Summary
The AAA ATPase p97/VCP regulates protein homeostasis using a diverse repertoire of cofactors to fulfill its biological functions. Here we use the allosteric p97 inhibitor NMS-873 to analyze its effects on enzyme composition and the ability of cells to adapt to its cytotoxicity. We found that p97 inhibition changes steady state cofactor-p97 composition, leading to the enrichment of a subset of its cofactors and polyubiquitin bound to p97. We isolated cells specifically insensitive to NMS-873 and identified a new mutation (A530T) in p97. A530T is sufficient to overcome the cytotoxicity of NMS-873 and alleviates p97 composition changes caused by the molecule, but not other p97 inhibitors. This mutation does not affect NMS-873 binding, but increases p97 catalytic efficiency through altered ATP and ADP binding. Collectively, these findings identify cofactor-p97 interactions sensitive to p97 inhibition and reveal a new on-target mechanism to suppress the cytotoxicity of NMS-873.
Introduction
The AAA ATPase p97/VCP is a hexameric enzyme involved in cellular protein homeostasis (Meyer, et al., 2012). Although its most well-understood function is in the processing of ubiquitin-modified proteins prior to their degradation by the proteasome (Meyer, 2012), p97 has a variety of other roles such as membrane fusion (Kondo, et al., 1997), autophagy (Bug and Meyer, 2012), protein complex remodeling (Maric, et al., 2014; Moreno, et al., 2014; Yen, et al., 2012), and endosomal trafficking (Ritz, et al., 2011). These rely on mechanical force provided by conformational changes in p97 driven by ATP hydrolysis and mostly involve ubiquitin (Richly, et al., 2005; Rouiller, et al., 2002).
The N-terminal domain (N-domain) of p97 interacts with proteins that help define its cellular functions (Yamanaka, et al., 2012). UBX domain containing proteins represent the largest class of these cofactors (Schuberth and Buchberger, 2008). They often contain ubiquitin binding motifs involved in substrate recognition and p97 recruitment (Kloppsteck, et al., 2012). Well-characterized cofactors include UFD1/NPL4 which recognizes ubiquitin modified proteins destined for degradation by the proteasome (Meyer, et al., 2000; Ye, et al., 2001; Ye, et al., 2003) and p47 which regulates ubiquitin-dependent membrane fusion (Kondo, et al., 1997; Otter-Nilsson, et al., 1999). In addition to substrate receptors, other interacting proteins provide enzymatic activities to p97 such as ubiquitin hydrolysis (e.g., deubiquitinating enzymes VCIP135 (Uchiyama, et al., 2002), ataxin-3 (Zhong and Pittman, 2006), and OTUD2 (Ernst, et al., 2009)) and ubiquitin ligation (e.g., UBE4B (Laser, et al., 2006), gp78 (Zhong, et al., 2004), HOIP (Schaeffer, et al., 2014), and HRD1 (Schuberth and Buchberger, 2005)).
Missense mutations in p97 are associated with a diverse class of genetic diseases collectively known as multisystem proteinopathy type 1 (MSP1) disorders (Meyer and Weihl, 2014). These diseases are associated with intracellular protein aggregates, supporting the major function of p97 in cellular protein homeostasis. Identified mutations mostly localize to the interface between the N domain and D1 ATPase domain and affect cofactor binding and the enzyme’s ATPase activity (Niwa, et al., 2012). Recent studies suggest these variants have altered sensitivity to activating (p37) or inhibiting (p47) cofactors (Zhang, et al., 2015).
P97 has emerged as a promising cancer therapeutic target. Several pre-clinical molecules have been described and one (CB-5083, (Zhou, et al., 2015)) is in Phase I clinical trials (shown in Figure 1A). These have different mechanisms of action including reversible ATP competitive (DBeQ, CB-5083), covalent ATPase targeted (NMS-859), and allosteric (NMS-873) (Anderson, et al., 2015; Chou, et al., 2011; Magnaghi, et al., 2013). NMS-873 is broadly cytotoxic on cancer cells (Deshaies, 2014; Magnaghi, et al., 2013) and binds to a newly discovered allosteric binding site in the D2 domain of p97, revealed upon ATP binding. This prevents ATP hydrolysis propagation by affecting interactions between the arginine finger of the NMS-873-bound subunit with the gamma phosphate of ATP bound to its neighboring subunit.
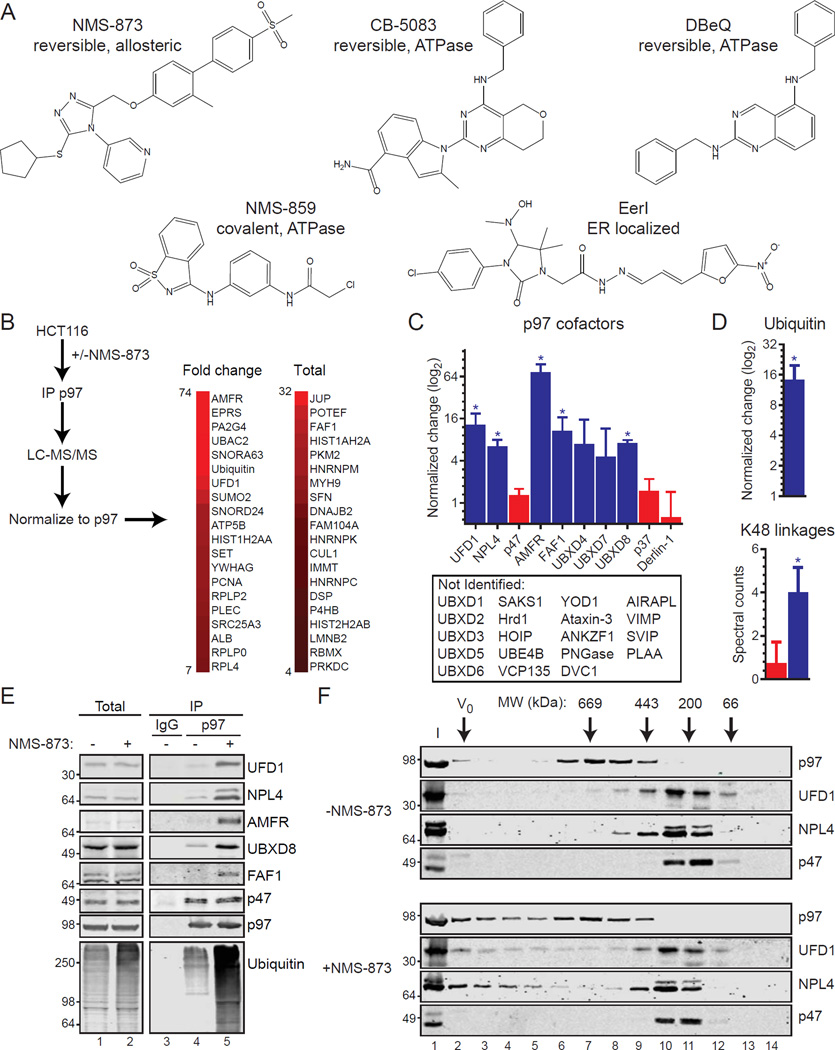
Figure 1. Allosteric inhibition alters cofactor and polyubiquitin binding to p97.
(A) Chemical structures of p97 inhibitors are shown.
(B) LC-MS/MS analyses of p97 complexes purified from HCT116 cells with or without 5 µM NMS-873 for 6 hours were performed. The top 20 NMS-873-associated interactors are shown based on protein spectrum count for fold change (left) or total (right). Extended data are in Supplemental Data File S1.
(C) The binding of several p97 cofactors is increased with NMS-873 (blue) while others do not (red). P97 cofactors not identified in LC-MS/MS analyses are also shown (below). Data represent the mean (fold change or total; n=4; *, p-value ≤ 0.05, log2 scale) and standard deviation (SD) error.
