Abstract
Background: Increased maternal body mass index (BMI) is a robust risk factor for later pediatric obesity. Accumulating evidence suggests that human milk (HM) may attenuate the transfer of obesity from mother to offspring, potentially through its effects on early development of the infant microbiome.
Objectives: Our objective was to identify early differences in intestinal microbiota in a cohort of breastfeeding infants born to obese compared with normal-weight (NW) mothers. We also investigated relations between HM hormones (leptin and insulin) and both the taxonomic and functional potentials of the infant microbiome.
Design: Clinical data and infant stool and fasting HM samples were collected from 18 NW [prepregnancy BMI (in kg/m2) <24.0] and 12 obese (prepregnancy BMI >30.0) mothers and their exclusively breastfed infants at 2 wk postpartum. Infant body composition at 2 wk was determined by air-displacement plethysmography. Infant gastrointestinal microbes were estimated by using 16S amplicon and whole-genome sequencing. HM insulin and leptin were determined by ELISA; short-chain fatty acids (SCFAs) were measured in stool samples by using gas chromatography. Power was set at 80%.
Results: Infants born to obese mothers were exposed to 2-fold higher HM insulin and leptin concentrations (P < 0.01) and showed a significant reduction in the early pioneering bacteria Gammaproteobacteria (P = 0.03) and exhibited a trend for elevated total SCFA content (P < 0.06). Independent of maternal prepregnancy BMI, HM insulin was positively associated with both microbial taxonomic diversity (P = 0.03) and Gammaproteobacteria (e.g., Enterobacteriaceae; P = 0.04) and was negatively associated with Lactobacillales (e.g., Streptococcaceae; P = 0.05). Metagenomic analysis showed that HM leptin and insulin were associated with decreased bacterial proteases, which are implicated in intestinal permeability, and reduced concentrations of pyruvate kinase, a biomarker of pediatric gastrointestinal inflammation.
Conclusion: Our results indicate that, although maternal obesity may adversely affect the early infant intestinal microbiome, HM insulin and leptin are independently associated with beneficial microbial metabolic pathways predicted to increase intestinal barrier function and reduce intestinal inflammation. This trial was registered at clinicaltrials.gov as NCT01693406.
Keywords: Gammaproteobacteria, breastfed, maternal obesity, metagenomics, pyruvate kinase
INTRODUCTION
Pregnancy and the early postnatal period are crucial windows of opportunity for the prevention of metabolic diseases (1). In the United States, 23% of children are already overweight or obese when entering kindergarten (2) and maternal obesity is consistently one of the most powerful predictors of pediatric obesity (3). There is abundant evidence that the microbiome has multiple effects on the physiology and immunology of both mothers and infants. Although the founding individual microbiota composition in newborns largely reflects the maternal transfer during birth, the postnatal assembly of the human microbiota plays an important role in resistance to pathogen invasion, immune stimulation, and other important developmental cues early in life (4–6). Alterations in intestinal microbiome composition and biodiversity have been correlated with the development of obesity (7, 8), asthma (9), allergies (10), inflammatory bowel disease (11) and, more recently, type 1 diabetes (12). Accumulating evidence suggests that exclusive breastfeeding is protective against pediatric obesity (13). However, the breastfeeding-derived protection against offspring obesity is not universal across all lactating mothers (14) and the mechanisms that account for this protection remain poorly characterized. Although it is well established that the intestinal microbiome of breast- and formula-fed infants differs dramatically, surprisingly little is known about how compositional differences in breast milk affect the development of the early infant gastrointestinal tract.
In addition to providing the infant with growth factors and satiety hormones, human milk (HM)15 may also exert indirect beneficial effects on the infant by virtue of its effects on the microbiome. These effects include the production of short-chain fatty acids (SCFAs) via colonic fermentation and regulation of host genes that regulate intestinal permeability, energy retention, and autoimmunity (15). In addition, hormones derived from HM, such as leptin and insulin, retain biological activity in the infant gastrointestinal tract and may be important additional factors that contribute to intestinal barrier function and the development of the intestinal mucosa, as well as the colonization patterns of the infant intestinal microbiome (16–18). Existing evidence shows that the probiotic and hormonal content of HM is correlated with maternal intestinal microbe colonization patterns (19) and differs between normal-weight (NW) and obese women (20); however, there is limited information that describes how HM leptin and insulin influence the development of the infant intestinal microbiome.
High-throughput sequencing has facilitated culture-independent strategies to investigate the contribution of intestinal microbes to the development of pediatric obesity (21), as well as the potential impact of maternal obesity on offspring adiposity, nutrition, and immune response (22–24). We hypothesized that maternal obesity plays an important role in the founding population of breastfed infants’ microbiome and that HM insulin and leptin are key hormones that shape the metabolic characteristics of the microbiome community.
METHODS
Participants
All of the women were healthy mothers, aged between 20 and 36 y, who planned to exclusively breastfeed through 4 mo postpartum and who delivered a term (>37 wk) singleton fetus between 2012 and 2015 at the University of Colorado Hospital (Aurora, Colorado). All aspects of this study were approved by the Colorado Multiple Institutional Review Board, and the study was registered at clinicaltrials.gov as NCT01693406. Women were excluded if they had type 1 or type 2 diabetes, pre-eclampsia, gestational diabetes, or a planned cesarean delivery, or if they or their infants were prescribed antibiotics in the 2 wk after birth. Nevertheless, 1 mother in this study gave birth via cesarean delivery; this infant was included in the study and tables given that our sensitivity analyses showed that the results were not altered when this infant was dropped from the analyses. This study included only mothers with a prepregnancy BMI (in kg/m2) <25 for the NW group and >30 for the obese group on the basis of self-reported prepregnancy weight. Study personnel visited mothers in the hospital within 48 h of delivery. Infant sex, birth weight, and mode of delivery were obtained from medical records. At 2 wk postpartum, participants traveled to Children’s Hospital Colorado (Aurora, Colorado) for a clinic visit during morning hours, and mothers fasted from midnight the previous night. At these visits, maternal height and weight were measured and venous blood and HM samples (detailed below) were collected. Infant weight was obtained and the percentage body fat was measured in duplicate by air-displacement plethysmography (PEA POD; COSMED).
Milk collection and biochemical analysis
At 2 wk postpartum, mothers were administered a modified version of the Infant Feeding Practices II questionnaire (25). Breastfeeding exclusivity was calculated as the percentage of feedings during the previous 7 d that consisted of breast milk. At this time, a midfeeding breast-milk sample was collected from mothers in a fasted state. In short, approximately halfway through a feeding (on the basis of normal feeding time and maternal sensation of milk removal and breast softening), the infant was removed, the nipple and areola were wiped clean by using a sterile gauze pad soaked in sterile water, and ∼20 mL was expressed by using a One-Hand Manual Breast Pump (Ameda). Milk was immediately placed on ice and transported back to the laboratory. Skim milk was generated by spinning milk at 10,000 × g at 4°C for 10 min. Aliquots of skim and whole milk were then stored at −80°C until analysis. The percentage of milk fat was measured by creamatocrit with the use of digital calipers, and fat (g/L) was estimated by using the following approximation: kcal/L = 290 + [66.8 × creamatocrit (%)] (26). Lactose was measured by enzymatic digestion and colorimetric detection of galactose (BioVision), and protein was measured by a modified version of the Bradford method (27). HM caloric density was calculated as the summation of fat, protein, and lactose assuming 9, 4, and 4 kcal/g, respectively. Leptin was measured in the skim fraction by using a high-sensitivity ELISA (R&D Systems), and insulin was measured in the skim fraction by using a radioimmunoassay (Millipore). Previous studies have shown that leptin may be retained in the lipid layer (28), whereas others have suggested that skim milk preparations are the most stable for HM leptin analyses (29). It is still possible that the leptin concentrations measured in this study were underestimated due to lost leptin in the lipid fraction; however, our skim milk preparation was applied consistently across all samples, allowing us to make valid comparisons within the cohort. The total antioxidant capacity (TAC) of whole milk was determined by using the TAC Assay Kit (Abcam). Clinical study data were collected and managed with the use of REDCap electronic data capture tools hosted at the University of Colorado Denver (30).
