Abstract
Background
Pine wilt disease (PWD) is a worldwide threat to pine forests, and is caused by the pine wood nematode (PWN) Bursaphelenchus xylophilus. Bacteria are known to be associated with PWN and may have an important role in PWD. Serratia sp. LCN16 is a PWN-associated bacterium, highly resistant to oxidative stress in vitro, and which beneficially contributes to the PWN survival under these conditions. Oxidative stress is generated as a part of the basal defense mechanism used by plants to combat pathogenic invasion. Here, we studied the biology of Serratia sp. LCN16 through genome analyses, and further investigated, using reverse genetics, the role of two genes directly involved in the neutralization of H2O2, namely the H2O2 transcriptional factor oxyR; and the H2O2-targeting enzyme, catalase katA.
Results
Serratia sp. LCN16 is phylogenetically most closely related to the phytosphere group of Serratia, which includes S. proteamaculans, S. grimessi and S. liquefaciens. Likewise, Serratia sp. LCN16 shares many features with endophytes (plant-associated bacteria), such as genes coding for plant polymer degrading enzymes, iron uptake/transport, siderophore and phytohormone synthesis, aromatic compound degradation and detoxification enzymes. OxyR and KatA are directly involved in the high tolerance to H2O2 of Serratia sp. LCN16. Under oxidative stress, Serratia sp. LCN16 expresses katA independently of OxyR in contrast with katG which is under positive regulation of OxyR. Serratia sp. LCN16 mutants for oxyR (oxyR::int(614)) and katA (katA::int(808)) were sensitive to H2O2 in relation with wild-type, and both failed to protect the PWN from H2O2-stress exposure. Moreover, both mutants showed different phenotypes in terms of biofilm production and swimming/swarming behaviors.
Conclusions
This study provides new insights into the biology of PWN-associated bacteria Serratia sp. LCN16 and its extreme resistance to oxidative stress conditions, encouraging further research on the potential role of this bacterium in interaction with PWN in planta environment.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2626-1) contains supplementary material, which is available to authorized users.
Keywords: Bursaphelenchus xylophilus, Catalase, Endophyte, Reactive oxygen species, OxyR, Serratia, Oxidative stress, Pine wilt disease, Plant defenses
Background
The prevalence of pine wilt disease (PWD) in European and Asian forestlands causes significant environmental and economical effects, which have encouraged vulnerable countries to strengthen pest control management policies [1, 2]. The primary pathogenic agent of PWD is the plant-parasitic nematode Bursaphelenchus xylophilus (pine wood nematode, PWN) [3, 4]. PWN infects coniferous trees, mostly Pinus sp., using an insect-vector, Monochamus sp., for tree-to-tree transmission [5]. In the last decade, the parasitism of B. xylophilus has been intensively investigated [6–10]. In 2011, Kikuchi et al. [11] published a draft genome sequence for PWN revealing its distinct and unique parasitism tools including enzymes for metabolism of the host cell wall and detoxification enzymes. Shinya and co-workers [12] investigated the PWN secretome and identified a range of secreted cell-wall degrading enzymes and host-defense evasion proteins, among which 12 antioxidant enzymes (PRX, peroxiredoxin; CAT, catalase; GPX, glutathione peroxidase; nucleoredoxin-like proteins; SOD, superoxide dismutase; TRX, thioredoxin) were identified. More recently, Vicente et al. [13] showed the importance of PWN catalases in H2O2 detoxification in vitro, and Espada et al. [14] identified novel proteins involved in the host-parasite interaction and provided clear evidence that PWN employs a multilayered detoxification strategy to overcome plant defenses.
PWN-associated bacteria have been suggested to play an important role in the development of PWD (detailed review in Nascimento et al. [15]). A dual role has been attributed to these bacteria due to their phenotypic plasticity, expressing both plant pathogenic and plant growth promoting abilities [16]. These nematode-associated bacteria were initially seen as putative PWN’s symbiotic partners in PWD [17–19], though lately Paiva et al. [20] has shown also in vitro nematicidal activity of some associated bacteria. In spite of the intricate detoxification system present in PWN, Cheng et al. [21] and Vicente et al. [22] have shown the potential of PWN-associated bacteria, respectively, in the xenobiotic degradation and in the neutralization of H2O2.
Reactive oxygen species (ROS) have important roles in plant physiological processes such as growth and development, response to biotic and abiotic stresses and programmed cell death [23]. In host-pathogen interactions, apoplastic ROS production, also known as the oxidative burst, is one of the earliest detectable events in plant basal defenses [24]. This production is biphasic: the first phase is non-specific, relatively weak and occurs within minutes of the plant detecting a potential pathogen while the second occurs after prolonged pathogen attack, resulting in establishment of plant defenses and may be accompanied by a hypersensitive response [25]. H2O2, hydrogen peroxide, is the most stable, and membrane diffusible ROS [24]. H2O2 has a variety of roles in plants; at low concentrations it serves as a signaling molecule for the plant (e.g., in defense gene activation) but, at high concentrations, can lead to oxidative stress and cell death [26]. Avirulent pathogens induce biphasic ROS. However, in the case of virulent pathogens or symbiotic partners, which can avoid or suppress host recognition, only the first ROS wave is detected [27]. In these situations, both plant and pathogen attempt to regulate intracellular and extracellular ROS accumulation by employing several enzymatic and non-enzymatic antioxidants, such as: ascorbate peroxidases, GPXs, SODs, CAT or KAT, PRXs and glutathione S-transferases (GSTs) [28].
