponi L, Vignolini F, Nobili F. Cytokine gene expression in intestine of rat during the postnatal developmental period: increased IL-1 expression at weaning. Life Sci 1996;59:1227–36. [DOI] [PubMed] [Google Scholar]
- 65.McCracken BA, Spurlock ME, Roos MA, Zuckermann FA, Gaskins HR. Weaning anorexia may contribute to local inflammation in the piglet small intestine. J Nutr 1999;129:613–9. [DOI] [PubMed] [Google Scholar]
- 66.Pié S, Lalles JP, Blazy F, Laffitte J, Seve B, Oswald IP. Weaning is associated with an upregulation of expression of inflammatory cytokines in the intestine of piglets. J Nutr 2004;134:641–7. [DOI] [PubMed] [Google Scholar]
- 67.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, Possemiers S, Van Holle A, Francois P, de Vos WM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011;60:2775–86. Corrected and republished from: Diabetes 2011;60:3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012;55:2285–94. [DOI] [PubMed] [Google Scholar]
- 70.Santacruz A, Collado MC, Garcia-Valdes L, Segura MT, Martin-Lagos JA, Anjos T, Marti-Romero M, Lopez RM, Florido J, Campoy C, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 2010;104:83–92. [DOI] [PubMed] [Google Scholar]
- 71.Karlsson CL, Onnerfalt J, Xu J, Molin G, Ahrne S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 2012;20:2257–61. [DOI] [PubMed] [Google Scholar]
- 72.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr 2008;88:894–9. [DOI] [PubMed] [Google Scholar]
- 73.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–84. [DOI] [PubMed] [Google Scholar]
- 74.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]