Stool collection and biochemical analysis
Infant stool samples were collected 24 h before the 2-wk visit by mothers with the use of a nonabsorbent liner placed in the infant’s diaper. When the diaper contained stool, mothers removed the liner, placed it in a plastic bag, and stored it at −20°C until the study visit. On receipt, microbial DNA was extracted from infant fecal samples by using standard protocols and the PowerFecal DNA Isolation Kit (Mo Bio). Stool samples were also subjected to SCFA extraction by mixing weighed frozen feces with acidified water (pH 2.5) containing 1 mmol ethylbutyric acid/L as an internal standard and sonicated for 10 min then incubated at room temperature for 10 min. Samples were centrifuged at 10,000 × g for 10 min at room temperature, and supernatant was filtered through a 0.45-μm nylon filter and stored at −80°C until analysis. The samples were analyzed by using a 6890 Series Gas Chromatographer with a flame ionization detector (Agilent). Samples were injected at a 10:1 split ratio; the inlet was held at 22°C and the transfer line was held at 230°C. Separation of SCFAs was achieved on a 30m TG-WAX-A column (0.25-mm ID, 0.25-μm film thickness; Thermo Scientific) by using a temperature program of 100°C for 1 min, increased by 8°C/min to 180°C, held at 180°C for 1 min, increased by 200°C at 20°C/min, and held at 200°C for 5 min. Helium carrier flow was held at 1.2 mL/min. Acetate, proprionate, and butyrate were quantified by using standards of commercially purchased compounds; and samples were adjusted for extraction efficiency differences by normalizing to the internal standard.
16S Amplicon library construction
Bacterial profiles were determined by broad-range amplification and sequence analysis of the V1V2 variable region in the 16S ribosomal RNA genes following our previously described methods (31). Illumina paired-end sequencing was performed on the MiSeq platform with version 2.3.0.8 of the MiSeq Control Software and version 2.3.32 of MiSeq Reporter by using a 600-cycle version 3 reagent kit (Illumina). As previously described (32), Illumina MiSeq paired-end sequences were sorted by sample with a python script. Assembled sequences were aligned and classified with SINA (1.2.11) (33) by using the 418,497 bacterial sequences in Silva 115NR99 (34) as a reference configured to yield the Silva taxonomy. Operational taxonomic units were produced by clustering 16S sequences with identical taxonomic assignments. This process generated 5,892,448 sequences for 30 samples (mean sample size: 196,415 sequences/sample; minimum sample size: 23,793; maximum sample size: 385,428). The present study restricted the analysis of operational taxonomic units to those taxa that were present in >20% of participants with a total relative abundance ≥5% at the family level.
Metagenomic library construction
Bulk fecal DNA samples prepared for 16S polymerase chain reaction were subjected to multiplexed shotgun sequencing by using the Nextera XT kit (Illumina) and the 600-cycle MiSeq Reagent Kit version 3 (Illumina). Raw, paired-end reads containing a mean of 998,198 high-quality reads/specimen were uploaded to the Metagenomics Rapid Annotation using Subsystem Technology (RAST) server [MG-RAST; http://metagenomics.anl.gov (accessed October 2014)] for automated sequence classification and analysis (35). Human sequences were removed by comparison to the Homo sapiens HG19 reference genome, and the remaining sequences were annotated by using the MD5 nonredundant database (36). Results are presented for annotations against the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologous (KO) hierarchy (37). The KO annotations presented in this study are structured according to a directed acyclic graph hierarchy with 4 levels (38). The top level (L1) consists of the following 5 categories: metabolism, genetic information processing, environmental information processing, cellular processing, and human diseases. The second level (L2) divides the 5 functional categories into finer subcategories. The third level (L3) corresponds directly to the KEGG pathways, and the fourth level (L4) consists of the leaf nodes, which are the functional terms. We mapped KO annotations using the MG-RAST API [http://api.metagenomics.anl.gov/api.html (accessed August 2014)]. Sequence annotations were downloaded and analyzed by using the R package “matR” version 1.0.0 (39). According to the default MG-RAST quality check, our analysis included 29,174,764 sequences that covered 30 samples (mean sequence length: 223 nucleotides; average sample size: 707,399 sequences/sample; minimum sample size: 7124; maximum sample size: 1,337,142). Metagenomics pathways included in the present analysis were limited to those KO pathways at the L2 level with a total relative abundance ≥1% among all participants.
Statistical analysis
All of the data presented were consistent with the assumptions of the statistical tests, and the analyses were implemented in the R statistical language version 3.1.1 (40) by using R-Studio version 0.98.501 (41). Descriptive statistics were generated for maternal and infant characteristics according to maternal BMI (NW, overweight/obese). The normality of each variable was tested by using the Shapiro-Wilk test, and nonparametric variables were log-transformed before utilization as covariates in multivariate models. We tested for differences in cohort characteristics and outcome variables across maternal BMI categories using Wilcoxon’s rank-sum test for continuous outcomes and chi-squared test of independence for categorical outcomes. P values <0.1 were reported as trends.
Microbial diversity indexes (Shannon-Wiener index and Simpson index) were calculated by using the “diversity” function in the vegan package (42), and values were formally compared across maternal BMI groups by using Wilcoxon’s rank-sum test. The “Adonis” function in the vegan R package (42) was used to conduct permutational multivariate ANOVA [PERMANOVA (43)] and to test for differences in the metagenomic composition of the infant microbiome according to maternal BMI. We used the “lmorigin” function in the ape R package (44) to conduct multivariable linear regression with permutation and to evaluate the association of HM leptin and insulin with microbial diversity and taxonomic composition. Metagenomic analysis with HM leptin and insulin was performed by using quasi-Poisson regression as implemented with the use of the “glm” function in the base R package (40). Metagenomic sequence counts at each KO level were standardized per individual to a relative proportion by using an offset in the quasi-Poisson regression model. Given the high degree of interpersonal variation observed in microbial taxonomy (11, 45), as well as the pilot nature of this study, P values related to microbial taxonomy and metagenome were not adjusted to control for multiple testing.
Multivariate models were used to test for associations of HM hormones with characteristics of the infant microbiome while controlling for potential confounders such as maternal BMI and other characteristics of HM. Given our sample sizes of infants born to NW (18) and obese (12) women, we had 80% power to detect differences between the groups of ∼1 SD on the basis of a type 1 error of 0.1 (46). For instance, our study was powered to detect a difference in milk insulin of ∼11 pg/mL and a difference in the relative abundance of Gammaproteobacteria of ∼20% between infants born to NW and obese mothers.