Vicente et al. [22] reported, for the first time, the high tolerance to oxidative stress of three PWN-associated bacteria (Serratia sp. LCN4, Serratia sp. LCN16, and Serratia marcescens PWN146), showing also the beneficial effect towards PWN under the same conditions. In the present work, we investigated the biology of Serratia sp. LCN16 through genome analyses, and further studied, using reverse genetics, the role of two genes directly involved in the neutralization of H2O2, namely the H2O2 transcriptional factor OxyR; and the H2O2 targeting enzyme, catalase (katA, hydroperoxidase II, HPII).
Results
Genome structure and general features
The draft genome of Serratia sp. LCN16 suggests a single chromosome of 5.09Mbp in size with an average GC content of 52.83 % (Fig. 1). Genome annotation predicts 4804 genes, of which 4708 were predicted protein coding sequences (CDS) and 96 were RNA genes (14 rRNA, 81 tRNA, and 1 tmRNA). Of the 4708 CDS, 4528 (96 %) were assigned InterPro entries, and 3413 (72 %) were assigned to Gene Ontology (GO) terms (Additional file 1: Table S1). Few mobile genetic elements (MGE) have been found to date: one transposase (LCN16_00783) and one transposon TN10 (LCN16_02368), and 35 putative phage sequences. No clustered regularly interspaced short palindromic repeats (CRISPR) were predicted in the Serratia sp. LCN16 genome.
Fig. 1.
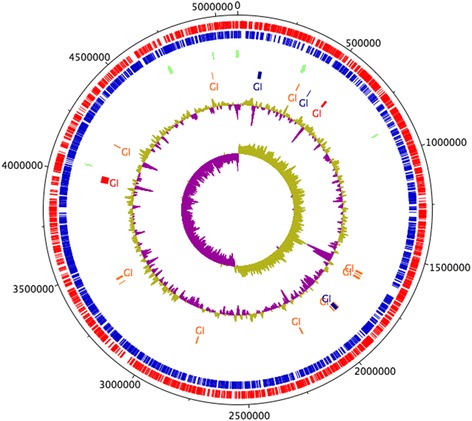
Circular representation of Serratia sp. LCN16. From the inner- to the outermost circle: circle 1, GC skew (positive GC skew in green and negative GC skew in purple); circle 2, GC plot; circle 3, predicted unique genomic regions of LCN16 known as genomic Islands (GI) [31]; circle 4, tRNA; circle 5, antisense strand (blue); and circle 6, sense strands (red). GIs in blue indicate prediction by IslandPath-DIMOB. GIs in orange indicate prediction following SIGI-HMM approach. GIs in red were predicted by both approaches
A phylogenetic analysis of Serratia sp. LCN16 based on the 16S rRNA gene and four housekeeping genes (rpoB, gyrB, dnaJ and atpD) is shown in Fig. 2. Two copies of the 16S rRNA gene (LCN16_00312 and LCN16_04450), sharing 99.3 % similarity, were found in the Serratia sp. LCN16 genome. Both copies clustered within the phytosphere Serratia complex (S. proteamaculans, S. grimessi and S. liquefaciens) [29, 30], grouping with S. proteamaculans 568 (99 % bootstrap support), and S. liquefaciens ATCC 27592 and S. grimessii AJ233430 (72 % bootstrap). Furthermore, the phylogeny based on the housekeeping genes reinforces the clustering of Serratia sp. LCN16 with S. proteamaculans 568 (99 % bootstrap support) within S. liquefaciens ATCC 27592 clade (100 % bootstrap support). Based on these observations, the Serratia sp. LCN16 genome was aligned to the genomes of S. proteamaculans 568 and S. liquefaciens ATCC 27592. The three genomes are highly syntenic with few rearrangements (Additional file 2: Figure S1). Serratia sp. LCN16 and S. liquefaciens ATCC 27592 share a unique region of similarity which is not present in S. proteamaculans 568, and which encode hypothetical and phage proteins, an MsgA protein (DNA-damage-inducible protein I, DinI) and a peptidase P60 (a bacterial cell wall-degrading enzyme) (Additional file 2: Figure S1). A total of 19 genomic islands (GI) were identified by at least one of SIGI-HMM or IslandPath-DIMOB in IslandViewer server [31] (Fig. 1, Additional file 3: Table S2). The 8 GIs predicted by both methods [32] have a size range between 4 kb and 39 kb, and are rich in hypothetical proteins, phage elements and ABC transporters (i.e., lipopolyssacharides). Genes potentially involved in the synthesis of antimicrobials, such as prnABD for the biosynthesis of the antifungal antibiotic pyrrolnitrin (LCN16_00326-27; LCN16_00329), kanB (LCN16_03985) for kanamycin biosynthesis, and mdtL (LCN16_03987) which is involved in chloramphenicol resistance (Additional file 3: Table S2) are also present.
Fig. 2.

