Abstract
The body surfaces of humans and other animals are colonized at birth by microorganisms. The majority of microbial residents on the human body exist within gastrointestinal (GI) tract communities, where they contribute to many aspects of host biology and pathobiology. Recent technological advances have expanded our ability to perceive the membership and physiologic traits of microbial communities along the GI tract. To translate this information into a mechanistic and practical understanding of host-microbe and microbe-microbe relationships, it is necessary to recast our conceptualization of the GI tract and its resident microbial communities in ecological terms. This review depicts GI microbial ecology in the context of 2 fundamental ecological concepts: (1) the patterns of biodiversity within the GI tract and (2) the scales of time, space, and environment within which we perceive those patterns. We show how this conceptual framework can be used to integrate our existing knowledge and identify important open questions in GI microbial ecology.
Animals have evolved on a planet predominated by microorganisms. To facilitate survival in this microbial world, animals have developed mechanisms for supporting vast communities of microorganisms (microbiota) on their body surfaces. The microbial cells residing on and within the adult human body are estimated to outnumber somatic and germ cells by an order of magnitude.1 Since the first documented observation of bacteria associated with the human body by Antonie van Leeuwenhoek in the late 1600s, human microbiota research has focused largely on microorganisms that could be cultured ex vivo.2 However, recent technological advances have helped reveal the deep biodiversity of the human microbiota and identified many functional contributions of the microbiota to our postnatal biology and pathobiology (reviewed by Bäckhed et al,3 Wostmann,4 Cheesman and Guillemin,5 and Kinross et al6). The collective genomes of our microbial residents (microbiome) encode physiologic capabilities that are absent from our own genomes. We can therefore consider each host and its associated microbiota as a “superorganism”—an emergent blend of host and microbial traits.7 The Human Microbiome Project and other interdisciplinary research initiatives are designed to characterize the composition and function of the human microbiome and to define the factors that pattern microbial life on the human body.8
The gastrointestinal (GI) tract harbors the majority of the microbial cells that reside in the adult human body (10–100 trillion microbial cells) and therefore is an important setting to investigate the relationships between host and microbiota. The constituency of the human GI microbiota includes members of all 3 domains of life (Bacteria, Archaea, and Eukarya) as well as viruses. Although >70 candidate bacterial phyla have been discovered on our planet, only 10 bacterial phyla (deep bacterial lineages, also known as divisions) have been observed in the human intestine (Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Cyanobacteria, TM7, Spirochaetes, and VadinBE97) and only 8 bacterial phyla have been observed in the stomach (Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, TM7, Deferribacteres, and Deinococcus-Thermus).3,9–13 Within these few deep bacterial lineages there exists an abundance of diversity at shallower phylogenetic resolution; there are estimated to be >15,000 species-level bacterial phylotypes associated with the human GI tract.10 The human GI tract also contains abundant and diverse viral and phage communities that can serve as predators of microbial cells and as reservoirs of genetic material that can expand microbial diversity.14–16 In contrast, the archaeal population in the human GI tract is dominated by a single species, Methanobrevibacter smithii, which contributes important metabolic activities to the intestinal ecosystem.11,17–19 There is also a low level of eukaryotic diversity in the GI tract, and the roles of these organisms within the larger microbial community remain to be defined.20,21
To understand the form and function of the GI microbiota, it is helpful to view this complex microbial community through the lens of ecology. Historically, ecological inquiry has focused on analysis of macroscopic organisms, yet there is considerable interest in applying relevant ecological concepts to the microbial world. An important challenge for future research is to determine whether ecological principles discovered by study of macroscopic organisms can be directly applied to microbial communities.22–25 There are 2 general factors that limit our ability to apply an ecological perspective to the human GI microbiota. First, ecological theories and methods of investigation have not yet successfully permeated into the field of GI microbiota research. Second, data and associated insights into the human microbiota are limited. Ongoing research initiatives are expected to expand this knowledge base in the coming years, providing data that will be essential for testing and adapting ecological concepts in human GI environments.
An important feature of the GI microbiota that distinguishes it from non–host-associated microbial communities is its residence within a living multicellular host organism that is sensitive and responsive to its microbial residents. The responses of a host to its microbial inhabitants can alter the selective pressures within the GI environment that act upon microbial traits, which can in turn alter the microbial community. This reciprocal relationship between host and microbial community adds a rich layer of complexity to the analysis of GI microbial ecology and requires that host responses to microbes be incorporated into the study of GI ecology.
In this review, we frame the field of GI ecology in terms of 2 fundamental ecological concepts: pattern and scale.26 Pattern is defined as the variation of biodiversity within an ecosystem, whereas scale is defined as the spatial, temporal, and environmental dimensions within which that variation is perceived. Pattern and scale are inextricably linked, because measurement of pattern occurs only within the context of a particular range of scales. A comprehensive understanding of the GI ecosystem will require us to appreciate patterns and their underlying causes along the continuum of scales.27 This review describes the different kinds of patterns that can be perceived within the GI ecosystem and then discusses the range of scales along which researchers can perceive those patterns. We then highlight how concepts of pattern and scale are being combined in the study of biogeography within the GI tract.
Defining Patterns in GI Microbial Ecology
Variation within an ecosystem such as the GI tract results in different kinds of patterns. These include patterns in the membership, function, and localization of the GI microbial community, as well as patterns of microbe-microbe and host-microbe relationships. Detection and description of patterns in the GI ecosystem are important prerequisites for understanding how patterns are established, how they change in response to different factors, and the consequences of pattern on microbial and host biology. In the following text we review the different types of patterns that have been described in the GI ecosystem.