(D) Normalized increase (log2 scale) of ubiquitin purifying with p97 after NMS-873 treatment (top) and spectral counts of p97-associated K48 ubiquitin linkages with (blue) and without (red) NMS-873. Data represent the mean (n=4) and SD error (*, p-value ≤ 0.05)
(E) P97 from cells treated with NMS-873 (5 µM, 6 hours) was subjected to immunoblotting (IB). Total: input cell extracts; IP: immunoprecipitation; IgG: non-specific antibody control purification.
(F) Cell extracts from control (-NMS-873, top) or NMS-873 treated (bottom) were separated by size exclusion chromatography and analyzed by IB. Calibration standards are shown. I: input extracts; V0: void.
Here, we used NMS-873 to examine the effects of p97 inhibition on the enzyme’s composition. We found that this allosteric inhibitor reversibly increases the binding of polyubiqutin and a subset of cofactors to p97. We identified a new mutation in p97 (A530T) from cells selected for decreased sensitivity to NMS-873 that specifically overcomes increased polyubiquitin and cofactor binding caused by the inhibitor. This alters the enzyme’s ATPase activity without affecting NMS-873 binding and maintaining CB-5083 potency. Our data provide new insight into the cellular responses to p97 inhibitors and how the enzyme can specifically adapt to overcome the cytotoxicity of allosteric inhibition by NMS-873 while maintaining sensitivity to other targeted inhibitors.
Results
Altered cofactor and polyubiquitin binding to p97 with allosteric inhibition
We purified p97 from HCT116 colorectal carcinoma cells treated with NMS-873 and analyzed samples by liquid chromatography tandem mass spectrometry (LC-MS/MS). Data were normalized to p97 by total spectrum count and interacting proteins were then ranked by fold-change or total count (see Supplemental Data File S1 for full dataset). Heat maps of the top 20 proteins by these analyses are shown in Figure 1B.
We identified 10 cofactors of which 5 (UFD1, NPL4, AMFR, FAF1, and UBXD8) significantly increased in their binding to p97 with NMS-873 (Figure 1C). In contrast, the binding of p47 (UBXN2C), p37 (UBXN2B), and Derlin-1 (DER1) were largely unaffected. We also found a 15-fold increase in bound ubiquitin (Figure 1D). These ubiquitin polymers are lysine 48 (K48)-linked as other ubiquitin linkages were not identified.
We performed immunoblots on cell extracts and p97 purifications to validate these findings. For all cofactors evaluated, NMS-873 had no effect on their steady state abundance in cell extracts while ubiquitin increased (Figure 1E, left panels). Consistent with our mass spectrometry data, we found NMS-873-dependent increases in UFD1, NPL4, AMFR, UBXD8, FAF1, and ubiquitin bound to p97, but not p47. To exclude the possibility that the difference in cofactors binding to p97 with NMS-873 is through differential dissociation during immunoprecipitation, we fractionated cell extracts by size exclusion chromatography and observed an NMS-873-dependent shift of p97, UFD1, and NPL4 to higher molecular weight fractions while p47 was unchanged (Figure 1F).
UFD1/NPL4 and polyubiquitin binding to p97 with transient NMS-873 exposure
The UFD1/NPL4 heterodimer has been linked to ubiquitin-dependent protein degradation by the proteasome through its association with p97 (Meyer, et al., 2000; Ye, et al., 2001; Ye, et al., 2003). Both subunits have enhanced binding to p97 after NMS-873 treatment (see Figure 1). P47 is involved in p97- and ubiquitin-dependent membrane fusion (Kondo, et al., 1997; Otter-Nilsson, et al., 1999). Unlike UFD1 and NPL4, its binding to p97 is largely unaffected by NMS-873.
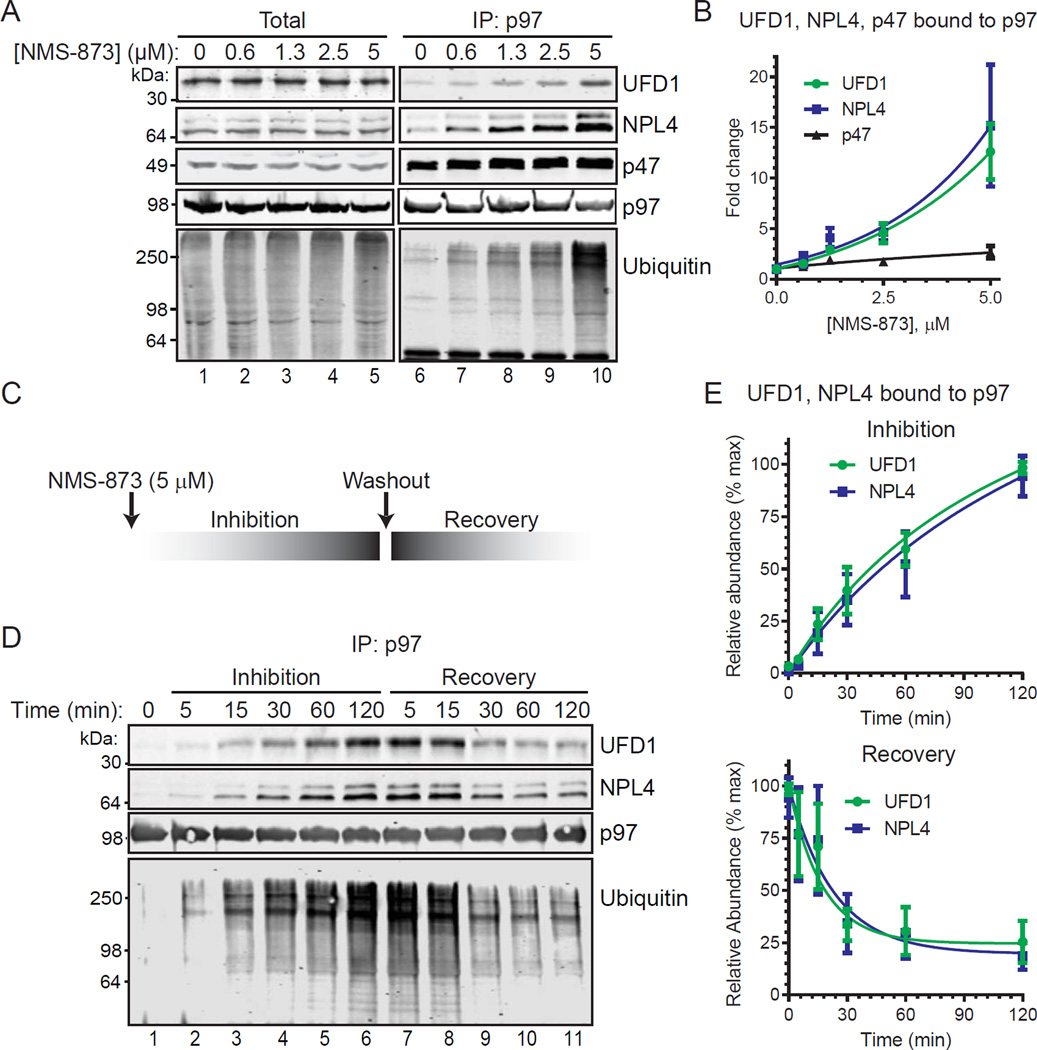
Cell extracts and purified p97 complexes from cells treated with increasing concentrations of NMS-873 were analyzed by immunoblotting (Figure 2A). Concentration-dependent increases in polyubiquitin were found in extracts and bound to p97. While the steady state abundance of cofactors in extracts did not change, UFD1 and NPL4 bound to p97 increased 12- to 15-fold in a concentration dependent manner while p47 increased only 2.5-fold (Figure 2B).