RESULTS
Cohort characteristics
Thirty women delivered infants between 2012 and 2015 and met the inclusion criteria at both birth and at 2 wk postpartum. We studied the infants born to 18 NW (maternal prepregnancy BMI: 21.0 ± 2.1) and 12 obese (maternal prepregnancy BMI: 32.1 ± 2.7) women at 2 wk of age (Table 1). All of the infants were born at ≥37 wk of gestation, and infant birth weight, percentage of body fat at 2 wk, and rate of weight gain from birth to 2 wk (g/d) did not differ between groups. The macronutrient composition (fat, protein, calories, lactose) and TAC of HM did not differ between obese and NW mothers (Table 2). HM insulin and leptin were significantly higher in obese than in NW women (P = 0.01 and P < 0.001, respectively) and were positively correlated with maternal prepregnancy BMI in the cohort as a whole (P < 0.01; HM insulin r2 = 0.14, HM leptin r2 = 0.24).
TABLE 1.
Maternal and infant characteristics1
| Normal-weight (n = 18) | Overweight/obese (n = 12) | P | |
| Maternal | |||
| Age, y | 31.2 ± 2.5 | 30.8 ± 4.6 | 0.97 |
| Prepregnancy BMI, kg/m2 | 21.0 ± 2.1 | 32.1 ± 2.7 | <0.001 |
| Gestational weight gain, kg | 15.5 ± 4.5 | 13.0 ± 7.5 | 0.24 |
| Gravidity,2 n (%) | 0.66 | ||
| Primigravida | 9 (50.0) | 5 (41.7) | |
| Multigravida | 9 (50.0) | 7 (58.3) | |
| Infant | |||
| Gestational age, wk | 39.7 ± 1.0 | 39.7 ± 1.0 | 0.96 |
| Birth weight, g | 3546.7 ± 452.4 | 3464.2 ± 578.2 | 0.90 |
| Mode of delivery,2 n (%) | 0.41 | ||
| Vaginal delivery | 17 (94.4) | 12 (100) | |
| Offspring sex,2 n (%) | 0.88 | ||
| Male | 10 (55.6) | 7 (58.3) | |
| Female | 8 (44.4) | 5 (41.7) | |
| Two-week PEA POD3 | |||
| Weight, g | 3663.4 ± 404.1 | 3633.1 ± 615.1 | 0.98 |
| Fat, % | 11.4 ± 4.2 | 9.8 ± 4.6 | 0.46 |
| Weight gain from birth to 2 wk, g/d | 7.7 ± 11.9 | 11.3 ± 13.8 | 0.85 |
Values are means ± SDs derived by using Wilcoxon’s rank-sum test unless otherwise noted.
Derived by using a general association test.
PEA POD, air-displacement plethysmography.
TABLE 2.
Human milk and infant stool characteristics1
| Normal-weight (n = 18) | Overweight/obese (n = 12) | P | |
| Human milk | |||
| Breastfeeding exclusivity, % | 98.9 ± 0.05 | 99.0 ± 0.03 | 0.39 |
| Milk | |||
| Fat, g/100 mL | 3.8 ± 1.2 | 3.2 ± 0.7 | 0.15 |
| Protein, g/100 mL | 1.2 ± 0.2 | 1.2 ± 0.2 | 0.42 |
| Calories, kcal/100 g | 2395.4 ± 3625.8 | 3027.8 ± 5566.4 | 0.44 |
| Lactose, g/100 mL | 6.9 ± 0.9 | 6.8 ± 1.1 | 0.73 |
| Insulin, μIU/mL | 11.8 ± 8.9 | 21.2 ± 13.3 | 0.01 |
| Leptin, pg/mL | 610.4 ± 675.4 | 1368.2 ± 547.2 | <0.001 |
| TAC, mmol/L | 0.53 ± 0.3 | 0.47 ± 0.2 | 0.50 |
| Stool metabolites, mmol · L−1 · g−1 | |||
| Total SCFAs | 36.8 ± 24.1 | 63.9 ± 52.5 | 0.06 |
| Acetate | 35.7 ± 22.7 | 60.4 ± 48.8 | 0.07 |
| Propionate | 0.8 ± 1.5 | 2.6 ± 4.7 | 0.53 |
| Butyrate | 0.3 ± 0.7 | 0.9 ± 2.4 | 0.82 |
Values are means ± SDs derived by using Wilcoxon’s rank-sum test. SCFA, short-chain fatty acid; TAC, total antioxidant capacity.
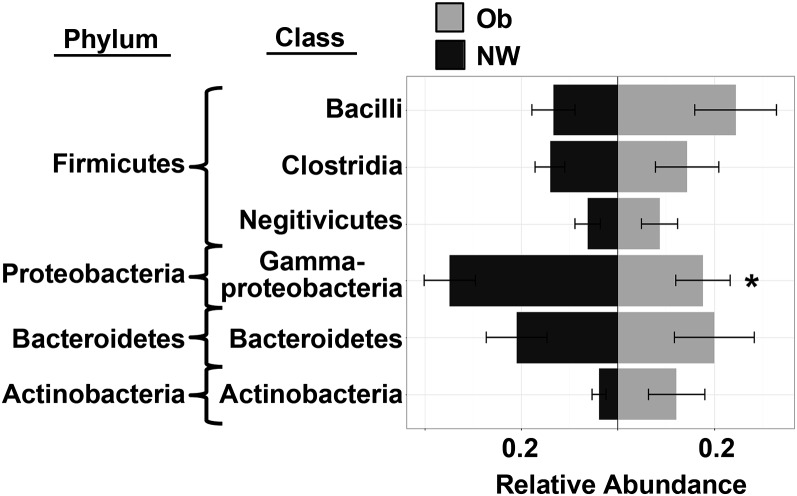
Infants of obese mothers have lower relative abundance of Gammaproteobacteria
We assessed the microbial diversity, taxonomic composition, and metagenomic profile of the infant intestinal microbiome at 2 wk postpartum and analyzed these data according to maternal BMI grouping. As expected, the taxonomic composition of the intestinal microbiota in all of the subjects was dominated by bacteria of the Firmicutes (30.5%), Proteobacteria (28.8%), Bacteroidetes (23.8%), and Actinobacteria (5.6%) phyla (Supplemental Table 1). However, no significant differences in microbial diversity (i.e., Shannon or Simpson indexes) or overall microbiome composition were observed between NW and obese groups (Supplemental Table 2). Nevertheless, infants born to obese mothers had a significantly lower relative abundance of the Proteobacteria phylum in the Gammaproteobacteria class (P = 0.03; Figure 1, Supplemental Table 1) than those born to NW mothers, which may have been driven by decreases in the Enterobacteriaceae family (P = 0.07; Supplemental Table 1). Metagenomic analyses did not show any significant differences in microbial metabolic pathways (KO classes) according to maternal BMI grouping (Supplemental Table 3).
FIGURE 1.
Taxonomic composition of infant intestinal microbiome at 2 wk according to maternal BMI. The relative abundance of intestinal microbes from infants born to NW (n = 18) and obese (n = 12) mothers is shown, with distance from the center line representing increased abundance. Maternal BMI in the obese range is associated with a significantly lower relative abundances of Proteobacteria at the phylum level and Gammaproteobacteria at the class level (P = 0.03). Relative abundance was measured by 16S sequencing including only taxa present in >20% of participants with a total relative abundance ≥5% at the family level. Results were derived by using Wilcoxon’s rank-sum test and are shown as means ± SDs. *P < 0.05. NW, normal-weight; Ob, obese.