Phylogenetic relationships between Serratia sp. LCN16 and other Serratia representatives. Green diamonds indicate the phytosphere Serratia complex. a Phylogeny based on 16S rRNA gene (1411 bp). b Phylogeny based on housekeeping genes (rpoB, gyrB, dnaJ and atpD) (8808 bp). Numbers above the clades are bootstrap values (1,000 replicates). Maximum likelihood (ML) trees were constructed using: (A) GTR + G + I, generalized time-reversible model with gamma distribution and proportion of invariable sites; and (B) K2 + G, Kimura 2-parameters with gamma distribution. Model determination and construction of ML trees were performed in MEGA 6 [67]
Serratia sp. LCN16, a putative plant-associated bacterium
The phylogenetic analysis places Serratia sp. LCN16 within a phytosphere group of Serratia (Fig. 2) [29, 30]. Thus, to understand if Serratia sp. LCN16 is able to live in a plant-environment, a set of 40 genes predicted to be important for endophytic behavior of 11 plant-associated bacteria [33] was searched for in the genome (Table 1). Serratia sp. LCN16 has 34 of these genes (85 %), S. proteamaculans 568 has 37 genes (90 %) while S. liquefaciens ATCC 27592 has 33 genes (83 %). Other sequences were found in Serratia sp. LCN16, supporting the idea that they have the ability to colonize plants (Additional file 4: Table S3) [34, 35]. These include genes encoding proteins for plant polymer degradation, such as glycoside hydrolases (endoglucanase, LCN16_00161) and pectin (galacturonate) degradation (uxaA and uxaC, LCN16_04263-64), fungal chitin degradation as chitinases (LCN16_00148, LCN16_01150, LCN16_02767, LCN16_03549), complete pathways for degradation of aromatic compounds (KEGG: ko00362), such as benzoate or catechol degradation; and genes involved in the synthesis of plant growth regulating compounds, such as indole-pyruvate decarboxylase (ipdC, LCN16_00911 and LCN16_03478) for indole acetic acid (IAA); genes for acetoin and 2,3-butanediol production via budABC (LCN16_03505-6, LCN16_02073), and genes for polyamine synthesis (plant volatiles, putrescine and spermidine). Additionally, common among plant-associated bacteria is the ability for iron acquisition via siderophore synthesis or by iron uptake transporters and siderophore receptors [36]. Serratia sp. LCN16 encodes the complete pathway for siderophore biosynthesis (entABEC, LCN16_03491-94; entF, LCN16_00890 and LCN16_03500; menF, LCN16_03345; and pchB, LCN16_03570), and iron uptake and transport systems (fhuDBC, efeBOU, and feoABC), including 24 genes in the iron complex transporter system (Additional file 4: Table S3). The main secretion systems (TSS) in Serratia sp. LCN16 are TSS1 (tolC, LCN16_04212; hasDEF, LCN16_01553-5) and TSS2, universal Sec-dependent (secretion) and Tat-independent (two-arginine translocation) proteins export systems, from which bacterial Type II toxins (membrane damaging) such as hemolysins (hpmA, LCN16_04426; hlyIII, LCN16_04002; tlh, LCN16_04200), phospholipase C (plcC, LCN16_04159), and serralysins (LCN16_00223; LCN16_02109; LCN16_03865) are secreted. In addition to the antimicrobial/antibiotic metabolism genes found in genomic islands described above, the Serratia sp. LCN16 genome encodes genes that could be involved in hydrogen cyanide synthesis (hcnABC, LCN16_01840-2), and complete gene sets for drug resistance such as beta-lactam (ampC) or macrolide (MacAB-TolC transporter), and multiple antibiotic resistance proteins (marC, LCN16_02155) (Additional file 4: Table S3).
Table 1.
List of predicted genes involved in bacterial endophytic behavior [38] in Serratia sp. LCN16 genome. Burkholderia phytofirmans PsJN (CP001052-54) was used as reference genome for orthologous search in Serratia sp. LCN16, Serratia proteamaculans 568 (Spro568, NC_009832) and S. liquefaciens ATCC 27592 (CP006252). The description presented is based on KEGG annotation [69]
| Gene Function | Description | Gene Identification | Orthologous genes | ||||
|---|---|---|---|---|---|---|---|
| Number | Copies | Name | PsJN | Spro568 | ATCC 27592 | ||
| Transporter | Arabinose operon regulatory | LCN16_02277 | 1 | araC | Bphyt_0033 | Spro_1385 | M495_06380 |
| Lysine exporter protein | LCN16_04024 | 1 | Bphyt_0034 | - | M495_06375 | ||
| High-affinity branched-chain amino acid transport | LCN16_00248 | 1 | livF | Bphyt_3906 | Spro_0232 | M495_01020 | |