Figure 2. Binding of UFD1/NPL4 and polyubiquitin to p97 with NMS-873 is concentration dependent and reversible.
(A, B) HCT116 cells were treated with NMS-873 for 1 hour and cell extracts (Total) or purified p97 (IP:p97) were analyzed by IB. Mean fold changes (n=3, SD error) for UFD1, NPL4, and p47 with NMS-873 are shown in (B).
(C) Experimental scheme to measure time dependent UFD1/NPL4 binding to p97 with NMS-873 inhibition and recovery. After 120 minutes with 5 µM NMS-873, cells were washed with PBS prior to addition of media without NMS-873.
(D, E) Purified p97 complexes (5 µM NMS-873) were analyzed by IB. Mean change (n=3, SD error) in relative abundance of bound UFD1 and NPL4 are shown in (E). Steady state levels of proteins in cell extracts are in Figure S1.
The binding of UFD1, NPL4, and ubiquitin to p97 was evaluated in response to transient NMS-873 exposure (Figure 2C). Immunoblot analyses found time-dependent increases in the binding of UFD1, NPL4, and ubiquitin to p97 during inhibition that decreases after inhibitor washout (Figure 2D). Non-linear regression analyses of the binding of UFD1 and NPL4 to p97 revealed a half-time (t1/2) of 39 min and 46 min during inhibition (Figure 2E, top graph). Decreases in NMS-873 dependent binding of these proteins during the recovery phase were notably faster (t1/2, 13 min and 17 min, respectively), although basal binding was not fully restored after 2 hours (Figure 2E, bottom graph).
Increased UFD1/NPL4 binding is linked to increased polyubiquitin binding to p97
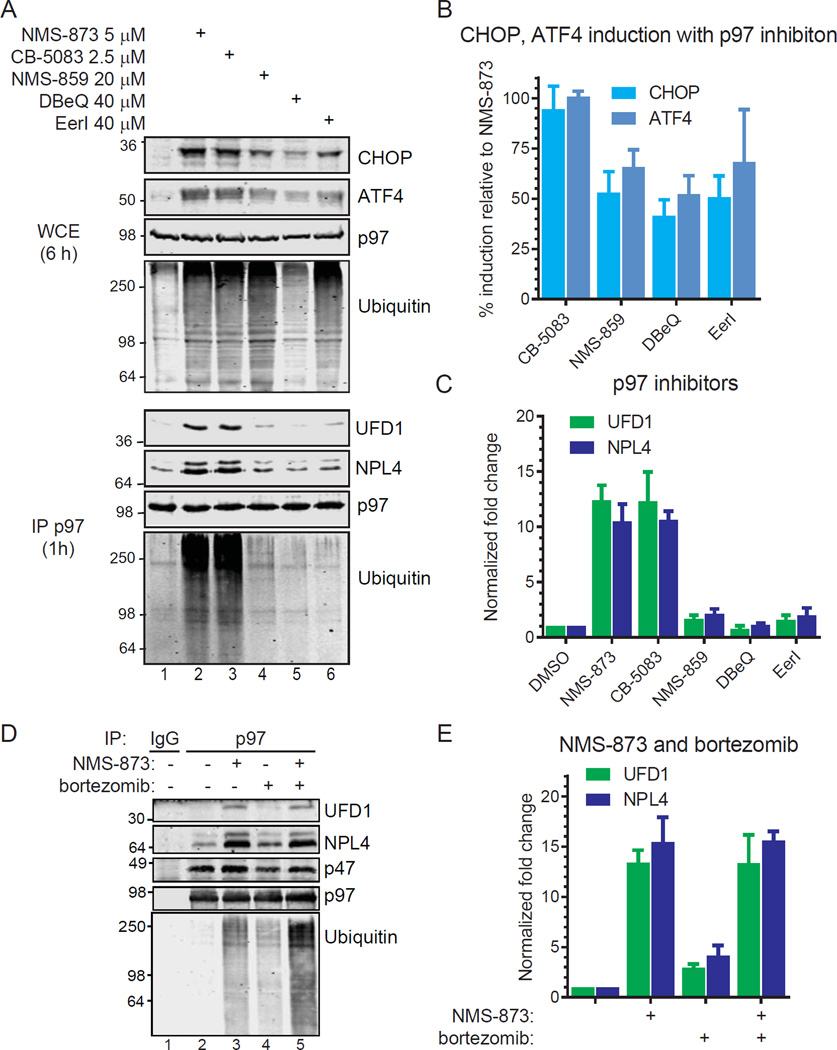
Since increased binding of UFD1 and NPL4 to p97 has not been previously linked to p97 inhibition, we sought to determine if this is specific to allosteric inhibition or a more general feature of p97 inhibition. We identified concentrations of p97 inhibitors that cause similar induction of CHOP and ATF4 (Figure 3A top panels, 3B), biomarkers of p97 inhibition (Magnaghi, et al., 2013). Although these all caused similar accumulation of polyubiquitin in cell extracts except DBeQ, the induction of CHOP and ATF4 suggest similar extents of p97 inhibition.
Figure 3. Enhanced UFD1/NPL4 and ubiquitin binding is a biomarker of p97 inhibition.
(A, B, C) HCT116 cells were treated with p97 inhibitors for 6 hours to analyze known biomarkers of p97 inhibition (top panels; CHOP, ATF4, ubiquitin) in cell extracts or 1 hour to analyze purified p97 for UFD1, NPL4, and ubiquitin (bottom panels). The % induction of CHOP and ATF4 relative to NMS-873 is in (B). The fold change for UFD1 and NPL4 bound relative to control p97 purifications is in (C). Data represent the mean (n=3, SD error).
(D, E) HCT116 cells were treated for 6 hours with NMS-873 (5 µM) and bortezomib (3 µM) prior to analyses. Steady state levels of proteins in cell extracts are in Figure S2. The amount of UFD1 and NPL4 bound to p97 were quantified (n=3, SD error).
We next tested if the inhibitors increase UFD1/NPL4 binding to p97 (Figure 3A bottom panels, 3C). While we observed slight increases in binding to p97 with NMS-859, DBeQ, and EerI (up to 2.1-fold), those caused by NMS-873 and CB-5083 were markedly greater (12-fold). These correlate to increased p97-bound polyubiquitin.
We examined p97 purified from cells treated with NMS-873 and the proteasome inhibitor bortezomib either as single agents or in combination (Figure 3D, E). While bortezomib caused a greater increase in steady state polyubiquitin conjugates in cells than NMS-873 (Figure S2), NMS-873 caused more bound to p97. The combination of both resulted in the amount of bound polyubiquitin greater than either inhibitor alone. If an increase in polyubiquitin bound to p97 is sufficient to cause increased UFD1/NPLF4 binding, the combination of NMS-873 and bortezomib should also result in more of this cofactor bound to p97 than NMS-873 alone. However, the binding of UFD1/NPL4 with both molecules together was similar to NMS-873 alone.
Cells with decreased sensitivity to NMS-873 contain a D2 domain mutation in p97
NMS-873 is cytotoxic to HCT116 cells and modulates biomarkers consistent with siRNA-mediated silencing of p97 (Magnaghi, et al., 2013). To study potential mechanisms of resistance to NMS-873 and how they affect cofactor interactions with p97, we grew HCT116 cells in the presence of 2 µM NMS-873 over 10 days. Although this resulted in almost complete cell death, foci ultimately developed consistent with cells acquiring resistance to NMS-873. Resistant clones were isolated and expanded.