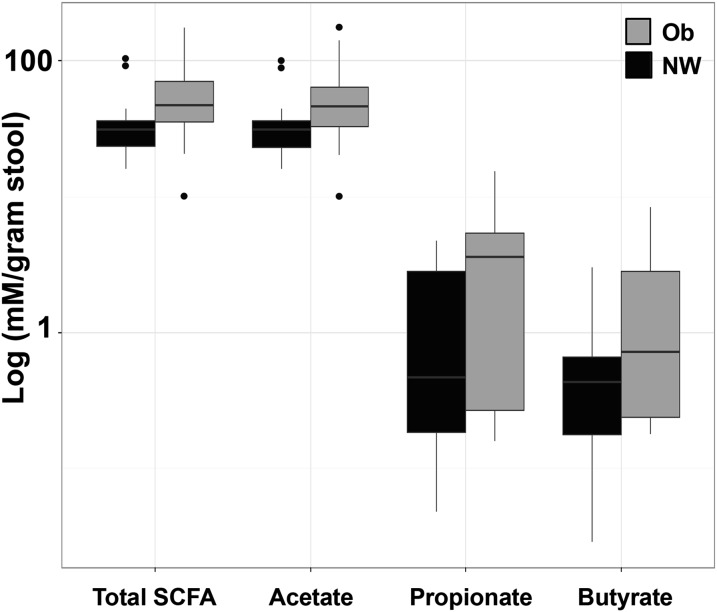
SCFAs are elevated in stool from infants born to obese mothers
To explore the association between maternal obesity and infant colonic fermentation, we measured fecal concentrations of the microbial-derived SCFAs acetate, propionate, and butyrate (Table 2). Figure 2 shows that stool samples from infants born to obese mothers were dominated by acetate (P = 0.07), which was a major contributor to the 74% increase in total SCFA concentration (P = 0.06). We did not find differences in concentrations of fecal propionate (P = 0.53) or butyrate (P = 0.82) according to maternal BMI status.
FIGURE 2.
Infant fecal SCFA profile at 2 wk according to maternal BMI. SCFAs, markers of colonic fermentation, in infants born to NW (n = 18) and obese (n = 12) women are shown (total SCFAs, P = 0.06; acetate, P = 0.07; propionate, P = 0.53; and butyrate, P = 0.82). In stool samples from infants born to obese women, concentrations of SCFA metabolites were 2-fold higher than those from NW mothers. SCFAs are represented as log (mmol/L per gram of stool) as measured by gas chromatography. Results were derived by using Wilcoxon’s rank-sum test and are presented as a box-and-whiskers plot with outliers indicated by individual points. NW, normal-weight; Ob, obese; SCFA, short-chain fatty acid.
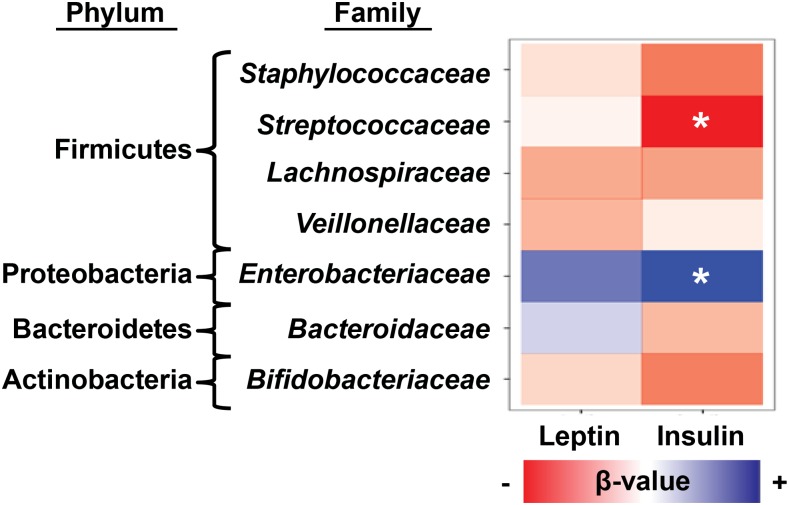
HM insulin is associated with microbial diversity and the composition of the infant microbiome
We next tested HM leptin and insulin concentrations for associations with microbial diversity and taxonomic features of the infant intestinal microbiome at 2 wk postpartum. After adjustment for maternal BMI, we found that concentrations of HM insulin were positively associated with microbial diversity indexes (P < 0.05; Supplemental Table 4) as well as phylum-level increases in Proteobacteria (P = 0.04; Supplemental Table 5). Consistent with these observations, HM insulin was positively associated with Gammaproteobacteria at the order level (Enterobacteriales, P = 0.04; Supplemental Table 5) and at the family level (Enterobacteriaceae, P = 0.04; Figure 3, Supplemental Table 5). In contrast, HM insulin was negatively associated with the relative abundance of the Streptococcaceae family (P = 0.05; Figure 3, Supplemental Table 5). We did not detect significant associations between HM leptin and the taxonomic composition of the infant microbiome (Supplemental Table 5).
FIGURE 3.
Influence of HM hormones on composition of the infant microbiome at 2 wk postpartum. When controlled for maternal BMI (n = 30), infant microbiome composition at the phylum and family levels correlated with insulin, but not leptin, concentrations in maternal milk. HM insulin was positively correlated (in blue) with Proteobacteria/Enterobacteriaceae (P = 0.04) and negatively correlated (in red) with Firmicutes/Streptococcaceae (P = 0.05) at the family level. Multivariable linear regression models with permutation were used to test for associations in which the degree of coloration indicates the strength of association as measured by β-value. *P ≤ 0.05. HM, human milk.