Patterns in Microbial Community Membership
Variation in the membership of microbial lineages within a community is the most common pattern used to describe and compare microbial communities. Traditional methods for analyzing microbial community membership relied largely on cultivation of microorganisms. The principal limitation of cultivation-based approaches is that approximately 80% of all microbial members of the GI microbiota currently cannot be cultivated outside the GI tract.11 However, advances in microbial culture methods are increasing the fraction of the community that can be cultivated,28 and novel high-throughput culture-based phenotyping methods can help rapidly identify microbial community members with specific functional traits.29 Although culture-based methods remain indispensable for isolating individual microorganisms for physiologic and molecular analyses, rapid advances in DNA sequence-based methods have dramatically expanded our ability to survey biodiversity in a microbial community. Gene sequences derived from small subunit ribosomal RNA (SSU rRNA; 16S rRNA in Bacteria and Archaea, and 18S rRNA in Eukarya) have been used extensively for phylogenetic analysis.30 SSU rRNA genes are encoded in the genomes of all cellular organisms and are highly conserved but contain regions of sequence variability sufficient to distinguish between specific microbial groups. SSU rRNA sequences can be clustered based on selected levels of sequence identity into operational taxonomy units to provide an operational measure of microbial phylogeny and diversity without culture bias.31 An important consideration in any taxonomic analysis is the definition of a species, because such criteria often serve as the basis for sorting organisms into genetic lineages. There is surprisingly little consensus regarding the definition of prokaryotic species, and current methods have not kept pace with the discovery rate of new prokaryotic diversity.32 DNA-DNA hybridization methods and multilocus sequence analysis are often used for defining species in culturable isolates; however, the relative ease and culture independence of SSU rRNA sequence analysis has caused this method to emerge as the predominant taxonomic metric. Species-level phylotypes are defined as operational taxonomy units sharing ≥97% or ≥99% identity, depending on the study. Although broadly utilized, this definition is debated due to genomic variation between members of a species-level phylotype. Comparison of 32 Escherichia coli and Shigella strain genomes33 and 6 Streptococcus agalactiae strain genomes34 revealed that many genes are found in only a subset of strains (the “dispensable genome”) while other genes are found in all tested strains (the “core genome”).35 One potential cause of this genetic variation is the high rate of lateral gene transfer observed in genomic analysis of enteric prokaryotes.36,37 This genetic exchange between community members serves as a potential source of genetic diversity and is believed to be a result of phage, mobile elements, and conjugal transfer.15,36,37 Despite these caveats, analysis of SSU rRNA sequences can provide important insights into the structure of a microbial community beyond the classification of observed phylotypes. Because most SSU rRNA sequence-based analyses of microbial communities do not reach saturation (ie, relatively rare community members are not detected), observed SSU rRNA sequences can be binned into operational taxonomy units to permit estimation of “true” biodiversity within that community using statistics such as richness (the number of phylotypes in an area), diversity (the number of phylotypes in an area weighted for relative abundance), and evenness (the relative abundance with which species are observed in an area).38,39 DNA sequences derived from SSU rRNA genes (or other genes) in different microbial communities can also be used to directly compare the phylogenetic structures of those microbial communities (reviewed by Lozupone and Knight39).
Genomic DNA extracted from a microbial community can be used for a spectrum of SSU rRNA sequence-based phylogenetic approaches to define microbial community composition (reviewed by Dethlefsen et al40 and Zoetendal et al41). These include rapid and cost-effective methods that provide a “fingerprint” of the microbial diversity within a community (eg, T-RFLP,42 ARISA,43 and DGGE/TGGE44), sensitive methods for detecting specific community members (eg, quantitative polymerase chain reaction45,46 and high-density SSU rRNA microarrays47–49), and SSU rRNA sequencing methods that provide maximum phylogenetic information (eg, Sanger sequencing of SSU rRNA clone libraries and pyrosequencing of amplified pools of SSU rRNA genes50,51). While culture-independent sequence-based methods for defining community composition have revolutionized our understanding of microbial diversity in the GI tract, there are caveats to these methods. Different microbial species can display distinct susceptibilities to specific cell lysis and DNA extraction methods,52 and the selection of polymerase chain reaction primers to amplify SSU rRNA genes can also impact on the observed diversity within a sample.53 Furthermore, it is important to recognize that different microbial genomes encode variable numbers of SSU rRNA genes, potentially leading to apparent phylotype abundances that do not accurately represent cellular abundance.54
Sequence-based phylogenetic surveys have revealed major patterns in the membership of the GI microbiota. The human intestinal microbiota at homeostasis is numerically dominated by members of just 2 bacterial phyla: Firmicutes and Bacteroidetes.9–11,51 The prevalence of Firmicutes and Bacteroidetes is not absolute along the length of the GI tract, because the stomach is instead predominated by members of the Proteobacteria phylum.12 Moreover, distinct alterations in the membership of the intestinal microbiota have been associated with a growing spectrum of human diseases (reviewed by Kinross et al6), including inflammatory bowel disease (reviewed by Sartor55 and Sokol et al56) and obesity (reviewed in the following text and by DiBaise et al57).
Patterns in Microbial Community Function
Microbial community membership continues to serve as the predominant basis for analyzing and comparing microbial communities; however, several new complementary approaches are extending our ability to perceive patterns of microbial community function in the GI tract. Whereas phylogenetic methods utilize microbial lineages as a measure of community membership, metagenomic methodologies utilize the microbiome and its encoded products as a culture-independent measure of the potential activities of a microbial community (reviewed by Zoetendal et al,41 Medini et al,58 and Turnbaugh and Gordon59). Most metagenomic analyses of the GI microbiota to date have focused on shotgun sequencing of genomic DNA isolated directly from the intestinal microbial community in humans and mice.51,60–62 These data are providing early insights into the gene content and physiologic potential of the intestinal microbiome, including enrichment of genes involved in metabolism of carbohydrates, amino acids, and xenobiotics, synthesis of vitamins, and methanogenesis.51,60 Patterns of in vivo microbial transcription and translation can also be analyzed by isolating and quantifying RNAs (metatranscriptomics63–67) and proteins (metaproteomics68). These methods provide important complementary perspectives to genomic DNA sequence analysis, because the patterns of bacterial gene expression can be strongly influenced by multiple factors in vivo, including host developmental stage,67 nutrient availability,63,66,67 and the presence of other microbial species.17,64
The in vivo biological activity of the human microbiota can also be monitored by isolating metabolites from specific anatomic compartments within the host and analyzing them using mass spectrometry and nuclear magnetic resonance methodologies.69 This strategy is based on the observation that the GI microbiome encodes enzymatic capabilities that are not encoded in our own genome51,60 and that identifiable microbial metabolites and cometabolites (proxies for microbial enzymatic activities) can be detected in multiple host compartments in correlation to the activities of the microbiome.70–72 For example, a genetic predisposition to impaired glucose homeostasis and nonalcoholic fatty liver disease in mice is associated with defects in choline metabolism, including low plasma phosphatidylcholine levels and microbiota-mediated urinary excretion of methylamines.70 The role of the microbiota on metabolite profiles in diverse host compartments was directly tested by comparing germ-free mice with those colonized with a normal microbiota. The microbiota was found to produce numerous metabolic changes in the intestine as well as extraintestinal compartments, including alterations in choline and bile acid metabolism.71 As our understanding of microbial metabolic diversity catches up with our new grasp of microbial phylogenetic and metagenomic diversity, the known microbial contribution to the host metabolome can be expected to expand.59
Patterns of Microbial Localization and Behavior
The spatial localization and dynamic behaviors of microorganisms in the GI tract can determine microbial community functions. In vitro studies have shown that spatial organization of simple microbial communities can promote beneficial interspecies relationships and adaptive responses.73–75 It is likely that spatial organization serves a similar role in the complex microbial communities along the GI tract. Traditional approaches of light and electron microscopy allow estimation of spatial organization as well as microbial abundance; however, it can be difficult to distinguish between microbial species with similar physical properties using these methods. Fluorescence in situ hybridization using probes targeting SSU rRNA or other microbial genes has emerged as a common technique to provide highly quantitative information on the number and spatial localization of microbial lineages within the GI tract.76 Fluorescence in situ hybridization has also been coupled with stable isotope labeling and mass spectrometry to assay metabolic properties of individual microbial cells within a community.77
Many in vitro analyses have shown that microbial cells are capable of a diversity of behaviors, including different forms of motility, chemotaxis, adhesion, quorum sensing, and formation of spores and multicellular biofilms.78–80 These microbial behaviors are likely to have strong effects on microbial proliferation, dispersal, and persistence in the GI tract, as they do in other environments.81 Despite this acknowledged behavioral diversity in microorganisms, our understanding of how microbial behavior impacts GI microbial ecology and host biology is remarkably limited. This can be largely attributed to the difficulties associated with in vivo monitoring of microbial localization and behavior in the GI tracts of humans and mammalian models. In vivo bioluminescence assays can reveal localization of specific microbial lineages within mammalian hosts, but their spatial resolution is not sufficient to view individual cells.82 The optical transparency of the zebrafish model system presents new opportunities to view microbial behavior in a living vertebrate intestine. This model was recently used to reveal that individual microbial cells within the intestine can display a diversity of behaviors in vivo, ranging from rapid motility to formation of biofilms to physical interaction with the intestinal mucosa.83 A comprehensive understanding of the GI ecosystem must incorporate the contribution of distinct microbial behaviors to community membership and function.