In cell viability experiments, resistant cells (873-R) were greater than 15-fold less sensitive to NMS-873 than the parental cells (873-S, Figure 4A, Figure S3). Both remained similarly sensitive to other p97 inhibitors CB-5083, NMS-859, DBeQ, and EerI, the proteasome inhibitors bortezomib and carfilzomib, and ER stress inducers brefeldin A, DTT, and tunicamycin (Figure 4A, B, C, Figure S3). These data suggest that the resistance mechanism in these cells is unique to NMS-873.
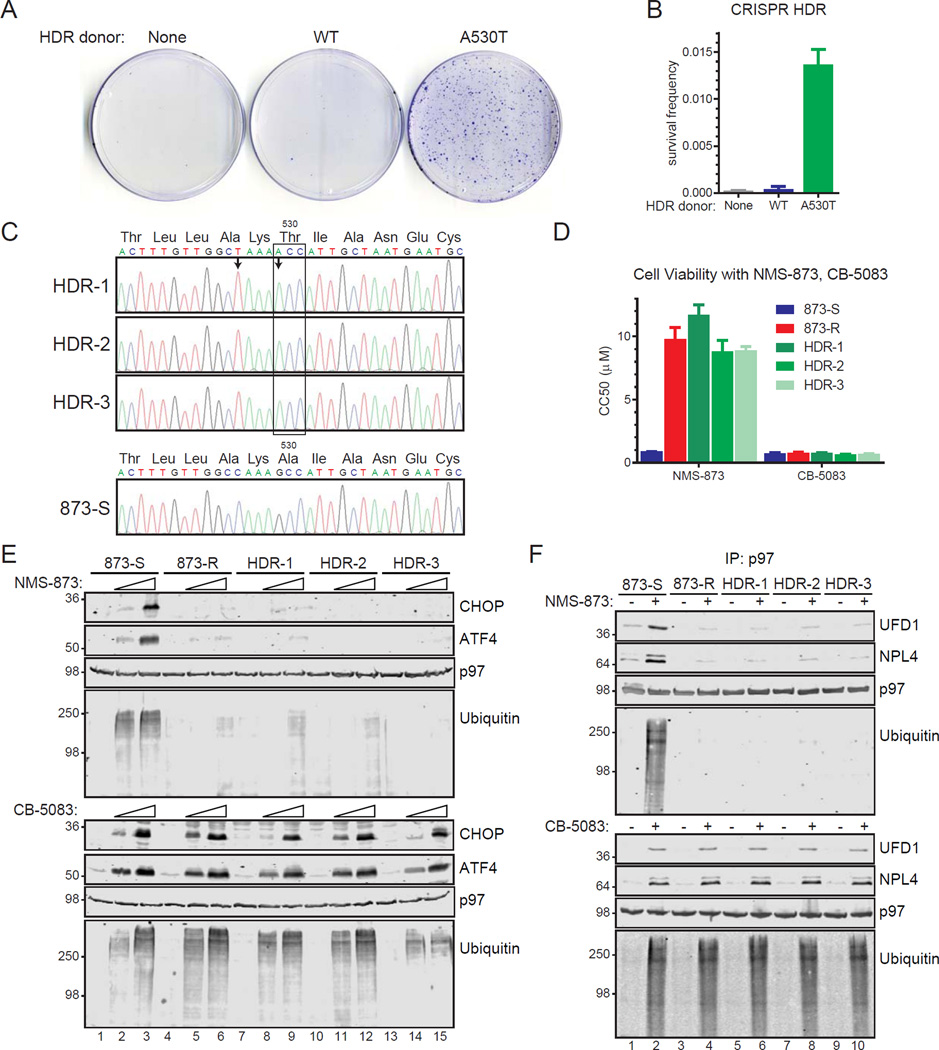
Figure 4. Cells insensitive to NMS-873 contain a D2 domain mutation.
(A, B, C) Summary of cell viability experiments testing p97 (A) and proteasome (B) inhibitors and ER stress inducers (C) on NMS-873-sensitive (873-S) and –resistant (873-R) cells. The CC50 values represent the mean (n=3 with standard error, SE) from non-linear regression analyses of data in Figure S3.
(D) Sequencing chromatograms of genomic DNA from 873-R show one allele of the p97 gene contains a G to A transition resulting in a missense mutation (top) not found in 873-S cells (bottom). Known p97 mutations associated with cancer (top) and multisystem proteinopathy type 1 disorders (MSP1, bottom) are shown below.
(E) 873-S and 873-R cells were treated with NMS-873 (2 and 5 µM) or CB-5083 (0.3 and 2.5 µM) for 6 hours and analyzed by IB for biomarkers of p97 inhibition (CHOP, ATF4, and ubiquitin).
(F, G, H) 873-R cells have attenuated cofactor and ubiquitin binding to p97 with NMS-873. A summary of cofactors identified by LC-MS/MS analyses of p97 purified from 873-S and 873-R cells (5 µM NMS-873, 6 hours) is in (F) with ubiquitin in (G). Expanded data sets (n=4, SD error) are in Supplemental Data File S2. IB experiments comparing cofactor and ubiquitin binding to purified p97 with NMS-873 and CB-5083 are in (H). Steady state levels of proteins in cell extracts are in Figure S3.
We identified a single heterozygous mutation in the p97 gene in 873-R cells (Figure 4D). This results in a missense mutation changing alanine 530 to threonine (A530T). Additional resistant cell lines (n=5) were heterozygous for the same mutation, suggesting it is the most prevalent in these conditions (Figure S3). A530 had not been previously identified as mutated in cancer or in multisystem proteinopathy type 1 (MSP1) disorders. We examined biomarkers of p97 inhibition by NMS-873 and CB-5083 on 873-S and 873-R cells (Figure 4E). Similar induction of CHOP, ATF4, and polyubiquitin was observed with CB-5083 treatment for both while 873-R had diminished responses to NMS-873.
We analyzed p97 composition from 873-R cells by mass spectrometry (see Supplemental Data File S2 for the full data set). We observed smaller increases in the binding of cofactors in response to NMS-873 for 873-R cells than 873-S (Figure 4F). These cells also had diminished increases in p97-bound ubiquitin (Figure 4G). These effects are specific for NMS-873 since 873-R cells had similar increases in UFD1/NPL4 and polyubiquitin binding to p97 with CB-5083 treatment as 873-S cells (Figure 4H).
A530T is sufficient to decrease cell sensitivity to NMS-873
We used homology-directed repair (HDR, see Experimental Procedures) to test if DNA repair donors corresponding to either wild-type (WT) or A530T sequences increase survival of HCT116 cells grown with 4 µM NMS-873 (Figure 5A, B). While similar survival frequencies were observed for cells transfected without a donor or WT sequences, those transfected with A530T had a greater than 30-fold increase.
Figure 5. A530T is sufficient to decrease cell sensitivity to NMS-873.
(A, B) HCT116 cells were co-transfected with a plasmid containing Cas9 and sgRNA targeting sequences adjacent to A530 in p97 and homology-directed repair (HDR) donor sequences. After 48 hours, NMS-873 (4 µM) was added and replenished every 72 hours for 6 days and then without NMS-873 for 5 days prior to crystal violet staining. Survival frequencies (n=3, SD error) are shown in B.
(C, D) Cell lines engineered by HDR to have A530T were identified by genomic DNA sequencing. Only cells homozygous for the mutation were obtained and contain the indicated silent mutation (C/T) designed to prevent Cas9-mediated cleavage. Cell lines were evaluated in viability experiments with NMS-873 and CB-5083 to obtain CC50 measurements (n=3, mean with SE) shown in (D). Dose response curves are in Figure S4.
(E, F) HDR cells homozygous for A530T have decreased induction of CHOP, ATF4, and polyubiquitin (E) and do not have increased binding of UFD1, NPL4, and polyubiquitin to p97 (F) with NMS-873 treatment while remaining sensitive to CB-5083. Cell treatments were for 6 hours.