| High-affinity branched-chain amino acid transport | LCN16_00245 | 1 | livH | Bphyt_3908 | Spro_3202 | M495_01005 | |
| NAD(P) transhydrogenase subunit beta | LCN16_02610 | 1 | pntB | Bphyt_4261 | Spro_2584 | M495_12845 | |
| ABC transporter related | LCN16_02535 | 1 | malk_1 | Bphyt_4584 | Spro_4470 | M495_22510 | |
| Metabolite:H+ symporter (MHS) family | LCN16_03836 | 1 | citA | Bphyt_5520 | Spro_3179 | M495_10135 | |
| Extracellular solute-binding protein | LCN16_01236 | 1 | modA | Bphyt_5521 | Spro_3180 | - | |
| Gluconate 2-dehydrogenase | LCN16_02150 | 1 | Bphyt_4638 | Spro_2138 | M495_10390 | ||
| Gluconate 2-dehydrogenase | LCN16_02151 | 1 | Bphyt_4639 | Spro_2137 | M495_10385 | ||
| Gluconate 2-dehydrogenase | LCN16_02152 | 1 | Bphyt_4640 | Spro_2136 | M495_10380 | ||
| Secretion and delivery system | TypeVI secretion protein | - | 0 | - | Bphyt_4913 | Spro_3003 | - |
| TypeVI secretion protein | - | 0 | - | Bphyt_4914 | Spro_3004 | - | |
| TypeVI secretion protein | - | 0 | - | Bphyt_4919 | Spro_3013 | M495_03685 | |
| RND family efflux transporter MFP subunit | LCN16_01039 | 1 | acrA | Bphyt_6992 | Spro_1127 | M495_04880 | |
| Plant polymer degradation/modification | Alpha/beta hydrolase family protein | LCN16_01434 | 1 | Bphyt_6134 | Spro_0990 | M495_12205 | |
| Alpha/alpha-trehalase | - | 0 | Bphyt_5350 | - | - | ||
| Cupin | LCN16_02559 | 2 | Bphyt_2288 | - | - | ||
| Peptidase M48 Ste24p | LCN16_04051 | 1 | loiP | Bphyt_3335 | Spro_3955 | M495_20655 | |
| Transcriptional regulator | HTH-type transcriptional regulator LrpC | LCN16_01418 | 1 | lrpC | Bphyt_0434 | Spro_1462 | M495_06820 |
| Regulator protein FrmR | LCN16_01244 | 1 | frmR | Bphyt_0109 | - | - | |
| AraC family transcriptional regulator | LCN16_02277 | 1 | araC | Bphyt_2287 | Spro_2540 | M495_12625 | |
| Transcriptional regulatory protein | LCN16_03523 | 1 | ompR | Bphyt_4604 | Spro_4621 | M495_23305 | |
| Transcriptional regulatory, DeoR family | LCN16_01600 | 1 | deoR | Bphyt_4951 | Spro_2259 | M495_11240 | |
| Transcriptional regulatory, LysR family | LCN16_02297 | 1 | ampR | Bphyt_5523 | Spro_3181 | M495_17720 | |
| LrgB family operon | - | 0 | Bphyt_5345 | Spro_1569 | M495_07365 | ||
| Flavoprotein WrbA | LCN16_01736 | 1 | wrbA | Bphyt_6351 | Spro_1813 | M495_08400 | |
| Detoxification | Glutathione S-transferase | LCN16_01390 | 7 | gst | Bphyt_1366 | Spro_3320 | M495_17060 |
| Short-chain dehydrogenase | LCN16_02779 | 1 | Bphyt_1098 | Spro_1971 | M495_09250 | ||
| S-(hydroxymethyl)-gluthathione dehydrogenase | LCN16_01515 | 1 | frmA | Bphyt_5114 | Spro_1557 | M495_07305 | |
| 2-hydropantoate 2-reductase | LCN16_00995 | 1 | panE | Bphyt_5159 | Spro_3174 | - | |
| Redox potential maintenance | Acetoacetyl-coa reductase | LCN16_01349 | 1 | phbB | Bphyt_5655 | Spro_3465 | M495_17855 |
| Acetaldehyde dehydrogenase | LCN16_02742 | 1 | adhE | Bphyt_1467 | Spro_3026 | M495_05210 | |
| Carbonate dehydratase | LCN16_00514 | 1 | cynT | Bphyt_2146 | Spro_1534 | M495_07235 | |
| Aldehyde dehydrogenase | LCN16_02563 | 3 | gabD | Bphyt_4023 | Spro_4305 | M495_21680 | |
| Malate/L-lactate dehydrogenase | LCN16_02031 | 1 | ybiC | Bphyt_5456 | Spro_2010 | M495_09840 | |
| 3-hydroxyisobutyrate dehydrogenase | LCN16_01348 | 1 | garR | Bphyt_5931 | Spro_1492 | M495_07025 | |
| Others | Amino-acid metabolite efflux pump | LCN16_01419 | 1 | eamA | Bphyt_0435 | Spro_1463 | M495_06825 |
| 2-isopropylmalate synthase | LCN16_00673 | 2 | leuA | Bphyt_0573 | Spro_1875 | M495_00910 | |
| Diaminopimelate decarboxylase | LCN16_03946 | 1 | lysA | Bphyt_7089 | Spro_3836 | M495_20025 | |
Serratia sp. LCN16 tolerance to oxidative stress
Serratia sp. LCN16 has been reported as highly tolerant to oxidative stress, exerting a beneficial effect towards B. xylophilus under stressful conditions in vitro [22]. Serratia sp. LCN16 encodes many antioxidant enzymes in its genome (Table 2), including 7 GSTs, 3 SODs, 2 KATs (HPII, katA, LCN16_03339; HPI, katG, LCN16_03210), 1 AHP (alkyl hydroperoxide), 3 GPXs, 3 GRXs (glutaredoxin), 2 TRXs and 2 TPXs. To examine the potential roles of some of these proteins in the neutralization of oxidant stressors (e.g., H2O2), two genes were selected for complete gene knockout, namely the H2O2 transcriptional factor oxyR (LCN16_04688), and the enzyme catalase katA (LCN16_03339).
Table 2.