Patterns of Microbe-Microbe and Host-Microbe Relationship
Understanding relationships between individual microbial cells, between groups of microbes, and between microbes and their host is fundamental to our understanding of the GI ecosystem. These relationships can be considered as emergent patterns generated by the membership, activity, and localization of the microbial community. Host-microbe and microbe-microbe relationships have traditionally been defined along a continuum ranging from pathogenic to commensal to mutualistic (see definitions in Table 1).23,25 The intricate relationship between a host and its GI microbiota is generally considered to be mutualistic because the microbiota gains a relatively stable nutrient-rich environment while the host gains extended digestive capabilities and exclusion of potential pathogens. This 2-dimensional concept of a pathogen-mutualist range can be operationally useful; however, the potential complexity of relationships and the physiologic/genomic flexibility of individual species demand a more sophisticated perspective. Any 2 organisms within an ecosystem are likely to interact in multiple ways via their spatial relationship within a habitat and also via their metabolic relationships within a niche. A relationship between 2 organisms (or groups of organisms) that is defined as pathogenic by one set of criteria might be defined as mutualistic by another. For example, the parasitic protozoan Toxoplasma gondii and the bacterium Helicobacter pylori can be classified as human pathogens using a specific set of criteria. However, recent evidence suggests that each of these organisms may have evolved as mutualists by providing protection from other pathogens and gastroesophageal reflux disease, respectively (personal communication, Laura Knoll, December 2008).12 How we define a relationship therefore depends on how we measure benefit and detriment as well as the breadth and accuracy of our perceptual abilities.
Table 1.
Glossary of Key Terms in GI Microbial Ecology
| Term | Definition |
|---|---|
| Biogeography | The study of patterns of biodiversity along spatial, temporal, and environmental scales |
| Commensalism | Relationship in which one partner benefits without detriment to the other |
| Conventionalized | Animals derived germ-free and later colonized with a microbiota harvested from conventionally raised donors |
| Conventionally raised | Animals raised under standard conditions in the presence of a normal microbiota |
| Germ-free | Animals raised in the absence of all microorganisms; also called axenic |
| Gnotobiotic | Animal or environment in which all microorganisms are excluded or known |
| Habitat | Physical location or “address” occupied by an organism within an ecosystem |
| Metabolomics | Identification and quantification of host and microbial metabolites in a particular host compartment |
| Metagenomics | Culture-independent measure of the gene content and physiologic potential of a microbial community |
| Microbiome | Collective genomes within a microbiota |
| Microbiota | Collective community of microorganisms within a habitat |
| Mutualism | Relationship in which both partners benefit; also called symbiosis |
| Niche | Function or “profession” of an organism within an ecosystem |
| Pathogenesis | Relationship in which one partner benefits to the detriment of the other; also called parasitism |
| Pattern | The variation of biodiversity within an ecosystem |
| Phylotype | Group of SSU rRNA gene sequences with ≥97%–99% sequence identity |
| Scale | The spatial, temporal, or environmental ranges within which variation in an ecosystem is perceived |
An important step toward elucidating relationships between organisms is to understand the different strategies used in those relationships. Ecological theory and in vitro models have been developed to describe social strategies used by microbes in competition, cooperation, cheating, and food web interactions,25,75,81,84 but these theories remain largely untested in the context of the GI tract. Analysis of gnotobiotic animal hosts colonized with simple microbial communities is providing early insights into the utilization of these social strategies by enteric microbes. Colonization of germ-free mice with a simple microbial community consisting of the bacterium Bacteroides thetaiotaomicron and the archaeon Methanobrevibacter smithii resulted in elevated colonization density and production of short-chain fatty acids compared with animals colonized with either microbial species alone.17 This cooperative microbe-microbe interaction also conferred an apparent benefit to the host by enhancing energy storage. Relationships between microbial species will be influenced by the degree of modularity and functional redundancy within the ecosystem. A recent analysis of glycoside hydrolases and glycosyltransferases in the genomes of gut-derived microbial species and non-gut microbes revealed a significant convergence of these genes in gut-derived species, indicating a high degree of functional redundancy among gut microbes for those enzymatic activities.36
As described previously, patterns of biodiversity in the GI tract can be perceived in multiple ways, and these patterns need to be integrated to fully understand the structure and function of the GI microbiota. For example, measurements of community membership combined with metagenomic analysis of the same community can build associations between dominant community members and specific gene content.51,61,62 Paired assessment of GI microbiota membership and host metabolomic profiles can also reveal novel associations between specific community members and specific metabolic activities.72 These integrated patterns of biodiversity in the GI tract will become increasingly important as we attempt to gain a mechanistic understanding of how patterns change between diverse host environments, between different spatial locations within a host, and over time.
Defining Scales in GI Microbial Ecology
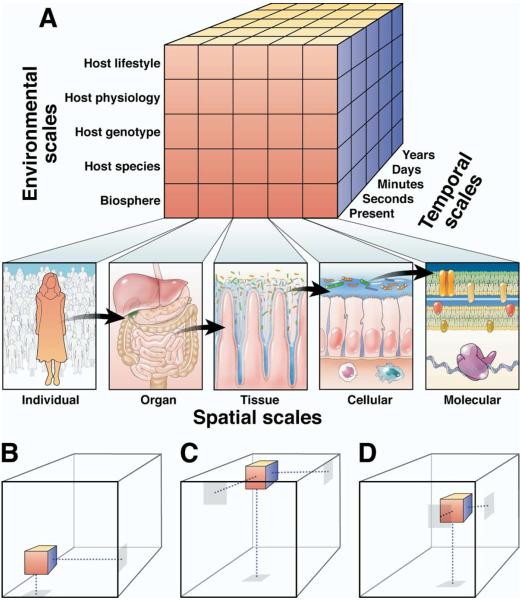
Any ecosystem, including the GI tract, exists along temporal, spatial, and environmental scales; these scales define 3 conceptual dimensions within which the GI ecosystem exists (Figure). However, any single analysis of an ecosystem is performed using a limited range of scales: “a low-dimensional slice through a high-dimensional cake.”26 This restricted range of scale is imposed by our experimental design as well as limitations in our perceptual capabilities. This is important because different scales might be subject to different selective processes. Fine temporal and spatial scales can generally provide greater detail yet be more susceptible to stochastic events, whereas coarser scales can be more regular and predictable. We must therefore recognize the biases associated with each scale and develop an understanding of the interaction among phenomena on different scales. It is helpful to first consider how temporal, spatial, and environmental scales apply to the GI ecosystem.