We next isolated engineered A530T cell lines (Figure 5C). In contrast with 873-R cells, these cells are all homozygous for the mutation and contain a second designed silent mutation that disrupts the protospacer adjacent motif (PAM) used for Cas9-mediated cleavage of genomic DNA. In cell viability experiments (Figure 5D), they were 10-fold less sensitive to NMS-873 than the parental HCT116 cells, but remained sensitive to CB-5083. They also had diminished induction of CHOP, ATF4, and polyubiquitin with NMS-873 treatment (Figure 5E) and did not have NMS-873-dependent increases in the binding of UFD1, NPL4, and polyubiquitin (Figure 5F). However, their responses to CB-5083 were indistinguishable from 873-S cells.
A530T decreases NMS-873 sensitivity in vitro without affecting its binding
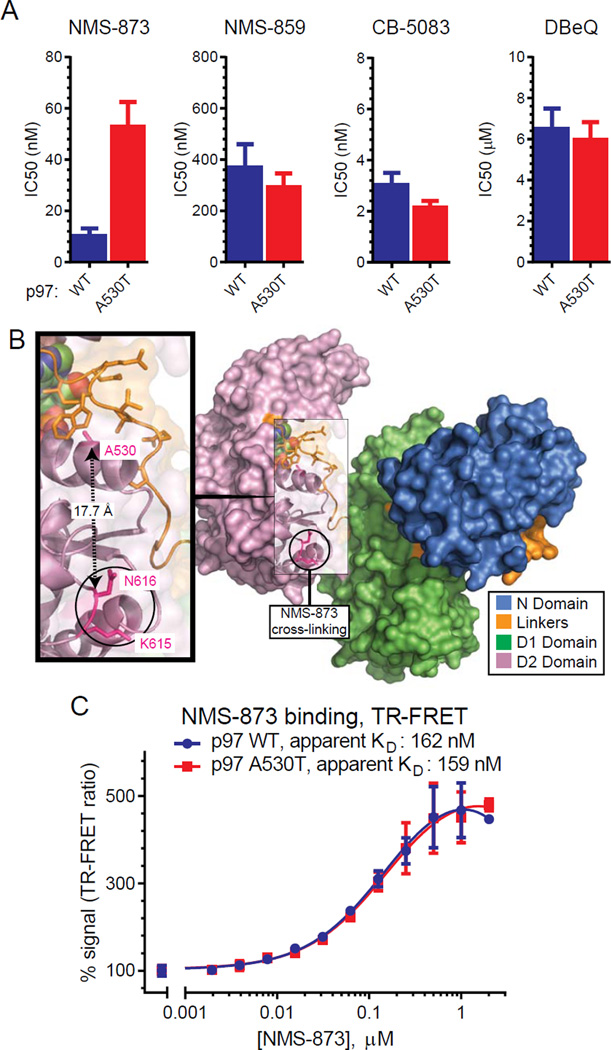
We generated recombinant p97 WT and A530T to directly evaluate the effects of inhibitors on ATPase activity in vitro (Figure 6A). As described in more detail below, we observed that catalytic efficiency of A530T was 9-fold higher than WT. We further found that NMS-873 was 4-fold less potent on A530T (IC50 52 nM, A530T; IC50 12 nM, WT). This difference is specific to NMS-873 since the potencies of other inhibitors were similar.
Figure 6. A530T decreases the potency of NMS-873 on p97 without affecting its binding.
(A) The potency of inhibitors on p97 activity was assessed in vitro. Dose response curves in Figure S5 were used to obtain IC50 values (n=3, mean with SE) shown.
(B) A model of a p97 monomer (PDB:3CF1) shows the positions of A530 relative to K615 and N616 which comprise the NMS-873 binding site. A530 is more than 17 Å from these residues and is buried through contacts with residues within the D1–D2 linker (orange).
(C) The apparent KD of NMS-873 to p97 was measured by TR-FRET between BODIPY-ATP and terbium-labeled anti-His tag antibody. The increase in signal is consistent with NMS-873 binding to p97. Data points represent the mean (n=4) with SD.
A530 is within an α-helix of the D2 ATPase domain (Figure 6B). This residue is more than 17 Å from the allosteric inhibitor binding site identified by UV cross-linking azido derivatives of NMS-873 to lysine 615 and asparagine 616 (Magnaghi, et al., 2013). A530 interacts with the linker between the D1 and D2 domains (“linker 2”) and is 10 Å from the nearest edge of the ATP binding pocket. Since A530 seems unlikely to directly participate in NMS-873 binding, the mutation could either indirectly affect NMS-873 binding and/or affect conformational changes necessary for activity inhibition.
To explore these possibilities, we used a time-resolved fluorescence resonance energy transfer (TR-FRET) assay to measure NMS-873 binding to p97. In NMS-873 titration experiments (Figure 6C), we observed similar concentration dependent increases in TR-FRET for both WT and A530T. Extrapolation of binding affinities (apparent Kd; WT: 162 nM; A530T: 159 nM) from non-linear regression analyses indicate that the A530T mutation does not affect NMS-873 binding.
A530T alters ATP/ADP binding and increases p97 catalytic efficiency
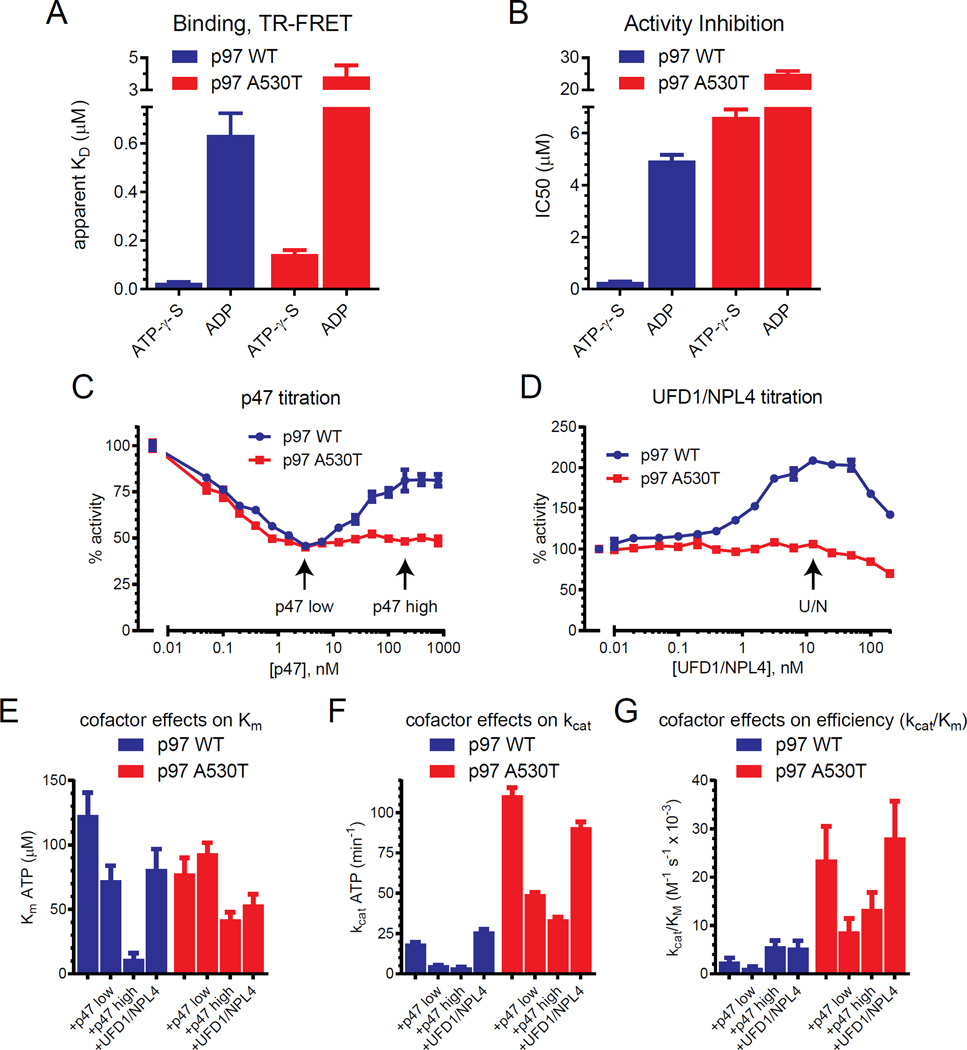
Since A530T does not affect NMS-873 binding, it could alter catalytic functions of the enzyme important for allosteric inhibition. We used TR-FRET to measure binding of ATP-γ-S (slowly hydrolyzed analog of ATP substrate) and ADP (Figure 7A). For both, we observed 5- to 6-fold increases in the apparent Kd for A530T (ATP-γ-S, WT: 25 nM, A530T: 140 nM; ADP, WT: 600 nM, A530T: 3.8 µM), suggesting that the mutation alters the D2 catalytic pocket to decrease ATP and ADP affinity. In examining the effect of these on ATPase activity (Figure 7B), we found A530T was markedly less sensitive to both (ATP-γ-S IC50, WT: 247 nM, A530T: 6.6 µM; ADP IC50, WT: 4.5 µM, A530T: 24.6 µM). While WT activity is 18-fold less sensitive to ADP than ATP-γ-S, A530T activity is only 3.7-fold less sensitive.