List of predicted genes involved in oxidative stress of Serratia sp. LCN16 genome. Genes descriptions based on KEGG [69]
| KEGG | EC | Description | Predicted gene | |
|---|---|---|---|---|
| K00799 | 2.5.1.18 | Glutathione S-transferase | gst | LCN16_01390 |
| LCN16_01491 | ||||
| LCN16_01648 | ||||
| LCN16_02242 | ||||
| LCN16_03108 | ||||
| LCN16_03382 | ||||
| LCN16_04377 | ||||
| K03782 | 1.11.1.21 | Catalase-peroxidase | katG | LCN16_03210 |
| K03781 | 1.11.1.6 | Catalase | katA | LCN16_03339 |
| K04565 | 1.15.1.1 | Superoxide dismutase Cu-Zn | sod1 | LCN16_02232 |
| K04564 | 1.15.1.1 | Superoxide dismutase Fe-Mn | sod2 | LCN16_00084 |
| K04564 | 1.15.1.1 | Superoxide dismutase Fe-Mn | sod2 | LCN16_02218 |
| K00432 | 1.11.1.9 | Glutathione peroxidase | gpx | LCN16_02167 |
| gpx | LCN16_02187 | |||
| gpx | LCN16_04689 | |||
| K00384 | 1.8.1.9 | Thioredoxin | trxB | LCN16_01650 |
| trx | LCN16_00969 | |||
| K00383 | 1.8.1.7 | Glutathione reductase | gorA | LCN16_04647 |
| K03674 | 1.20.4.1 | Glutaredoxin | grxA | LCN16_01610 |
| K07390 | 1.20.4.2 | grxD | LCN16_02220 | |
| K03675 | 1.20.4.3 | grxB | LCN16_02844 | |
| K03386 | 1.11.1.15 | Alkyl hydroperoxide | ahpD | LCN16_03826 |
| K04761 | - | Hydrogen-peroxide transcriptional regulator | oxyR | LCN16_04688 |
| K11065 | 1.11.1.- | Thiol peroxidase | tpx_1 | LCN16_02660 |
| tpx_2 | LCN16_03583 | |||
| - | - | Organic hydroperoxide resistance transcriptional regulator | ohrR | LCN16_00141 |
| - | - | Organic hydroperoxide resistance protein | ohrB | LCN16_00142 |
| K13892 | - | Glutathione ABC transporter | gsiA | LCN16_01509 |
| K13889 | - | gsiB | LCN16_01510 | |
| K13890 | - | gsiC | LCN16_01511 | |
| K13891 | - | gsiD | LCN16_01512 | |
| K01919 | 6.3.2.2 | Glutamate-cysteine ligase | gshA | LCN16_00781 |
| K18592 | 2.3.2.2 | Gamma-glutamyltranspeptidase | ggt_1 | LCN16_00968 |
| ggt_2 | LCN16_02503 | |||
| K00430 | 1.8.-.- | Thiol-disulfide oxidoreductase | ykuV | LCN16_04070 |
The nucleotide sequence of the LCN16 oxyR gene (LCN16_04688) is 918 bp long and is located between fabR (LCN16_4686), a predicted HTH-type transcriptional repressor protein, and LCN16_04691, a predicted glutathione peroxidase-like protein. The oxyR sequence encodes a 34 kDa unstable protein (305 a.a.) with a predicted LysR-type HTH domain (PROSITE: PS50931). The OxyR protein was 100 % identical to the orthologous sequences from S. proteamaculans 568 and S. grimessii CR62_05005, 99 % identical to the S. liquefaciens ATCC 27592, and shares 88 % identity with the E. coli K-12 protein. The Serratia sp. LCN16 catalase (katA) gene (LCN16_03339) is 1437 bp long, and is located between LCN16_03338, a predicted yfaZ precursor, and a cluster of genes menECBHDF (LCN16_03339-03345), presumably involved in the menaquinone (vitamin K12) biosynthesis. This gene encodes a 54 kDa stable protein (478 a.a.) with a catalase_3 domain (PROSITE: PS51402), and shares 99 % identity with S. grimessii CR62_05005, 98 % with S. proteamaculans 568 and S. liquefaciens ATCC 27592, and only 42 % identity with E. coli K. 12 [37].
Using TargeTron® (Sigma-Aldrich, MO, St. Louis), a mobile group II intron was modified (retargeted) to be specifically inserted into oxyR and katA genes in Serratia sp. LCN16. The insertion of the retargeted introns (with an approximately size of 2Kb) in the predicted positions, Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808), were confirmed by PCR (Fig. 3a and b). As foreseen in S. proteamaculans 568 genome through OperonDB and OperonDetection tools [38, 39], we also predict that, in Serratia sp. LCN16, both genes are independently transcribed (not included in an operon-like structure), which may indicate that the Serratia sp. LCN16 mutants’ phenotype are only due to these mutated genes.
Fig. 3.