Figure 1.
Scales in GI microbial ecology. (A) The GI ecosystem can be conceptualized as a 3-dimensional space defined by variation along environmental, spatial, and temporal scales. Environmental scales (y-axis) are defined by different types of variation affecting the GI environment, including variation in biosphere, host species, host genotype, host physiology, and host lifestyle. Spatial scales (x-axis) are defined by the spatial resolution at which the GI ecosystem is perceived. The upper macroscopic level consists of the individual host and progresses down through the levels of organ system, tissue, cell, and molecule. Note that the molecular spatial scale is illustrated here by molecules within a gram-negative bacterium, although this same scale can be applied to molecules of any host or microbial origin. Temporal scales (z-axis) are defined by the time over which variation in the GI ecosystem is perceived, beginning with the present and progressing into seconds, minutes, days, and years. B–D show how this conceptual frame of reference can be used to provide context for 3 recent studies of the GI ecosystem. B depicts a phylogenetic comparison of the microbial community in the feces (spatial scale: organ) of different mammalian host species (environmental scale: host species) at a single time point (temporal scale: present).98 C depicts a phylogenetic comparison of the microbial community in the feces (spatial scale: organ) of individual healthy humans at multiday intervals (temporal scale: days) before and after treatment with the antibiotic ciprofloxacin (environmental scale: host lifestyle).50 D depicts a comparison of bacterial cell behavior (spatial scale: cells) in zebrafish intestines at different stages of development (environmental scale: host physiology) using real-time in vivo imaging (temporal scale: seconds).83
Temporal Scales
Patterns within the GI ecosystem and their underlying selective forces can change over the course of seconds, minutes, hours, days, and years (Figure 1). Moreover, different communities may display varied levels of stability and resilience. The temporal distance between sampling time points in an experiment can therefore impact the resulting observations and interpretations. Many of the recent analyses of the GI microbiota have focused on either a single time point in individual hosts10,11,60,71,72,85 or multiple time points taken from the same individual host over the course of days or weeks.9,50,51 Some of these recent studies have provided insights into the temporal scales over which the GI microbiota changes in response to different types of perturbation. When obese people were switched to a low-calorie diet, their fecal microbiotas displayed a gradual phylum-wide decrease in the ratio of Firmicutes to Bacteroidetes over the course of 1 year.9 In contrast, treatment of healthy people with ciprofloxacin for only 3–5 days resulted in rapid and marked individualized reductions in diversity across all predominant bacterial phyla. Remarkably, the pretreatment pattern of bacterial diversity was largely restored by 4 weeks after the end of ciprofloxacin treatment, indicating that the GI microbiota is highly resilient.50 These results show the wide range of temporal scales over which patterns of community membership can change and highlight the need to extend analysis of the GI ecosystem to finer temporal scales (ie, hours, minutes, and seconds). It will be instructive to correlate patterns of microbial community membership with patterns of microbial activity and localization at these finer temporal scales.
Spatial Scales
Beginning at the “top” of the spatial scale, we can perceive patterns of variation between individual hosts (Figure 1). Recent analysis of the GI ecosystem has been biased toward this spatial scale, in which individual hosts are compared using samples obtained from a common anatomic site such as biopsy of the intestinal mucosa,10,11 feces,9,11,50,51,60 intestinal contents,61,62,85–87 or whole intestinal segments.71,87 The selection of analytic method can have a profound impact on our ability to perceive patterns in GI microbial ecology at the scale of individual hosts. 16S rRNA sequence-based comparisons of human fecal microbiota have revealed high levels of interindividual variation9,11,50,88 and have indicated that the number of species-level phylotypes shared across individuals is exceedingly rare or nonexistent.51 In contrast, metagenomic analyses of gene content in the fecal microbiome of different human hosts revealed a wide array of microbial gene families that are shared between individuals. These data indicate that a “core microbiome” shared between human hosts exists at the level of gene content but not at the level of microbial lineages. This implies the existence of significant functional redundancy between dominant members of the GI microbiota and suggests that each host environment exerts selective pressures on microbial traits rather than on microbial taxonomy.51
The proximal-distal axis of the alimentary tract provides microbes with a range of physical habitats and metabolic niches determined by salient differences in anatomy, physiology, and nutrient availability. Selective pressures that act on the microorganisms at different proximal-distal locations along the alimentary tract can result in compositionally distinct microbial communities. This is evident in the increasing density of microbial cells proceeding from the proximal small intestine (103 cells/g contents) to the colon (1011 cells/g contents),1 the significant compositional differences between the bacterial communities in the stomach (usually predominated by Proteobacteria12) and intestine (usually predominated by Firmicutes10), as well as relatively minor differences in microbial community composition between different regions of the large intestine.11 To understand the mechanisms that underlie community structure and function at any point along the proximal-distal axis of the GI tract, we must also consider the radial axis that spans the lumen, mucus layer, and epithelium of the GI tract. A prominent feature of the radial axis is the mucus layer covering the epithelium. The mucus layer provides a dynamic physical barrier that separates enteric microbes from the host epithelium76,89 while simultaneously serving as a diffusion barrier for antimicrobial peptides and other factors released by host cells.90 O- and N-linked glycans associated with mucins also provide a nutrient source for enteric microbes,67 which can serve as an important determinant of persistence within the GI tract habitat.66 Selective pressures that act at distinct locations along the radial axis are therefore likely to influence the organization, composition, and activity of the local microbial community. For example, the fecal microbiota and colonic mucosal-adherent microbiota from the same individual contain distinct microbial lineages.11 Moreover, liquid and particulate fractions of human feces contain distinct patterns of microbial diversity.91 Further culture-independent analysis is required to describe the distribution of microbial lineages along the proximal-distal and radial axes of the GI tract and the consequences of these microbial localization patterns to GI ecology.
The emergent properties of the microbiota and its contribution to host biology are a result of the interactions between microbial cells and host cells. By perceiving the GI ecosystem on the spatial scale of cellular interactions, we can begin to discern the mechanisms that underlie the functional relationships between different microbial and host cells. Microbe-microbe interactions can be perceived at several levels, including interactions between operational groups of microbes, between different microbial species, and between members of the same microbial species. As described previously, microbial cells can utilize a variety of behaviors and communication mechanisms to establish relationships with other microbes in their habitat.79–81 Elucidating relationships between microbial cells in the context of the complex normal GI microbiota poses daunting challenges. It can therefore be useful to experimentally reduce the complexity of the microbial community. Gnotobiotic animal models, in which all microbial life can be defined or excluded, provide opportunities to control the complexity of the GI microbial community and thereby study specific relationships between microbial cells. Analysis of simple communities consisting of only a few defined microbial lineages in gnotobiotic hosts is beginning to reveal the nature of microbial relationships in heterogeneous ecosystems.17,64,66 In vitro culture systems that mimic conditions within the GI tract can also be used to study interactions between normal members of the GI microbiota,92 although microbes can display important phenotypic differences between in vitro and in vivo environments.63,66 An often-overlooked factor in microbial relationships is the issue of phenotypic heterogeneity. Even within a community consisting of only a single bacterial species, there is marked phenotypic heterogeneity because of stochastic changes in transcription and translation in individual microbial cells.93 The contribution of this phenotypic heterogeneity to the form and function of complex microbial communities like the GI microbiota remains unclear.