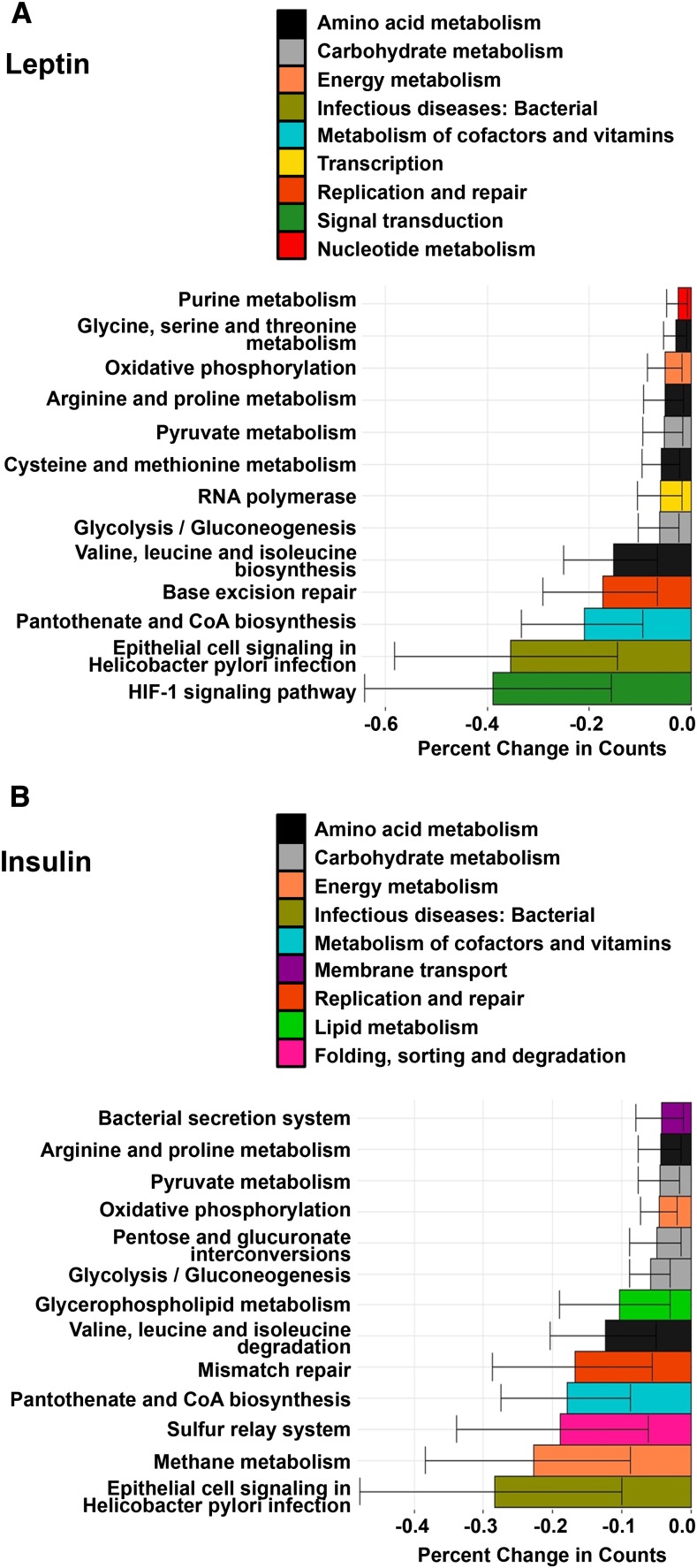
HM leptin and insulin are negatively associated with microbial functional pathways implicated in intestinal inflammation
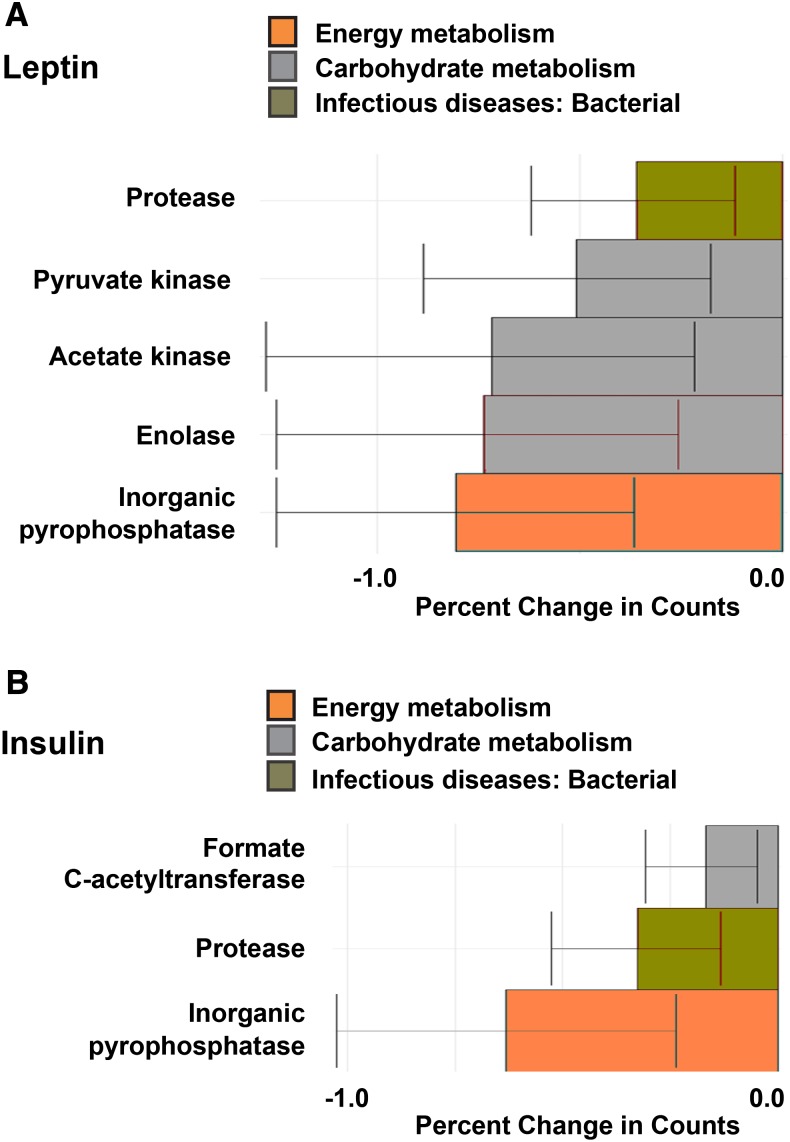
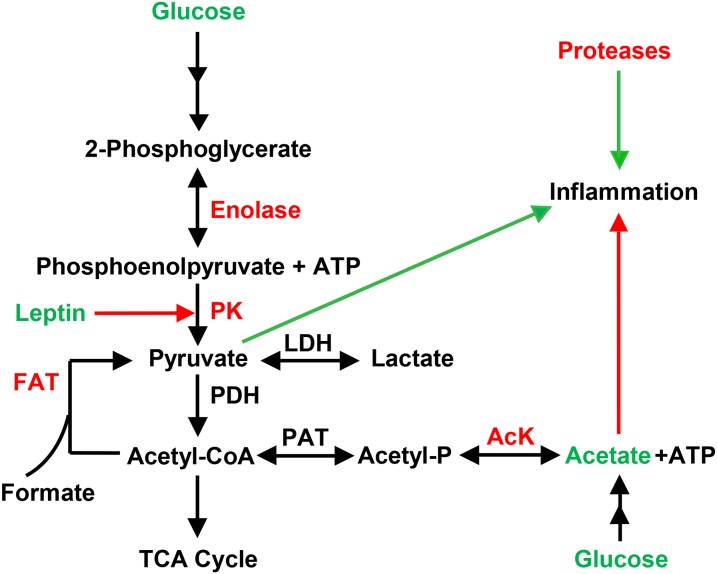
Both HM leptin and insulin were inversely associated with functional KO annotations at the L2 level that represented a variety of KEGG pathways, including bacterial amino acid, carbohydrate, vitamin, and energy metabolism (P < 0.01; Figure 4, Supplemental Table 6). We next interrogated the L3-level KO annotations significantly associated with concentrations of HM leptin and insulin and identified individual genes (i.e., KO L4) implicated as pediatric biomarkers of gastrointestinal health (47, 48). Specifically, we found that HM leptin and insulin were inversely associated with a protease of unknown catalytic function (K08303) as well as inorganic pyrophosphatase (K01507), a critical regulator of pyruvate metabolism. Figure 5A shows that HM leptin was inversely associated with genes that regulate pyruvate formation, including pyruvate kinase (K00873), acetate kinase (K00925), and enolase (K01689). Similarly, HM insulin (Figure 5B) was inversely associated with genes that regulate pyruvate formation, including formate C-acetyltransferase (K00656). Fecal bacterial protease and pyruvate kinase have previously been implicated as biomarkers of gastrointestinal permeability (48) and inflammation (49). Collectively, these results suggest that early exposure of the offspring to HM leptin and insulin was associated with changes in the metagenomics profile of the infant intestinal microbiome that may promote gastrointestinal barrier function and attenuate intestinal inflammation, as shown in Figure 6.