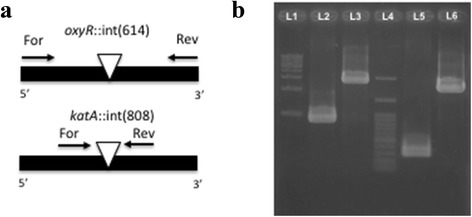
Colony PCR results indicating that group II intron-based vectors successfully targeted the Serratia sp. LCN16 oxyR and kat genes. a Introns were inserted in the position 614|615 of oxyR gene, oxyR::int(614); and in the position 808|809 of katA gene, katA::int(808). For both genes, the arrows indicate the position of forward and reverse primers designed to infer intron insertion. b L1 and L4 indicate 100-bp and 1-kb molecular markers, respectively. L2 and L5 correspond to the oxyR (846 bp) and katA (292 bp) fragments. L3 and L6 correspond to mobile group II intron integrated in oxyR (about 3Kb) and katA (about 2-3Kb), respectively
The growth of Serratia sp. LCN16 WT and mutants is shown in Fig. 4a. Slightly changes were observed in wild-type and mutants’ generation times. The generation time of Serratia sp. LCN16 WT was 1.2 h, and the generation times of Serratia sp. LCN16 katA::int(808) and oxyR::int(614) were, respectively, 1.1 h and 1.4 h. The tolerance to H2O2 was considerably affected in the oxyR and katA mutants (Fig. 4b, c). The H2O2 (30 %, w/v; 9.79 M) inhibition was higher in Serratia sp. LCN16 oxyR::int(614) than in Serratia sp. LCN16 katA::int(808) (Fig. 4b). Both mutants were statistically different (P < 0.01) than the wild-type. At 50 mM and 100 mM H2O2, both mutants were completely inhibited while Serratia sp. LCN16 WT grew easily (Fig. 4c). Consequently, both failed to protect B. xylophilus Ka4 after 24 h exposure to 50 mM H2O2 (Table 3). No statistical differences were seen between B. xylophilus Ka4 and B. xylophilus Ka4_LCN16 oxyR::int(614)/LCN16 katA::int(808) (P > 0.05), with mortality ranging between 94 and 99 %. Only B. xylophilus Ka4_LCN16 WT could reduce significantly (P < 0.01) B. xylophilus Ka4 mortality to 0.2 %.
Fig. 4.
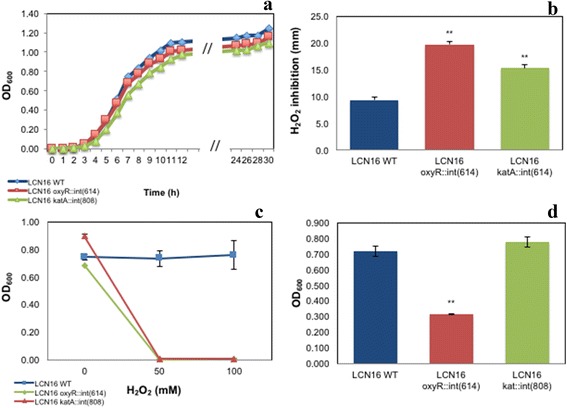
Characterization of H2O2-sensitive Serratia sp. LCN16 (oxyR::int(614) and katA::int(808)) and resistant Serratia sp. LCN16 wild-type (WT): growth curves (a); H2O2 inhibition (mm) (b); growth in 24 h exposure to H2O2 (c); and biofilm production (d). H2O2 inhibition was determined by measuring the diameter of the halo surrounding the H2O2 (30 %, v/v) spot-inoculation. Biofilm production was determined as described in [56]. Error bars indicate standard deviation. Asterisk (*, **) on the top of the columns denotes statistical differences at 95–99 % confidence level by Students T-test (EXCEL version 15.14), when compared with wild-type Serratia sp. LCN16
Table 3.
Mortality of Bursaphelenchus xylophilus Ka4, alone or in association with Serratia sp. LCN16 WT and mutants (Serratia sp. LCN16 oxyR::int(614) and Serratia sp. kat::int(808)), in H2O2 conditions (0 and 50 mM). Statistical differences between treatment Ka4 and the other treatments (Ka4_LCN16 WT; Ka4_LCN16 oxyR::int(614); and Ka4_LCN16 kat::int(808)) were calculated using Students T-test (EXCEL version 15.14). Asterisk (**) denotes statistical differences at 99 % confidence level
| Treatment | H2O2 (mM) | Mortality | P-value | |
|---|---|---|---|---|
| Mean | S.D. | |||
| Ka4 | 0 mM | 0.01 | 0.00 | |
| Ka4 | 50 mM | 0.99 | 0.01 | |
| Ka4_LCN16 WT | 0.02** | 0.00 | 0.00 | |
| Ka4_LCN16 oxyR::int(614) | 0.94 | 0.03 | 0.06 | |
| Ka4_LCN16 kat::int(808) | 0.96 | 0.07 | 0.49 | |
Biofilm production (Fig. 4d) was only compromised in Serratia sp. LCN16 oxyR::int(614), with a significant reduction (P < 0.01) in comparison with Serratia sp. LCN16 WT. No significant differences (P > 0.05) were seen between Serratia sp. LCN16 WT and Serratia sp. LCN16 katA::int(808). In terms of swimming and swarming abilities (Fig. 5), only Serratia sp. oxyR::int(614) swimming trait was improved.
Fig. 5.