At the “bottom” of the spatial scale, we can analyze the molecules that comprise the microbial cells, host cells, and interstitial spaces within the GI ecosystem. Microbial genome sequencing projects have revealed patterns of genome organization and the evolutionary history of microbes adapted to the GI tract18,37,62,94 while providing valuable references for interpreting metagenomic data sets.8,51 Metagenomic and metabolomic approaches described previously are providing unprecedented insights into the gene content and physiologic capabilities of GI microbiomes.59 A long-term goal for microbiome research is to empirically define the functions of identified microbial genes as well as the processes and metabolic networks in which they participate. It will be equally important to understand how the molecular biology of different host cell types is altered as a function of microbial community composition and activity. Tools and concepts cultivated in the field of systems biology will be especially useful for modeling and analyzing genetic and metabolic networks in the GI ecosystem.27 Successful implementation of these systems biological approaches will require an understanding of the activity of genes and gene products encoded in the microbiome and the host genome as well as the transcriptional and translational mechanisms that regulate their expression. Genetic and molecular analysis in representative cultivatable members of the GI microbial community are beginning to reveal the function of specific gene products,66,95–97 but many additional microbial genes remain to be functionally characterized in the context of the GI ecosystem.
Environmental Scales
To understand the principles that govern the ecosystem within the GI tract, it can be helpful to observe how the ecosystem changes as the host environment changes in different ways. Here we define “environment” from the perspective of the GI microbial community: all features of the habitat within the host as well as the features of the host's biosphere together comprise the environment in which the GI microbial community exists. This environment can be modified in different ways, including variation in the local biosphere in which hosts reside,85,87 host phylogeny,87,98 host genotype,99 host physiology (eg, age, metabolism, immunology, pathobiology),100 and host lifestyle (eg, diet, exposure to antibiotics and pathogens) (Figure 1).50,62,101,102 As described in the following text, variation along these environmental scales can alter the selective pressures within the GI ecosystem and can impact on microbe-microbe and host-microbe relationships.
Patterns Within Scales: Biogeography of the GI Ecosystem
The field of biogeography was founded more than 250 years ago by Carl Linnaeus and his colleagues, who studied the patterns of plant and animal biodiversity in diverse terrestrial environments. Recent advances in molecular phylogenetics, metagenomics, and metabolomics have permitted the application of biogeography to the biodiversity of the microbial world.27,103 Because biodiversity can be defined by taxonomy as well as functional traits, the modern definition of biogeography could be broadened to the study of biological patterns within a range of scales. A comprehensive understanding of GI biogeography will therefore require knowledge of the many patterns within the GI ecosystem along the full range of spatial, temporal, and environmental scales. This integrated perspective must account not only for the microbiota and its component members, but also for the biology of the host and the reciprocal relationship between host and microbiota.
The biogeography of the GI tract is an important frontier of medical research because microbial dysbioses have been associated with a growing number of human diseases, including inflammatory bowel disease and obesity (reviewed by Kinross et al,6 Sartor,55 Sokol et al,56 and DiBaise et al57). When a disease is associated with an alteration in GI microbiota composition or activity, a fundamental challenge is to determine whether the observed changes are causes and/or consequences of the respective disease. If microbial dysbiosis is found to be a consequence of host disease, it could be possible to develop new diagnostic and predictive tools for monitoring human health. If microbial dysbiosis is found to contribute to the etiology of disease, specific antibiotic/prebiotic/probiotic/postbiotic approaches could be used to control abundance or activity of the implicated community members.104,105
Host Habitat Effects on the Microbial Community
A fundamental goal in biogeography research is to understand how the local environment influences biodiversity. The Baas-Becking hypothesis proposes that all microbial life is distributed worldwide but that the local environment selects upon, and is therefore in part responsible for, the variation in microbial biodiversity between different environments.106,107 Initial tests of the Baas-Becking hypothesis within the context of the GI tract have focused on the relative contribution of the local biosphere (eg, differences in the local microbial community available to colonize the host or the composition of the parentally transmitted microbial community) to GI microbiota composition. The composition of the fecal microbiota in babies delivered by cesarean section is often compositionally distinct from babies delivered vaginally,108–110 although the infant fecal community is also susceptible to significant stochastic effects.88 Phylogenetic comparisons of the cecal microbiota from mouse pups and their mothers revealed that the pups' microbial community resembled that of their mother.85 These results show that the microbial community within the local biosphere can influence the composition of the GI microbiota. However, specific selective pressures might act within the habitat of the host GI tract (host habitat effects; eg, host immunity, physiology, and diet) to further shape the diversity of the GI microbiota. A role for host habitat effects in GI microbial diversity was tested by colonizing germ-free mice and zebrafish recipients with intestinal microbiotas obtained from conventionally raised zebrafish and mouse donors, respectively. Phylogenetic analysis revealed that when a microbial community is transferred to a new host, the relative abundance of the bacterial lineages in the transplanted microbial community change to resemble the normal intestinal microbial community composition of the recipient host.87 For example, colonization of germ-free mice with a zebrafish intestinal microbiota predominated by Proteobacteria resulted in an output community with an amplification of the Firmicutes phylum that dominates the conventional mouse intestinal microbiota. The GI habitat within different hosts therefore helps shape microbial community composition in distinctive ways.
It is not clear which factors within a host habitat are responsible for selecting a specific microbial community; however, experiments in animal models are beginning to elucidate the contribution of host genotype and diet to GI microbial diversity. For example, mice with a homozygous knockout of Myd88, which encodes an adaptor protein for almost all Toll-like receptors (TLRs), consistently displayed reduced ratios of Firmicutes to Bacteroidetes and increased representation of specific families within these phyla (ie, Lactobacillaceae, Rikenellaceae, and Porphyromonadaceae) in their cecal microbiota compared with wild-type mice.111 It remains unknown how loss of Myd88 function causes this shift in community composition, but it is possible that selective pressures caused by Myd88-dependent production of antimicrobial proteins by Paneth cells help to shape microbial community structure.112 Phylogenetic analysis of the cecal microbiota in mice with a homozygous mutation in the gene encoding leptin receptor (ob/ob) and their wild-type siblings revealed that the obesity phenotype linked to the ob/ob genotype was associated with a phylum-wide increase in the ratio of Firmicutes to Bacteroidetes.85 Intriguingly, human obesity is also associated with a phylum-wide increase in the Firmicutes/Bacteroidetes ratio within the fecal microbiota, indicating that obesity might similarly alter selective pressures on GI ecology in both humans and mice.9 In contrast, a mouse model of diet-induced obesity showed an increased Firmicutes/Bacteroidetes ratio in the cecal microbiota that was caused by a marked amplification of the Mollicutes class within the Firmicutes phylum together with a phylum-wide suppression of Bacteroidetes.62 Metagenomic analysis of cecal microbiomes from these obese mouse models revealed that their increased Firmicutes/Bacteroidetes ratios were associated with increased abundance of gene categories involved in metabolism of complex polysaccharides and other carbohydrates.61,62 The amplification of specific enteric bacterial taxa in the context of obesity could therefore be due to their enhanced capacity for nutrient harvest and/or their ability to thrive under other physiologic or immunologic conditions associated with the respective host obesity phenotype.