Figure 7. A530T increases p97 catalytic efficiency and affects ATP/ADP binding.
(A, B) The binding of ATP-γ-S and ADP to p97 (A) and their effects on ATPase activity (B) were evaluated. The apparent KD and IC50 measurements were obtained from dose-response experiments shown in Figure S6 (n=4, SD)
(C, D) The effect of p47 (C) or UFDl1/NPL4 (D) on p97 ATPase activity was measured. The concentrations of p47 (p47 low, 3 nM; p47 high, 200 nM) that had different effects only for WT are indicated as is UFD1/NPL4 that gave the largest increase in activity (U/N, 12.5 nM).
(E, F, G) Steady state kinetic analyses of p97 were performed with p47 or UFD1/NPL4. The effects of cofactors on Km (D), kcat (E), and kcat/Km (F) are based on Michaelis Menten curves shown in Figure S6. Bars represent SD (n=4).
We next evaluated the effects of p47 and UFD1/NPL4 on the enzymes’ ATPase activity. Consistent with recently published studies (Zhang, et al., 2015), we observed a biphasic response with respect to p47 concentration for WT (maximum inhibition at 3.1 nM with recovery at 200 nM (Figure 7C). While A530T had a similar inhibition phase, no recovery was observed at the highest p47 concentrations tested, similar to that observed for MSP1 variants containing R155H, L196W, or A232E (Zhang, et al., 2015). We found that increasing UFD1/NPL4 concentration caused up to a 2-fold increase in the ATPase activity of WT (Figure 7D). In contrast, A530T was largely insensitive over the concentration range tested. These data suggest UFD1/NPL4 is an activating cofactor for p97 similar to p37 (Zhang, et al., 2015).The loss of sensitivity to UFD1/NPL4 activation of A530T was similar to that observed for the MSP1 variants with p37.
We next measured the steady state kinetic constants of these enzymes with respect to ATP concentration. A530T had a lower KM (Michaelis Menten constant, ATP concentration for half maximal activity) than WT (Figure 7E, Figure S6). With respect to the maximum rate of ATP turnover (kcat) and catalytic efficiency (kcat/KM), these were all markedly higher for A530T than WT (Figure 7F, 7G).
Discussion
P97 inhibition has emerged as a promising anti-cancer strategy through the development of small molecule inhibitors with distinct mechanisms of action. Using the allosteric inhibitor NMS-873, we analyzed its effects on cellular p97 composition and examined the ability of cells to adapt to its cytotoxicity. Since cofactors and other interacting proteins have well appreciated roles in determining the diverse cellular functions of p97 (Meyer, et al., 2012), establishing how these may be affected by the enzyme’s inhibition is critical to understand associated biological responses and their contributions to its observed cytotoxicity on cancer cells. The identification and characterization of mechanisms that attenuate the cytotoxicity of p97 inhibitors such as NMS-873 are also important as they reveal unexpected vulnerabilities of these molecules and cellular changes that can occur in response to them to support the essential functions of p97.
Our observation that p97 inhibition causes an increase in the co-purification of the enzyme with a subset of its cofactors and polyubiquitin provides new insight into the cellular responses to these inhibitors. Conventional biomarkers of p97 inhibition such as the induction of polyubiquitin, CHOP, and ATF4 in cell extracts are indirect, can occur through numerous mechanisms independent of p97, and require prolonged treatment to detect. Changes to p97 composition are sensitive to inhibitor concentration and appreciably more rapid. These are readily detected with both NMS-873 and CB-5083 treatment, but are much more modest with NMS-859, DBeQ, and EerI. This could reflect the relatively poor potency and/or selectivity of these latter inhibitors as considerably higher concentrations of them are required to obtain similar extents of CHOP and ATF4 induction found with the more potent and selective inhibitors NMS-873 and CB-5083.
NMS-873 cell treatment increases the binding of the ubiquitin ligase AMFR (gp78 (Zhong, et al., 2004)), ubiquitin binding cofactors UFD1/NPL4 (Meyer, et al., 2000), FAF1 (Besche, et al., 2009), UBXD4 and UBXD8 (Alexandru, et al., 2008), and UBXD7 which binds NEDD8-modified cullins (den Besten, et al., 2012), and K48-linked polyubiquitin to p97. One plausible explanation for these observations is that p97 inhibition blocks the enzyme’s ability to process ubiquitinated substrates. This leads to an increase in bound cofactors involved in aspects of polyubiquitin recognition and modification. Another possibility is that p97 inhibition traps the enzyme in a conformation that causes increased binding or prevents the dissociation of these cofactors. Since the cofactors all appear involved in aspects of ubiquitination, their increased binding could cause a corresponding increase in bound polyubiquitin. Although we currently cannot distinguish between these possibilities (and they are not necessarily mutually exclusive), it is clear from our data that there is a direct relationship between increased UFD1/NPL4 and polyubiquitin binding to p97 with NMS-873 as they have similar concentration- and time-dependent increases.
The validity of changes in the composition of p97 as an accurate and specific read-out of cellular p97 inhibition is supported by our characterization of 873-R cells. These cells are more than 10-fold less sensitive to NMS-873 than the parental HCT116 cells and have attenuated induction of CHOP, ATF4, and polyubiquitin in response to the molecule. The underlying changes in the resistant cells are specifically attributable to overcoming NMS-873 cytotoxicity since they remain similarly sensitive to other p97 inhibitors and have similar concentration dependent increases in CHOP, ATF4, and polyubiquitin with CB-5083. While the composition of p97 complexes purified from 873-R cells appears largely unaffected by NMS-873, inhibitor-dependent increases in polyubiquitin and UFD1/NPL4 binding still occur with CB-5083. This suggests that while subpopulations of cells could have intrinsically lower levels of cofactor binding to p97 that may be fortuitously enriched upon the clonal isolation of 873-R cells, this is unlikely responsible for reduced cofactor binding found in 873-R cells with NMS-873.