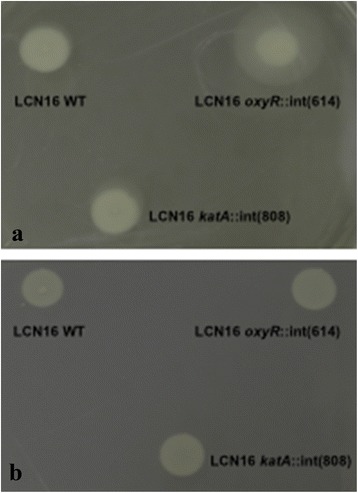
Swimming (a) and Swarming (b) behaviors of H2O2-sensitive mutants Serratia sp. LCN16 oxyR::int(614) and katA::int(808) and resistant Serratia sp. LCN16 WT. Only Serratia sp. LCN16 oxyR::int(614) change the swimming behavior as it grown further in the semisolid medium. No effects were seen in swarming behaviors of mutants and wild-type (WT)
Relative gene expression of oxyR, katA, katG was analyzed in mid-log phase for the Serratia sp. LCN16 WT and mutants (Fig. 6). In terms of the relative expression of oxyR (stress versus non-stress conditions), only Serratia sp. LCN16 WT showed a 2.2-fold induction, while for LCN16 oxyR::(614) and LCN16 katA::(808) oxyR the expression levels remained unchanged. Statistical differences (P < 0.05) were only detected comparing relative expression of oxyR of WT and LCN16 katA::(808). For the relative expression of katA, only Serratia sp. LCN16 WT showed a 2.7-fold induction. The relative expression of katA in LCN16 oxyR::(614) was almost similar between stress and non-stress conditions with a slight induction of 0.4-fold. For the LCN16 katA::(808) mutant, katA expression was almost null indicating successful mutation of this gene. Statistical differences (P < 0.05) were only seen between WT and LCN16 katA::(808). The relative gene expression of katG was considerably high in LCN16 WT and LCN16 katA::(808), respectively 22.6-fold and 11.0-fold. Since katG is under regulation of OxyR, its expression is supposed to be comprised in LCN16 oxyR::(614) mutant. Thus, the relative expression of katG in LCN16 oxyR::(614) was equal between stress and non-stress conditions and statistically different (P < 0.01) from WT.
Fig. 6.
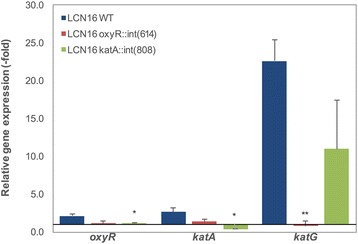
Relative gene expression of oxyR, katA and katG genes in wild-type Serratia sp. LCN16 (WT) and Serratia sp. LCN16 mutants (oxyR::int(614) and katA::int(808)), after H2O2-shock induction. The x-axis was positioned for 1.0, which indicates that the level of gene expression between stress and non-stress conditions is similar. Values were normalized using reference gene gyrA, and analyzed with ΔΔCT method. Error bars indicate standard deviation. Asterisk (*, **) on the top of the columns denotes statistical differences at 95–99 % confidence level by Students T-test (EXCEL version 15.14), when compared with wild-type Serratia sp. LCN16
Discussion
Forest trees harbor population densities of endophytic bacteria ranging between 101 and 106 CFU (colony forming unit) per gram of sample, most of which are host-specific [40, 41]. Genomes of plant-associated bacteria reflect a wide spectrum of life style adaptations [36]. Therefore, we cannot dismiss the potential of bacteria when we are trying to understand a particular ecosystem. In the present study, we characterized the biology of Serratia sp. LCN16 analyzing, in detail, its genome content, and investigated the role OxyR and KatA in the extreme oxidative stress resistance of this PWN-associated bacteria.
Members of the genus Serratia are ecological generalists found inhabiting water, soil, plants, animals and humans [24]. Belonging to the phytosphere Serratia group [24], also known as Serratia liquefaciens complex [29], Serratia sp. LCN16 is phylogenetically closest to the poplar endophyte S. proteamaculans 568 [36] and S. liquefaciens strain ATCC 27592 [42]. However, this taxonomical identification may be incomplete due to the lack of complete genome sequence for other Serratia such as S. grimessi strain A2 [43], which is also included in the S. liquefaciens complex [30]. Serratia sp. LCN16 shares many features with endophytic bacteria, such as plant polymer degrading enzymes, iron uptake/transport, siderophore and phytohormone (i.e., IAA) synthesis, and aromatic compound degradation [33], some of which are supported by the previous phenotypic characterization [16]. Wilted pine trees are a rich source of fungi, which in the late-stages of PWD are consumed by PWN [2]. Serratia sp. LCN16, isolated from the cuticle of fungi-cultivated B. xylophilus [44], harbors genes involved in the production of antifungal agents (i.e., pyrrolnitrin) and chitinases, which may explain its survival and persistence in B. xylophilus lab-culture. Genes encoding other antimicrobial compounds were found in Serratia sp. LCN16, which can give a fitness advantage to this bacterium in a more complex environment such as host pine trees [45]. Few MGIs were found in Serratia sp. LCN16 suggesting a more stable genome probably adapted to a less broad environment as described elsewhere [34, 46, 47].
Endophytes have highly elaborate detoxification mechanisms to counter-attack host ROS [35, 48]. Vicente et al. [22] showed that Serratia sp. LCN16 is highly H2O2-tolerant bacterium. A total of 16 antioxidant enzymes were found in the Serratia sp. LCN16 genome, which is within the range described in other endophytes (from 8 in Azospirillum sp. B510 to 21 in Burkholderia phytofirmans PsJN) [35]. Serratia sp. LCN16 is a copious siderophore producer [16], rich in iron uptake and transport systems (i.e., multiple copies for iron III complex transporter) and, recently, Li et al. [49] reported the importance of siderophore synthesis and iron uptake systems in the resistance against oxidative stress of insect-gut Elizabethkingia anophelis NUHP1. The relation between oxidative stress and siderophore synthesis has also been explored in the fungus Alternaria alternata [50] and in Aspergillus nidulans [51].