Taken together, these studies support the Baas-Becking hypothesis within the context of the GI environment and identify host genotype and diet as 2 important factors that govern the membership and physiologic potential of the GI microbial community. This notion is further supported by a recent comparison of fecal microbiotas from humans and 59 other mammalian species that identified diet and host phylogeny as key determinants of mammalian intestinal microbiota composition.98 The mechanistic bases for these distinct effects of host diet, genotype, and phylogeny on GI microbiota composition and activity have yet to be empirically determined. Importantly, previous studies have been focused on the spatial scale of individual hosts (Figure 1), and additional analysis will be required to measure these host habitat effects at finer spatial and temporal scales.
Microbial Effects on the Host
In the analysis of GI biogeography, it is important to identify the mechanisms by which microbes communicate with their hosts and how host cells perceive and respond to these microbial cues. Alterations in the GI microbial community have been associated with a spectrum of disease states, although empirical effects of compositionally distinct microbial communities on host biology have only been defined in a few cases. The transcription factor T-bet is encoded by one of a group of genes implicated in the control of intestinal inflammation.55 Mice that lack functional T-bet are immunocompromised and display increased susceptibility to colitis. Interestingly, the colitis associated with T-bet deficiency was found to be communicable to T-bet-sufficient hosts, indicating that the absence of T-bet resulted in formation of a colitigenic microbial community.99 As described previously, an increased ratio of Firmicutes to Bacteroidetes is observed in mice that become obese due to deletion of the leptin receptor gene85 or consumption of a high-fat diet.62 Remarkably, introduction of intestinal microbial communities isolated from the obese mice into wild-type mice or mice fed a control diet, respectively, resulted in increased fat deposition.61,62 These studies illustrate the reciprocal relationship between microbiota and host: an alteration in the host habitat (ie, host genotype or diet) alters microbial community, which in turn alters host physiology. Although we have a working knowledge of the many host biological processes that are affected by enteric microbes (reviewed by Bäckhed et al,3 Wostmann,4 Cheesman and Guillemin,5 and Blaut and Clavel113), we understand relatively little about the microbial signals and host signal transduction mechanisms that mediate these effects. As summarized in the following text, significant advances have recently been made toward elucidating the host molecules and cells that facilitate microbial effects on host immunity and nutrient metabolism.
Members of the GI microbiota stimulate a program of homeostatic immune responses in intestinal epithelial cells as well as multiple populations of associated immune cells.105 Microbes are detected by pattern recognition receptors that include the transmembrane TLR and the intracellular nucleotide-binding and oligomerization domain-like receptor families. TLRs and nucleotide-binding and oligomerization domain-like receptors recognize conserved microbe-associated molecular patterns produced by bacteria, parasites, fungi, and viruses.114,115 Microbe-associated molecular patterns such as lipopolysaccharide (a major component of the gram-negative bacterial outer membrane) and flagellin (the major structural subunit of the bacterial flagellar filament) are detected by specific members of the TLR family (TLR4 and TLR5, respectively).105,115 Upon binding to their respective microbe-associated molecular pattern, TLRs act together with the Myd88 adapter protein to induce intracellular signaling events that converge upon the nuclear factor κB (NF-κB) and mitogen-activated protein kinase pathways to regulate expression of genes involved in epithelial barrier fortification and inflammation.55,114,115 Hosts have evolved a range of mechanisms to mitigate their own innate immune responses to enteric microbes (reviewed by Neish105). For example, lipopolysaccharide produced by the microbiota stimulates production of intestinal alkaline phosphatase in the intestinal epithelium of both zebrafish and mice. Intestinal alkaline phosphatase acts at the brush border of intestinal epithelial cells to detoxify lipopolysaccharide and thereby reduce the proinflammatory potential of the microbiota.116,117
The importance of the TLR and NF-κB pathways in host-microbe relationships has been determined in genetically engineered animal models. Analysis of Myd88-null mice, which have an increased susceptibility to colitis in the presence of the microbiota, indicates that microbial signals perceived by TLRs are required to prevent intestinal inflammation.118 Mice that lack TLR5 develop spontaneous colitis, but this phenotype is rescued in animals that lack both TLR5 and TLR4.119 Therefore, different TLRs can have opposing roles in intestinal homeostasis despite having common downstream effector pathways such as NF-κB. Experimental manipulation of the NF-κB pathway has revealed both anti-inflammatory120 and proinflammatory121 roles for this transcriptional control pathway in the intestine, raising questions about the spatial and temporal patterns in which TLR and NF-κB pathways are activated in response to distinct microbial signals. Mice with intestinal epithelial-specific knockout of the genes encoding NF-κB essential modulator/Ikkγ122 or Ikkβ,123 which are 2 components of the IKK complex responsible for activation of NF-κB, were found to be susceptible to chemically induced colitis or develop spontaneous intestinal inflammation, respectively. Similarly, intestinal epithelial-specific deletion of RelA/p65, which encodes a primary subunit of the NF-κB transcription factor, caused elevated epithelial cell proliferation and apoptosis as well as increased susceptibility to colitis.124 These results indicate that NF-κB activation in the intestinal epithelium promotes anti-inflammatory responses to microbial signals. This anti-inflammatory role for NF-κB in the intestinal epithelium is consistent with in vivo analysis of NF-κB activation during intestinal inflammation. Induction of experimental colitis in mice that express the marker green fluorescent protein under control of NF-κB cis-elements revealed initial transient NF-κB activation in epithelial cells followed by induction of NF-κB in lamina propria cells.125 Study of cell type–specific knockout animals is beginning to reveal the roles of TLR and NF-κB signaling pathways in specific cell types. For example, loss of Myd88 function specifically in dendritic cells established that TLR/Myd88-mediated MAMP recognition activates dendritic cells to produce proinflammatory cytokines and promote T-helper responses.126 Recent analysis of mice lacking Myd88 function specifically in Paneth cells revealed that intestinal bacteria are detected by Paneth cells through a cell-autonomous Myd88-dependent mechanism, resulting in production of antimicrobial proteins and fortification of the mucosal barrier.112 It will be important to continue to correlate specific TLR and NF-κB functions with the spatial and temporal patterns in which these pathways respond to microbial cues at intestinal and extraintestinal sites.