A530T is a newly discovered, heterozygous mutation in p97 that appears to be the most prevalent in HCT116 cells surviving cytotoxic concentrations of NMS-873. This residue is within the D2 ATPase domain and has not been previously associated with cancer, MSP1 disorders, or responses to p97 inhibition. Cell lines engineered to be homozygous for A530T respond similarly to NMS-873 as heterozygous 873-R cells, suggesting that a single mutated allele provides sufficient p97 function to overcome NMS-873 cytotoxicity. This is in contrast with recently described homozygous mutations found in cells rendered resistant to CB-5083 (Anderson, et al., 2015) and could reflect the different mechanisms of action of the inhibitors on p97 and how treatment emergent mutations affect enzyme function.
Molecular models indicate that A530 is within an α helix in the D2 domain close to the ATP binding site and interacts with other hydrophobic residues (V474 and I479) at the linker between D1 (see Figure 6B). This residue is largely buried in available structures and is more than 17 Å from residues critical for NMS-873 binding (K615 and N616) on a different surface of the p97 monomer (Magnaghi, et al., 2013). The mutation of A530 to threonine decreases the potency of NMS-873 through a mechanism that does not affect inhibitor binding to p97. Instead, it alters the enzyme’s affinity for both its substrate ATP and ADP product. This is associated with an increase in the enzyme’s catalytic efficiency and altered sensitivity to cofactor-associated changes in ATP hydrolysis found with p97. Although the physiological relevance of cofactor effects on p97 activity is currently unclear (Zhang, et al., 2015), these may reflect conformational changes in the enzyme required for cofactor interactions and could represent a mechanism involved in substrate recognition and processing.
Recent structural studies on p97 have provided new insight into how conformational changes are driven by nucleotide binding and how they may be affected by an allosteric inhibitor known as UPCDC30245 (Banerjee, et al., 2016). This work suggests that the sequential binding of ATP to the D2 domain and then D1 are involved in cooperative conformational changes required for enzyme function. UPCDC30245 binding has been proposed to block conformational changes associated with ATP binding to D2, thereby preventing subsequent ATP binding to D1 and the extended positioning of the N-domain. Although the binding site of UBCDC30245 is distinct from that for NMS-873 as it does not appear to involve K615 and N616 (Banerjee, et al., 2016; Magnaghi, et al., 2013), it is likely that A530T allows for these sequential conformational changes to occur even with NMS-873 binding. The proximity of A530 to the D2 nucleotide binding site suggests that observed decreases in nucleotide binding to A530T are most likely attributable to this domain, although it is plausible that it could also have an effect on nucleotide binding to D1. Future mechanistic studies examining the relevance of this newly described model to cofactor binding and the precise contributions of D1 and D2 to the increased catalytic efficiency of p97 A530T should be able to address these important issues.
Collectively, our study provides new insight into the effects of p97 inhibition and the suppression of the cytotoxicity of an allosteric inhibitor by a mutation in the enzyme that does not affect its binding. These will facilitate further investigations into the molecular basis of the responses of cancers to p97 inhibition and help guide the development of improved inhibitors to treat the disease.
Methods
Cell lines
HCT116 cells were purchased from the American Type Culture Condition (ATCC). These were grown in McCoy’s 5A with 10% fetal bovine serum. Resistant cells (873-R), isolated as described below, were maintained in 2 µM NMS-873, replenishing inhibitor-containing media every 72 hours.
873-R cells were isolated by culturing HCT116 cells (3.6×105 cells per 10 cm2 plate) with 2 µM NMS-873 for 10 days. Emergent colonies were isolated and expanded. The mutation for A530T in p97 was identified by sequencing cDNAs and confirmed by sequencing genomic DNA. Five cell lines with the same mutation were identified.
Homology-directed repair (HDR) was used to assess the potential of A530T to support cell survival in the presence of NMS-873 and generate cell lines engineered with A530T. Cells were co-transfected with plasmid PX458, a gift from Feng Zhang (Ran, et al., 2013), containing a sgRNA targeting the antisense strand of 35,060,422 to 35,060,403 of the p97 gene and PCR-generated repair donor sequences of 35,060,791 to 35,060,042 containing sequences for either A530 or the A530T mutation. The donors also contained a C/T silent mutation to alter the PAM used by the CRISPR/Cas9 system shown in Figure 5E. Full sequences will be provided upon request. Transfected cells were cultured for 6 days in the presence of 4 µM NMS-873 and then 5 days in its absence prior to crystal violet staining. The survival frequencies were measured by particle counting scanned plates using ImageJ and dividing the number of foci by the total input cells. To isolate engineered A530T cells without NMS-873 treatment, individual GFP positive cells were isolated and expanded, and clones containing the A530T and PAM mutations were identified by DNA sequencing genomic DNA. Only cells homozygous for the engineered mutations were identified.
Short-tandem repeat (STR) genotyping was performed using the PowerPlex 16 System (Promega) on 873-S and 873-R cells. These analyses resulted in 97% and 90% threshold scores respectively relative to existing HCT116 STR profiles provided by the Children’s Oncology Group Cell Culture and Xenograft Repository (www.cog.cell.org).
Reagents and antibodies
NMS-873 and NMS-859 were purchased from Xcessbio, CB-5083 was purchased from Active Biochem, DBeQ and Eeyarestatin I (EerI) were purchased from EMD Millipore, bortezomib and carfilzomib were purchased from LC Laboratories, brefeldin A and tunicamycin were purchased from Sigma, and DTT from AMRESCO. The purity of all compounds was greater than 95% as judged by liquid chromatography/mass spectrometry.
Antibodies were from commercial sources: anti-p97 (sc-57492) and anti-ubiquitin (sc-8017) from Santa Cruz Biotechnology, anti-UFD1 (#13789), anti-NPL4 (#23489), anti-FAF1 (#4932), anti-AMFR (#9590), anti-CHOP (#2895), and anti-ATF4 (#11815) from Cell Signaling, anti-p47 (ab95963) from Abcam, and anti-UBXD8 (SAB4200475) from Sigma.
Purification of p97 and mass spectrometry analyses
HCT116 cells (873-S or 873-R) were lysed in 25 mM HEPES pH 7.6, 300 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 5 mM MgCl2, 1 mM DTT, and protease inhibitor cocktail. After clarification, extracts were incubated with 2 µg of p97 antibody for 1 hour at 4°C followed by 1 hour w ith 20 µl of protein A/G Ultra Resin (Thermo Scientific). After washing with lysis buffer, proteins were eluted in sample buffer. Samples were analyzed by immunoblotting or LC-MS/MS.
LC-MS/MS was performed as described (Brill, et al., 2009). For statistical analyses, 4 replicates (2 biological replicates, 2 technical replicates) were analyzed. Protein spectral counts were normalized to protein molecular weight (Yang, et al., 2014), followed by p97 normalization. Total spectrum count intensities were averaged and identified proteins were ranked by fold-change and total counts.
Size exclusion chromatography
Extracts (500 µl, 10 mg/ml) from HCT116 cells (untreated or with 2 mM NMS-873 for 6 hours) were prepared in 25 mM HEPES pH7.6, 150 mM NaCl, 0.2% Triton X-100, 1 mM EDTA, 10 mM MgCl2, 1 mM DTT, and protease inhibitor cocktail and were separated on a Superose 6 10/300 GL column (GE Healthcare) calibrated with a protein standard kit (Mr 29,000 to 700,000, Sigma) in the same buffer lacking detergent. Fractions (500 µl) were collected from 29,000 to the column void volume.