FIGURE 4.
HM leptin and insulin associated with changes in infant microbiome metagenome expression at the L2-level ontology. After adjustment for maternal BMI (n = 30), HM leptin (A) and insulin (B) were negatively associated with functional KO annotations from 2-wk infant microbiome composition representing pathways involved in bacterial amino acid, carbohydrate, vitamin, and energy metabolism, as well as with genes involved in cellular homeostasis and host intestinal barrier function. Outcomes presented are KO pathways from the L2 level with a total relative abundance ≥1% in all of the participants with the use of quasi-Poisson regression analysis (P < 0.01). Values are means ± SEs. HM, human milk; KO, KEGG Ortholog.
FIGURE 5.
HM bioactive components associated with changes in infant microbiome metagenome expression at the L4-level ontology. After adjustment for maternal BMI (n = 30), HM leptin and insulin were tested for associations with functional KO annotations at the L4 level from 2-wk infant microbiome composition. (A) HM leptin was negatively associated with a protease of unknown catalytic function (K08303), inorganic pyrophosphatase (K01507), pyruvate kinase (K00873), acetate kinase (K00925), and enolase (K01689). (B) HM insulin was negatively associated with a protease of unknown catalytic function (K08303), inorganic pyrophosphatase (K01507), and formate C-acetyltransferase (K00656). Outcomes presented are KO pathways from the L4 level with a total relative abundance ≥1% in all of the participants with the use of quasi-Poisson regression analysis (P < 0.01). Values are means ± SEs. HM, human milk; KO, KEGG Ortholog.
FIGURE 6.
Hypothesized model relating HM leptin and insulin to the suppression of intestinal microbial pathways associated with intestinal inflammation. Increased acetate production, reduced host pyruvate metabolism, and suppressed bacterial protease activity may suppress intestinal inflammation. Our results support a hypothesized mechanism whereby exposure to HM leptin and insulin during the first 2 wk of life is associated with improved intestinal barrier function and reduces intestinal inflammation by reducing the relative abundance of bacterial protease and pyruvate metabolism encoded by the infant intestinal microbiome. HM leptin and insulin are associated with reduced concentrations of enolase, PK, FAT, AcK, and bacterial proteases in breastfed offspring of normal-weight and obese mothers. Pyruvate metabolism in prokaryotic organisms [adapted and simplified from the KEGG pyruvate metabolism pathway in Enterobacter spp. (ent638)]. Red signifies underrepresented DNA or known inhibitory effect; green signifies overrepresentation or stimulatory effect. Acetyl-P, acetyl phosphate; AcK, acetate kinase; FAT, formate C-acetyltransferase; HM, human milk; LDH, lactate dehydrogenase; PAT, phosphoacetyl transferase; PDH, pyruvate dehydrogenase; PK, pyruvate kinase.
DISCUSSION
The intestinal microbiome in newborns and infants plays a critical role in intestinal health; however, studies that examined specific hormones found in HM and their effects on the infant intestinal microbiome are currently lacking. The data from this study are the first, to our knowledge, to suggest that differences in the early infant microbiome are present in exclusively breastfed infants born to obese compared with NW mothers and that exposure to HM leptin and insulin correlates with the composition and markers of function of the neonatal microbiome.
Notably, we identified a significant reduction in abundance of one of the earliest pioneering bacteria, Gammaproteobacteria, in infants born to obese mothers, with no group differences in overall bacterial diversity or abundance. The natural transition from a neonatal to a more mature microbiota is governed in part by the induction of Gammaproteobacteria, a dominant class established during the first week of life in newborns that diminishes over time (50). Other studies in infants as young as 12 d of age have suggested that the composition and temporal pattern of the intestinal microbiota were sequentially dominated by Proteobacteria, gram-positive bacteria (Firmicutes and Actinobacteria), and Flexibacter-Cytophaga-Bacteroides (51), which subsequently give rise to the Bacteroidetes phylum (50, 52). Our cohort fits into this generalized pattern, exhibiting a relatively high abundance of Proteobacteria (>35% in infants born to NW mothers) along with a dominating prevalence of Firmicutes (>40%) at 14 d.
Gammaproteobacteria have been associated with adult irritable bowel syndrome (53) and gastric bypass patients (54). However, in newborn mice, Gammaproteobacteria are essential for driving early inflammation necessary for protection against excessive inflammatory and autoimmune gastrointestinal disorders later in life (55). Consistent with these findings, the production of LPS by Gammaproteobacteria provides a hormetic stimulation that improves early development and function of innate immune responses (56, 57), sometimes referred to as a “priming” or “trained immunity” effect (58). It is intriguing to speculate that an early reduction in this pioneering, hormetic bacteria could contribute to long-term health risks of allergies, autoimmunity, and possibly even the metabolic syndrome in infants born to obese mothers (59).
Consistent with previous observations, we found that insulin and leptin were significantly elevated in HM of obese mothers (60, 61). HM insulin plays a critical role in normal intestinal development and maturation (18, 62) and can exert direct physiologic effects on the infant via binding with insulin receptors that are localized along the intestinal tract (63). Our data showed that HM insulin at 2 wk of age was independently and positively associated with microbial diversity and the relative abundance of Gammaproteobacteria (Enterobacteriaceae) and negatively associated with Lactobacillales (Streptococcaceae). The potential mechanism or mechanisms linking HM insulin to infant intestinal microbial colonization patterns, independent of maternal BMI, may include the following: 1) a direct role of insulin to regulate enterocyte maturation (64) and/or 2) the ability of oral insulin to increase glucose concentrations in the gastrointestinal lumen (64, 65). Enterobacteriaceae are a family of glucose metabolizers, which suggests that HM insulin may act as a positive selective force to enhance Gammaproteobacteria (Enterobacteriaceae) colonization. As noted above, our results showed that infants born to obese mothers had reduced concentrations of Gammaproteobacteria, despite the higher concentrations of insulin in HM from obese mothers.
HM leptin has been reported to regulate infant body weight control (28) and, in rodents, to induce changes in the composition of the intestinal microbiota (66). Our analyses did not detect associations between leptin and specific infant intestinal microbiota taxa abundance; however, this may not be surprising given that leptin receptors in the neonatal intestine are localized to the duodenum (17) and the microbiota composition of the duodenum does not overlap with colonic microbiota estimated by stool samples (67). In addition, TAC in HM is associated with concentrations of HM leptin (68), suggesting that TAC could be involved in microbial colonization due to changes in reactive oxygen species production that selectively control genes in aerobic bacteria (69). Our investigation of TAC activity in HM did not find a significant difference in TAC between NW and obese mothers despite higher concentrations of leptin in HM from obese mothers.
We also examined the total bacterial load in HM and tested several storage, collection, and DNA extraction methods to selectively enrich for HM bacteria. Regardless of protocol, we failed to detect bacterial 16S DNA concentrations above the background concentration of negative controls.