The GI microbiota is also an important regulator of dietary nutrient metabolism. The metabolic diversity encoded in the intestinal microbiome improves digestion efficiency of nutrients and permits the host to digest many nutrients that would be otherwise inaccessible. The encoded products of the human genome are insufficient for digestion of complex plant polysaccharides such as xylan-, pectin- and arabinose-containing carbohydrates. These complex polysaccharides enter the colon, where the microbiota produce various glycoside hydrolases, lyases, and esterases that aid the degradation of glycans into short-chain fatty acids and monosaccharides.60,61 Short-chain fatty acids are absorbed by the host, where they regulate colonocyte growth and differentiation,57 serve as an energy source for host tissues such as skeletal and heart muscles, and act as a substrate for lipogenesis in adipose tissue.127 Monosaccharides liberated through microbial fermentation are also absorbed and transported to the liver, where they induce de novo hepatic lipogenesis.86 The intestinal microbiota also contributes to host metabolism by deconjugating bile salts,71,128,129 salvaging urea, and synthesizing essential amino acids as well as K and B vitamins.127,130 These contributions of the microbiota to processing and uptake of dietary nutrients are accompanied by alterations in host energy balance. In contrast to the innate immune responses described previously, our understanding of the host pathways and processes that are impacted by these different products of microbial metabolism is much more limited. Colonization of germ-free mice with a normal microbiota results in increased serum glucose and insulin levels as well as suppression of intestinal epithelial expression of a peptide hormone called fasting-induced adipose factor (Fiaf/Angptl4).86 Fiaf synthesized in the intestinal mucosa is secreted into circulation, where it directly inhibits the activity of lipoprotein lipase to prevent fat deposition and also promotes fatty acid oxidation in muscle.86,131 Furthermore, germ-free mice homozygous for a Fiaf-null allele display higher lipoprotein lipase activities than their germ-free wild-type littermates and body fat content equivalent to wild-type conventionally raised littermates.86 These results support a model in which microbial activities regulate host energy balance by suppressing transcription of Fiaf in the intestinal epithelium, thereby promoting lipoprotein lipase activity and fat storage in peripheral tissues. The microbial factors and host signal transduction mechanisms that regulate intestinal expression of Fiaf are unknown but represent attractive targets for controlling host fat storage and energy balance.
Future Directions
The community of microorganisms that resides in the GI tract is a potent environmental factor contributing to human health and disease. The composition and activity of the GI microbiota could be used in diagnostic and prognostic measures of human health. Furthermore, reagents that target specific microbial lineages, gene products, and metabolic networks might be developed into new therapies to promote human health. Our ability to design accurate predictive measurements as well as safe and effective therapeutics will depend on our comprehension of patterns in GI ecology along spatial, temporal, and environmental scales. Each microorganism experiences the GI ecosystem on a unique range of scales, which together comprise the adaptive landscape in which it responds and evolves. There is no single correct scale in which to analyze GI microbial ecology. However, this does not mean that all scales are equally important and does not exclude the possibility that selective pressures acting on organisms change at different scales. Observations of variability and predictability in the GI ecosystem are interpretable only if we reference the range of scales that are relevant to the organisms or processes being examined.
The Human Microbiome Project and other research efforts are rapidly expanding our knowledge of microbial biodiversity on the landscape of the human body. However, recent molecular analyses of the GI ecosystem have generally focused on coarse temporal and spatial scales and have been largely limited to comparison between individuals. Analysis of GI ecology along finer spatial and temporal scales is therefore an important goal for future research. These efforts should be coupled with an increased appreciation for the reciprocal interactions between members of the microbial community and the host. Understanding the complexity of the GI ecosystem demands an integrated multidisciplinary approach that combines the fields of gastroenterology, physiology, nutrition, immunology, microbiology, ecology, evolutionary biology, and systems biology. This approach will not only depend on development and implementation of molecular analytic methods, but also on in vitro culture systems that accurately mimic GI tract environments as well as experimentally tractable mammalian and non-mammalian animal model systems that permit reductionist analyses of the GI ecosystem. To navigate and integrate the diverse patterns emerging from this multidisciplinary field, we will need a clear vision of the different spatial, temporal, and environmental scales within which the GI ecosystem operates.
Acknowledgments
The authors are grateful to two anonymous reviewers for helpful comments on this manuscript.
The authors regret that many excellent references relevant to the topics discussed in this review could not be included due to space limitations.
Funding J.G.C. was supported by National Institutes of Health (NIH) grant T32 HD046369, and M.K. was supported by NIH grant T32 GM007092. Work in the authors' laboratory is supported by NIH grants DK073695 and DK081426, the University of North Carolina Center for Gastrointestinal Biology and Disease (NIH grant P30 DK034987), the University of North Carolina Center for Environmental Health and Susceptibility (NIH grant P30 ES010126), and a Pew Scholar Award to J.F.R.
Abbreviations used in this paper
- FIAF
fasting-induced adipose factor
- GI
gastrointestinal
- NF-κB
nuclear factor κB
- SSU rRNA
small subunit ribosomal RNA
- TLR
Toll-like receptor
Biographies




Footnotes
Conflicts of interest The authors disclose no conflicts.