Cell viability measurements
Cells were seeded at 2000 (72 hour inhibitor treatment: NMS-859, NMS-873) or 4000 (48 hour inhibitor treatment: DBeQ, EerI, bortezomib, carfilzomib, DTT, brefeldin A, tunicamycin) cells per well of a 96-well plate. After 24 hours, inhibitors were added (3-fold serial dilutions from 20 µM for NMS-859, NMS-873, CB-5083, DBeQ, EerI, amd DTT, 2.22 µM for tunicamycin and brefeldin A, and 0.5 µM for bortezomib and carfilzomib in 1% DMSO final. HDR cell lines testing NMS-873 used 2-fold serial dilutions from 20 µM. Cell Titer-Glo (Promega) was used to assess cell viability. Percent viability was determined by dividing resulting values by the largest measurement (highest amount of ATP, typically DMSO or lowest inhibitor concentration. Data were analyzed in GraphPad Prism using the equation Y=Bottom + (Top-Bottom)/(1+10^((LogCC50-X)*HillSlope)) and constraining the range from 0 (Bottom) to 100% (Top) to obtain CC50 values. Experiments were performed in triplicate with S.D. error.
Recombinant proteins
P97 (wild-type and A530T), UFD1, NPL4, and p47 were expressed using baculovirus expression in High Five insect cells (Thermo Fisher). Open-reading frames were PCR amplified from the MegaMan Transcriptome library (Agilent) and cloned into pFastBac1 (Thermo Fisher) to express proteins with a C-terminal hexahistidine tag (p97), N-terminal hexahistidine tag (NPL4, p47) or untagged (UFD1). The mutation for A530T was introduced by QuikChange (Agilent). UFD1/NPL4 was generated by co-infection. All proteins were purified by Ni-NTA (Qiagen) chromatography and desalted into 25 mM HEPES pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 10% glycerol. A530T and wild-type forms equivalent amounts of hexamer as assessed by native gel.
ATPase activity assays
Assays were performed in 50 mM HEPES pH 7.5, 10 mM MgCl2, 2 mM DTT, 0.2% BSA, and 0.005% Tween 20. Compounds were dispensed into 1536-well pates for 2-fold serial dilutions, followed by 18 nM WT p97 monomer, 1.98 nM A530T p97 monomer, or no-enzyme control. After 30 minute pre-incubation at 25°C, 40 µM ATP was added to initiate the 75 minute reaction at 25°C. The reaction was q uenched with PiColorLock reagent, and read on a PHERAstar FS at OD620. For cofactor titration studies, increasing concentrations of UFD1/NPL4 or p47 were mixed with 40 µM ATP, followed by p97. For kinetic characterization studies, increasing concentrations of ATP were dispensed into the plate, followed by 12.5 nM UFD1/NLP4, 3.1 nM p47, 200 nM p47, or buffer, and 21 nM wild-type p97 monomer or 2.35 nM A530T p97 monomer for 120 minutes.
TR-FRET Binding Assays
Assays were performed in 25 mM HEPES pH 7.5, 1 mM MgCl2, 100 mM NaCl, 0.5 mM TCEP, and 0.005% Tween-20. NMS-873 (2 µM maximum, 2-fold serial dilutions or ATP-γ-S and ADP (200 µM maximum, 2-fold serial dilutions) were mixed with 9 nM p97 monomer and 1 nM terbium-labeled anti-His antibody followed by the addition of 40 nM BODIPY-FL-ATP. After 1 hour at 25°C, plates were re ad on a PHERAstar FS using LANTHASCREEN. Reactions without p97 were used as a background control.
Supplementary Material
Significance.
This study provides new insight into the effects of targeted small molecule inhibitors on the interactions between p97, its cofactors, and polyubiquitin. It also describes an adaptive mechanism involving a newly discovered mutation in p97 (A530T) that changes enzyme function to specifically decrease sensitivity to an allosteric inhibitor without affecting its binding. Our data are consistent with A530T affecting conformational changes in p97 important for allosteric inhibition by increasing the enzyme’s catalytic efficiency and altering the enzyme’s affinity for both ATP and ADP. Since p97 has emerged as a promising cancer therapeutic target, our work provides a mechanistic understanding of how targeted molecules affect the enzyme and can be overcome by adaptive changes. This will facilitate further investigations into the molecular basis of the responses of cancers to p97 inhibition and help guide the development of improved inhibitors to treat the disease.
Highlights.
Allosteric inhibition by NMS-873 changes p97-cofactor interactions
Cofactor and polyubiquitin binding to inhibited p97 are linked
The cytotoxicity of NMS-873 is suppressed by a p97 mutation, A530T
A530T changes p97 ATPase activity without affecting NMS-873 binding
Acknowledgments
We thank Brian James for STR genotyping, Tony Pinkerton for sourcing and evaluating compounds, and Dieter Wolf for helpful discussions. This work was supported in part by the National Cancer Institute of the National Institutes of Health (R01 CA180150, R21 CA182178, R01 CA185300, and Cancer Center Support Grant P30 CA030199), and the American Cancer Society (RSG-11-224-01-DMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, N.H., J.I.T., E.S., and M.D.P.; Methodology, J.I.T., N.H., Y.W., C.M., K.M., E.S.; Investigation, N.H., J.I.T., C.M., Y.W.; Writing – Original Draft, Review & Editing, M.D.P.; Resources, J.I.T., Y.W., M.D.P., Supervision, M.D.P.; Funding Acquisition, M.D.P.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, et al. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016 doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Motamedchaboki K, Wu S, Wolf DA. Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods. 2009;48:311–319. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug M, Meyer H. Expanding into new markets--VCP/p97 in endocytosis and autophagy. J Struct Biol. 2012;179:78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol. 2012;19:511–516. S511. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R, Mueller B, Ploegh HL, Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppsteck P, Ewens CA, Forster A, Zhang X, Freemont PS. Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim Biophys Acta. 2012;1823:125–129. doi: 10.1016/j.bbamcr.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Laser H, Conforti L, Morreale G, Mack TG, Heyer M, Haley JE, Wishart TM, Beirowski B, Walker SA, Haase G, et al. The slow Wallerian degeneration protein, WldS, binds directly to VCP/p97 and partially redistributes it within the nucleus. Mol Biol Cell. 2006;17:1075–1084. doi: 10.1091/mbc.E05-04-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P, D'Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H. p97 complexes as signal integration hubs. BMC Biol. 2012;10:48. doi: 10.1186/1741-7007-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J Biol Chem. 2012;287:8561–8570. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter-Nilsson M, Hendriks R, Pecheur-Huet EI, Hoekstra D, Nilsson T. Cytosolic ATPases, p97 and NSF, are sufficient to mediate rapid membrane fusion. EMBO J. 1999;18:2074–2083. doi: 10.1093/emboj/18.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- Schaeffer V, Akutsu M, Olma MH, Gomes LC, Kawasaki M, Dikic I. Binding of OTULIN to the PUB domain of HOIP controls NF-kappaB signaling. Mol Cell. 2014;54:349–361. doi: 10.1016/j.molcel.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Yang CC, Chung A, Ku CY, Brill LM, Williams R, Wolf DA. Systems analysis of the prostate tumor suppressor NKX3.1 supports roles in DNA repair and luminal cell differentiation. F1000Res. 2014;3:115. doi: 10.12688/f1000research.3818.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JL, Flick K, Papagiannis CV, Mathur R, Tyrrell A, Ouni I, Kaake RM, Huang L, Kaiser P. Signal-induced disassembly of the SCF ubiquitin ligase complex by Cdc48/p97. Mol Cell. 2012;48:288–297. doi: 10.1016/j.molcel.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gui L, Zhang X, Bulfer SL, Sanghez V, Wong DE, Lee Y, Lehmann L, Lee JS, Shih PY, et al. Altered cofactor regulation with disease-associated p97/VCP mutations. Proc Natl Acad Sci U S A. 2015;112:E1705–E1714. doi: 10.1073/pnas.1418820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- Zhong X, Shen Y, Ballar P, Apostolou A, Agami R, Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J Biol Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
- Zhou HJ, Wang J, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, et al. Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083) J Med Chem. 2015;58:9480–9497. doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.