Our metagenomic analyses showed that HM leptin and insulin were both associated with suppressing genes that encode putative bacterial proteases. Bacterial proteases have been linked to inflammatory bowel disease pathophysiology by increasing intestinal permeability (48), which suggests that HM leptin and insulin may improve intestinal integrity indirectly by negatively regulating bacteria that can increase permeability. Our results also showed that leptin was associated with suppressing gene sets that regulate pyruvate metabolism, including the novel biomarker of intestinal inflammation, pyruvate kinase. Pyruvate kinase is a critical enzyme in the glycolytic pathway that catalyzes the conversion of phosphoenolpyruvate into pyruvate and ATP, and fecal pyruvate kinase is a biomarker of low-grade inflammation in pediatric irritable bowel disease (47). The increased SCFA acetate in the stool of infants of obese mothers may also enhance the anti-inflammatory potential in the infant by preventing overgrowth of pH-sensitive pathogenic bacteria (70) and by providing a readily available fuel source for colonocytes (71). Data from humans suggest that systemic SCFAs regulate intestine-derived satiety hormones and lipid metabolism in obesity (72); however, the relevance of such findings in early infant metabolism is largely unknown. Taken together, our metagenomic and SCFA results support a hypothesizedmechanism whereby exposure to leptin and insulin in HM during the first 2 wk of life is associated with improved intestinal barrier function and reduced intestinal inflammation, in part by suppressing pyruvate metabolism, acetate production, and proinflammatory mediators encoded by the infant intestinal microbiome (Figure 6).
Our study has several important strengths, including a well-characterized, well-matched cohort of obese and NW maternal-infant pairs with HM analysis measured at the time of stool collection. In addition, metagenomic sequencing provided a more in-depth view of the compositional and functional changes in the early infant gastrointestinal tract in exclusively breastfed infants. The limitations of this study include a modest sample size that limited the detail to which we could compare differences in bacterial composition and the number of statistical tests we could perform. In addition, it is possible that our estimates of milk fat and total calories were either an under- or overestimation on the basis of our midfeeding sampling; however, leptin and insulin concentrations in milk do not change over the course of a feeding (73, 74), suggesting our HM insulin and leptin analyses were not affected by our sampling methodology. This study was also cross-sectional in nature and thus long-term implications of differences detected at 2 wk require further investigation. Finally, our analyses did not include measurements of oligosaccharides and other prebiotic and probiotic features of HM that have been shown to contribute to the developing microbiome (75). Despite these limitations, to our knowledge, this is the first study of its kind to document that HM insulin and leptin correlate with estimates of neonatal intestinal microflora and colonic fermentation in exclusively breastfed infants. Long-term studies will be necessary to identify whether these early effects on the microbiome contribute to future metabolic and disease risk in infants of mothers with obesity.
Acknowledgments
We thank Folker Meyer and his team at Argonne National Laboratory for his prompt and patient help in accessing MG-RAST data. We thank Catherine Chartier-Logan, Nicole Hirsch, and Rebecca Lander for implementation of the study. We appreciate the help of Rachel C Janssen with preparation of the manuscript.
The authors’ responsibilities were as follows—BEY, TLH, NFK, LAB, and JEF: designed the research; BEY, TLH, BAdlH, DI, ZWP, and TW: performed the research; CER and DNF: contributed new reagents/analytic tools; DJL, ACT, TKS, MCR, SAS, and DNF: analyzed the data; DJL, BEY, PRB, and JEF: wrote the manuscript; JEF: had primary responsibility for the final content; and all authors: read and approved the final manuscript. The authors declared no conflicts of interest.
Footnotes
Abbreviations used: HM, human milk; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, KEGG Orthologous; MG-RAST, Metagenomics Rapid Annotation using Subsystem Technology; NW, normal weight; RAST, Rapid Annotation using Subsystem Technology; SCFA, short-chain fatty acid; TAC, total antioxidant capacity.
REFERENCES
- 1.Dattilo AM, Birch L, Krebs NF, Lake A, Taveras EM, Saavedra JM. Need for early interventions in the prevention of pediatric overweight: a review and upcoming directions. J Obes 2012;2012:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the united states, 2011-2012. JAMA 2014;311:806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poston L, Harthoorn LF, Van Der Beek EM; Contributors to the ILSI Europe Workshop. Obesity in pregnancy: implications for the mother and lifelong health of the child—a consensus statement. Pediatr Res 2011;69:175–80. [DOI] [PubMed] [Google Scholar]
- 4.Verdam FJ, Fuentes S, De Jonge C, Zoetendal EG, Erbil R, Greve JW, Buurman WA, de Vos WM, Rensen SS. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–15. [DOI] [PubMed] [Google Scholar]
- 5.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh C-S. Peripheral education of the immune system by the colonic commensal microbiota. Nature 2011;478:250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006;118:511–21. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Ley RE, Sogin ML, Jones WJ, Roe BA, Jason P, Egholm M, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiBaise JK, Frank DN, Mathur R. Impact of the gut microbiota on the development of obesity: current concepts. Am J Gastroenterol 2012;1(Suppl):22–7. [Google Scholar]
- 9.Karvonen AM, Hyvärinen A, Rintala H, Korppi M, Täubel M, Doekes G, Gehring U, Renz H, Pfefferle PI, Genuneit J, et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy 2014;69:1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol 2012;129:434–40. [DOI] [PubMed] [Google Scholar]
- 11.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, Peet A, Tillmann V, Pöhö P, Mattila I, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015;17:260–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J, Liu L, Zhu Y, Huang G, Wang PP. The association between breastfeeding and childhood obesity: a meta-analysis. BMC Public Health 2014;14:1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo JG, Martin LJ. Does breastfeeding protect against childhood obesity? Moving beyond observational evidence. Curr Obes Rep 2015;4:207–16. [DOI] [PubMed] [Google Scholar]
- 15.Jeurink PV, van Bergenhenegouwen J, Jiménez E, Knippels LMJ, Fernández L, Garssen J, Knol J, Rodríguez JM, Martín R. Human milk: a source of more life than we imagine. Benef Microbes 2013;4:17–30. [DOI] [PubMed] [Google Scholar]
- 16.Casabiell X, Piñeiro V, Tomé MA, Peinó R, Diéguez C, Casanueva FF. Presence of leptin in colostrum and/or breast milk from lactating mothers: a potential role in the regulation of neonatal food intake. J Clin Endocrinol Metab 1997;82:4270–3. [DOI] [PubMed] [Google Scholar]
- 17.Barrenetxe J, Villaro AC, Guembe L, Pascual I, Muñoz-Navas M, Barber A, Lostao MP. Distribution of the long leptin receptor isoform in brush border, basolateral membrane, and cytoplasm of enterocytes. Gut 2002;50:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huo YJ, Wang T, Xu RJ, Macdonald S, Liu G, Shi F. Dietary insulin affects leucine aminopeptidase, growth hormone, insulin-like growth factor I and insulin receptors in the intestinal mucosa of neonatal pigs. Biol Neonate 2006;89:265–73. [DOI] [PubMed] [Google Scholar]
- 19.Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ, Donnet-Hughes A. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 2007;119:e724–32. [DOI] [PubMed] [Google Scholar]
- 20.Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 2012;96:544–51. [DOI] [PubMed] [Google Scholar]
- 21.Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may. Am J Clin Nutr 2008;87:534–8. [DOI] [PubMed] [Google Scholar]