References
- 1.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai M. Are biologists in “future shock”? Symbiosis integrates biology across domains. Nat Rev Microbiol. 2008;6:789–792. doi: 10.1038/nrmicro1982. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 4.Wostmann BS. Germfree and gnotobiotic models: background and applications. CRC Press; Boca Raton, FL: 1996. [Google Scholar]
- 5.Cheesman SE, Guillemin K. We know you are in there: conversing with the indigenous gut microbiota. Res Microbiol. 2007;158:2–9. doi: 10.1016/j.resmic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Kinross JM, von Roon AC, Holmes E, et al. The human gut microbiome: implications for future health care. Curr Gastroenterol Rep. 2008;10:396–403. doi: 10.1007/s11894-008-0075-y. [DOI] [PubMed] [Google Scholar]
- 7.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 10.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 14.Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinsdale EA, Edwards RA, Hall D, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 16.Danovaro R, Dell'Anno A, Corinaldesi C, et al. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature. 2008;454:1084–1087. doi: 10.1038/nature07268. [DOI] [PubMed] [Google Scholar]
- 17.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuel BS, Hansen EE, Manchester JK, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conway de Macario E, Macario AJ. Methanogenic archaea in health and disease: a novel paradigm of microbial pathogenesis. Int J Med Microbiol. 2009;299:99–108. doi: 10.1016/j.ijmm.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Scupham AJ, Presley LL, Wei B, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 22.Prosser JI, Bohannan BJ, Curtis TP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 23.Little AE, Robinson CJ, Peterson SB, et al. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 24.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin SA. The problem of pattern and scale in ecology. Ecology. 1992;73:1943–1967. [Google Scholar]
- 27.Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 28.Zengler K, Toledo G, Rappe M, et al. Cultivating the uncultured. Proc Natl Acad Sci U S A. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingham CJ, Sprenkels A, Bomer J, et al. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci U S A. 2007;104:18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox GE, Magrum LJ, Balch WE, et al. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane DJ, Pace B, Olsen GJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gevers D, Cohan FM, Lawrence JG, et al. Opinion: re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 33.Willenbrock H, Hallin PF, Wassenaar TM, et al. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 2007;8:R267. doi: 10.1186/gb-2007-8-12-r267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medini D, Donati C, Tettelin H, et al. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Lozupone CA, Hamady M, Cantarel BL, et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci U S A. 2008;105:15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Mahowald MA, Ley RE, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes JB, Hellmann JJ, Ricketts TH, et al. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dethlefsen L, Eckburg PB, Bik EM, et al. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21:517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 42.Marsh TL. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr Opin Microbiol. 1999;2:323–327. doi: 10.1016/S1369-5274(99)80056-3. [DOI] [PubMed] [Google Scholar]
- 43.Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol. 1999;2:317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 45.Rinttila T, Kassinen A, Malinen E, et al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 46.Matsuki T, Watanabe K, Fujimoto J, et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70:7220–7228. doi: 10.1128/AEM.70.12.7220-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeSantis TZ, Brodie EL, Moberg JP, et al. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 48.Harrington CR, Lucchini S, Ridgway KP, et al. A short-oligonucleotide microarray that allows improved detection of gastrointestinal tract microbial communities. BMC Microbiol. 2008;8:195. doi: 10.1186/1471-2180-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loy A, Lehner A, Lee N, et al. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol. 2002;68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scupham AJ, Jones JA, Wesley IV. Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J Appl Microbiol. 2007;102:401–409. doi: 10.1111/j.1365-2672.2006.03094.x. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe K, Kodama Y, Harayama S. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods. 2001;44:253–262. doi: 10.1016/s0167-7012(01)00220-2. [DOI] [PubMed] [Google Scholar]
- 54.Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 56.Sokol H, Lay C, Seksik P, et al. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis. 2008;14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 57.DiBaise JK, Zhang H, Crowell MD, et al. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460–469. doi: 10.4065/83.4.460. [DOI] [PubMed] [Google Scholar]
- 58.Medini D, Serruto D, Parkhill J, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 59.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 60.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 62.Turnbaugh PJ, Bäckhed F, Fulton L, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnenburg JL, Xu J, Leip DD, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 64.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reigstad CS, Hultgren SJ, Gordon JI. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J Biol Chem. 2007;282:21259–21267. doi: 10.1074/jbc.M611502200. [DOI] [PubMed] [Google Scholar]
- 66.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 68.Verberkmoes NC, Russell AL, Shah M, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 69.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 70.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claus SP, Tsang TM, Wang Y, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li M, Wang B, Zhang M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen SK, Rainey PB, Haagensen JA, et al. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 74.Kim HJ, Boedicker JQ, Choi JW, et al. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kerr B, Riley MA, Feldman MW, et al. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 76.Swidsinski A, Sydora BC, Doerffel Y, et al. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis. 2007;13:963–970. doi: 10.1002/ibd.20163. [DOI] [PubMed] [Google Scholar]
- 77.Behrens S, Losekann T, Pett-Ridge J, et al. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol. 2008;74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 79.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 80.Verstraeten N, Braeken K, Debkumari B, et al. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 81.West SA, Griffin AS, Gardner A, et al. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 82.Dennis A, Kudo T, Kruidenier L, et al. The p50 subunit of NF-kappaB is critical for in vivo clearance of the noninvasive enteric pathogen Citrobacter rodentium. Infect Immun. 2008;76:4978–4988. doi: 10.1128/IAI.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rawls JF, Mahowald MA, Goodman AL, et al. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci U S A. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balagadde FK, Song H, Ozaki J, et al. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rawls JF, Mahowald MA, Ley RE, et al. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer C, Bik EM, Digiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 91.Walker AW, Duncan SH, Harmsen HJ, et al. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 92.Molly K, Vande Woestyne M, Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- 93.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 94.Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 95.Coyne MJ, Reinap B, Lee MM, et al. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 96.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 97.Fujiya M, Musch MW, Nakagawa Y, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 98.Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lupp C, Robertson ML, Wickham ME, et al. Host–mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 102.Kuehl CJ, Wood HD, Marsh TL, et al. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect Immun. 2005;73:6952–6961. doi: 10.1128/IAI.73.10.6852-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Green JL, Bohannan BJ, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 104.Jia W, Li H, Zhao L, et al. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 105.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baas-Becking LGM. Geobiologie of inleiding tot de milieukunde. Vand Stockkum & Zoon; p. 1934. [Google Scholar]
- 107.de Wit R, Bouvier T. “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 108.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 109.Hallstrom M, Eerola E, Vuento R, et al. Effects of mode of delivery and necrotising enterocolitis on the intestinal micro-flora in preterm infants. Eur J Clin Microbiol Infect Dis. 2004;23:463–470. doi: 10.1007/s10096-004-1146-0. [DOI] [PubMed] [Google Scholar]
- 110.Gronlund MM, Lehtonen OP, Eerola E, et al. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28:19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 111.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr. 2007;137:751S–755S. doi: 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- 114.Viswanathan VK, Sharma R, Hecht G. Microbes and their products—physiological effects upon mammalian mucosa. Adv Drug Deliv Rev. 2004;56:727–762. doi: 10.1016/j.addr.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 115.Albiger B, Dahlberg S, Henriques-Normark B, et al. Role of the innate immune system in host defence against bacterial infections: focus on the Toll-like receptors. J Intern Med. 2007;261:511–528. doi: 10.1111/j.1365-2796.2007.01821.x. [DOI] [PubMed] [Google Scholar]
- 116.Bates JM, Akerlund J, Mittge E, et al. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goldberg RF, Austen WG, Jr, Zhang X, et al. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc Natl Acad Sci U S A. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 119.Vijay-Kumar M, Sanders CJ, Taylor RT, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Neurath MF, Pettersson S, Meyer zum Buschenfelde KH, et al. Local administration of antisense phosphorothioate oligonucleotides to the p65 subunit of NF-kappa B abrogates established experimental colitis in mice. Nat Med. 1996;2:998–1004. doi: 10.1038/nm0996-998. [DOI] [PubMed] [Google Scholar]
- 121.Greten FR, Eckmann L, Greten TF, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 122.Nenci A, Becker C, Wullaert A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 123.Zaph C, Troy AE, Taylor BC, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 124.Steinbrecher KA, Harmel-Laws E, Sitcheran R, et al. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 125.Kim SC, Tonkonogy SL, Karrasch T, et al. Dual-association of gnotobiotic Il-10−/− mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 126.Hou B, Reizis B, DeFranco AL. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 128.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 129.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 130.Stevens CE, Hume ID. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev. 1998;78:393–427. doi: 10.1152/physrev.1998.78.2.393. [DOI] [PubMed] [Google Scholar]
- 131.Bäckhed F, Manchester JK, Semenkovich CF, et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]