- 22.Walker WA, Iyengar RS. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res 2015;77:220–8. [DOI] [PubMed] [Google Scholar]
- 23.Newburg DS, Morelli L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res 2015;77:115–20. [DOI] [PubMed] [Google Scholar]
- 24.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 2011;474:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fein SB, Labiner-Wolfe J, Shealy KR, Li R, Chen J, Grummer-Strawn LM. Infant feeding practices study II: study methods. Pediatrics 2008;122(Suppl 2):S28–35. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence RA, Lawrence RM. Breastfeeding: a guide for the medical professional. 7th ed. Philadelphia: Saunders; 2010. [Google Scholar]
- 27.Spector T. Refinement of the Coomassie blue method of protein quantitation: a simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal Biochem 1978;86:142–6. [DOI] [PubMed] [Google Scholar]
- 28.Miralles O, Sánchez J, Palou A, Picó C. A physiological role of breast milk leptin in body weight control in developing infants. Obesity (Silver Spring) 2006;14:1371–7. [DOI] [PubMed] [Google Scholar]
- 29.Eilers E, Ziska T, Harder T, Plagemann A, Obladen M, Loui A. Leptin determination in colostrum and early human milk from mothers of preterm and term infants. Early Hum Dev 2011;87:415–9. [DOI] [PubMed] [Google Scholar]
- 30.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)—a metadata driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 2012;189:3805–14. [DOI] [PubMed] [Google Scholar]
- 32.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, Von Bergen M, Mccoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013;339:1084–8. [DOI] [PubMed] [Google Scholar]
- 33.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilke A, Harrison T, Wilkening J, Field D, Glass EM, Kyrpides N, Mavrommatis K, Meyer F. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinformatics 2012;13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005;21:3787–93. [DOI] [PubMed] [Google Scholar]
- 39.Braithwaite DT, Keegan KP. matR: metagenomic analysis tools for R. R package version 1.0.0. Vienna (Austria): R Foundation for Statistical Computing; 2013. [Google Scholar]
- 40.R Core Team. R: a language and environment for statistical computing [Internet]. Vienna (Austria): R Foundation for Statistical Computing; 2013. [cited Jun 2015]. Available from: www.r-project.org.
- 41.RStudio Team. RStudio: integrated development environment for R [Internet]. RStudio; 2012. [cited Jun 2015]. Available from: http://www.rstudio.com/.
- 42.Oksanen J, Blanchet G, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. R package version 2.2-1. CRAN. Vienna (Austria): R Foundation for Statistical Computing; 2015. [Google Scholar]
- 43.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001;26:32–46. [Google Scholar]
- 44.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004;20:289–90. [DOI] [PubMed] [Google Scholar]
- 45.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 2011;17:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann EL. Nonparametrics: statistical methods based on ranks. Revised ed. New York: Springer; 2006. [Google Scholar]
- 47.Pang T, Leach ST, Katz T, Day AS, Ooi CY. Fecal biomarkers of intestinal health and disease in children. Front Pediatr 2014;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll IM, Maharshak N. Enteric bacterial proteases in inflammatory bowel disease— pathophysiology and clinical implications. World J Gastroenterol 2013;19:7531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czub E, Nowak JK, Moczko J, Lisowska A, Banaszkiewicz A, Banasiewicz T, Walkowiak J. Comparison of fecal pyruvate kinase isoform M2 and calprotectin in acute diarrhea in hospitalized children. Sci Rep 2014;4:4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eggesbø M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, Bushel PR, Sekelja M, Rudi K. Development of gut microbiota in infants not exposed to medical interventions. APMIS 2011;119:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 2007;5:1556–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF, Björkstén B, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014;63:559–66. [DOI] [PubMed] [Google Scholar]
- 53.Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, De Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011;141:1792–801. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Dibaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 2009;106:2365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014;5:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannone PJ, Schanbacher BL, Bauer JA, Reber KM. Effects of prenatal lipopolysaccharide exposure on epithelial development and function in newborn rat intestine. J Pediatr Gastroenterol Nutr 2006;43:284–90. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, McCusker C. Neonatal exposure with LPS and/or allergen prevents experimental allergic airways disease: development of tolerance using environmental antigens. J Allergy Clin Immunol 2006;118:143–51. [DOI] [PubMed] [Google Scholar]
- 58.Netea MG, Quintin J, van der Meer JWM. Trained immunity: a memory for innate host defense. Cell Host Microbe 2011;9:355–61. [DOI] [PubMed] [Google Scholar]
- 59.Hooper LV, Littman DR, Macpherson AJ, Program MP. Interactions between the microbiota and the immune system. Science 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weyermann M, Brenner H, Rothenbacher D. Adipokines in human milk and risk of overweight in early childhood: a prospective cohort study. Epidemiology 2007;18:722–9. [DOI] [PubMed] [Google Scholar]
- 61.Ley SH, Hanley AJ, Sermer M, Zinman B, O’Connor DL. Associations of prenatal metabolic abnormalities with insulin and adiponectin concentrations in human milk. Am J Clin Nutr 2012;95:867–74. [DOI] [PubMed] [Google Scholar]
- 62.Shamir R, Shehadeh N. Insulin in human milk and the use of hormones in infant formulas. Nestle Nutr Inst Workshop Ser 2013;77:57–64. [DOI] [PubMed] [Google Scholar]
- 63.Shehadeh N, Sukhotnik I, Shamir R. Gastrointestinal tract as a target organ for orally administered insulin. J Pediatr Gastroenterol Nutr 2006;43:276–81. [DOI] [PubMed] [Google Scholar]
- 64.Buts JP, De Keyser N, Marandi S, Maernoudt A-S, Sokal EM, Rahier J, Hermans D. Expression of insulin receptors and of 60-kDa receptor substrate in rat mature and immature enterocytes. Am J Physiol 1997;273:G217–26. [DOI] [PubMed] [Google Scholar]
- 65.Shulman RJ. Effect of enteral administration of insulin on intestinal development and feeding tolerance in preterm infants: a pilot study. Arch Dis Child Fetal Neonatal Ed 2002;86:F131–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandoval D. Old dog, new trick: a direct role for leptin in regulating microbiota composition. Endocrinology 2014;155:653–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep 2011;1:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta R, Petrova A. Is variation in total antioxidant capacity of human milk associated with levels of bio-active proteins? J Perinatol 2014;34:220–2. [DOI] [PubMed] [Google Scholar]
- 69.Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys 2012;525:161–9. [DOI] [PubMed] [Google Scholar]
- 70.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 2001;81:1031–64. [DOI] [PubMed] [Google Scholar]
- 71.Clausen MR, Mortensen PB. Kinetic studies on colonocyte metabolism of short chain fatty acids and glucose in ulcerative colitis. Gut 1995;37:684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 2015;11:577–91. [DOI] [PubMed] [Google Scholar]
- 73.Whitmore TJ, Trengove NJ, Graham DF, Hartmann PE. Analysis of insulin in human breast milk in mothers with type 1 and type 2 diabetes mellitus. Int J Endocrinol 2012;2012:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cannon AM, Kakulas F, Hepworth AR, Lai CT, Hartmann PE, Geddes DT. The effects of leptin on breastfeeding behaviour. Int J Environ Res Public Health 2015;12:12340–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 2012;22:1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]