Mobile group II introns are retroelements, which through retrohoming mechanism, can be inserted into a DNA target site in a site-specific manner via the activity of an associated intron-encoded enzyme protein [52]. Wang et al. [53] could efficiently disrupt the acid production pathways in Clostridium beijinrinckii by the insertion of group II intron specifically retargeted to pta (encoding phosphotransacetylase) and buk (encoding butyrate kinase). Using this gene knockout system, we obtained two mutants, Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808), disrupting respectively, oxyR and katA genes. Both genes have been investigated for their direct involvement in oxidative stress in several organisms [54–59]. The OxyR is a global regulator of peroxide stress response that maintains intracellular H2O2 homeostasis, and it is known to regulate several genes involved in H2O2 detoxification (i.e., katG, grxA, ahpCF and trxC) [54, 55]. The Serratia sp. LCN16 oxyR::int(614) was severely impaired in its ability to survive in high H2O2 conditions in vitro, highly affected in biofilm production (production decreased) and swimming abilities. Our results are corroborated by Shank et al. [56], which previously observed that Serratia marcescens OxyR mediated oxidative stress response, biofilm formation and surface attachment. The high sequence similarity between Serratia sp. LCN16 OxyR and E. coli K12 (88 %) suggests that Serratia sp. LCN16 utilizes a similar mechanism of regulation as E. coli. Under high H2O2 stress conditions, OxyR (reduced/inactive form) is oxidized triggering conformational changes (OxyR oxidized/active form) leading to the regulation of several proteins including the expression of KatG [55]. This effect was clearly seen in our results. The expression of katG was significantly down regulated in Serratia sp. LCN16 oxyR::int(614) (Fig. 6), suggesting its OxyR-dependent regulation. The importance of KatG has been observed in several studies. Jamet et al. [57] observed the role of katG as a mediator of bacteria-plant interaction for Sinorhizobium meliloti, reporting its constitutive expression and considering katG to be a housekeeping gene. Mycobacterium tuberculosis, an opportunistic bacterial pathogen, also relies on KatG for its resistance to high oxidative stress environments [58]. Serratia sp. LCN16 encodes also other catalase, katA (mono-functional), which is homologous with E. coli katE. Serratia sp. LCN16 katA::int(808) was affected in H2O2 resistance but not in biofilm production nor swimming and swarming traits. The expression of the katA in Serratia sp. LCN16 WT was slightly induced when compared with katG, indicating that its expression in exponential phase is reduced. In oxyR mutant, katA expression was relatively closest to the expression level showed in wildtype, suggesting to be regulated in a OxyR-independent manner. In E. coli, katE is transcriptionally regulated in the stationary phase by RpoS [55]. As for KatG, KatA was also found important in bacteria-plant interaction [60].
Conclusions
The present study revealed the potential of PWN-associated bacteria Serratia sp. LCN16 to live in a plant-environment, and also that its high tolerance to oxidative stress is OxyR- and KatA-dependent. In the PWD context, we showed that Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808) failed to protect PWN against H2O2 oxidative conditions. As previously hypothesized [22], PWN-associated bacteria may opportunistically assist the nematode in the disease by amelioration of oxidative burst of pine defenses. Through this study, we have set the proper conditions to explore bacteria-nematode association in planta environment.
Methods
Bacterial strain
Serratia sp. LCN16 was isolated from the cuticle of lab-culture Bursaphelenchus xylophilus isolate Bx153-3A (Setubal Peninsula, Portugal) [16]. This bacterium belongs to the bacterial culture collection of NemaLab/ICAAM (Évora, Portugal), and is maintained in 30 % (w/v) glycerol stocks at −80 °C. For all experiments, Serratia sp. LCN16 was recovered from long-term stock and grown in LB (Luria-Bertani) for 1 day at 28 °C.
Genome sequencing, annotation and analysis
A single colony of Serratia sp. LCN16 was used to inoculate 10 ml of LB and incubated overnight at 28 °C with shaking. Genomic DNA was extracted from the overnight culture using the QIAGEN Genomic DNA Purification kit (Qiagen), following the manufacturer’s instructions. This DNA was sequenced on the Roche Titanium 454 platform at the Centre for Genomic Research, University of Liverpool, with large-insert 3 kb paired end libraries. This gave HYMXIQB02 (ENA accession ERS980300) and HYMXIQB03.sff (ENA accession ERS980301) with in total 607,360 sequences, mean length 497.0 bp, median length 465 bp. Initial assemblies were performed with Roche “Newbler” gsAssembler [61], and MIRA v4.0.2 [62]. This data was supplemented with 169,073 paired reads of mean length 132.6 bp from an Illumina MiSeq commissioning test run at the James Hutton Institute (ENA accession ERS980302), as one of 11 barcoded samples. Again, multiple assemblies were evaluated. The final assembly selected was a hybrid 454 and MiSeq assembly using the MIRA v4.0.2, which resolved some of the homopolymer errors detected in the initial 454 assemblies. The genome sequence of Serratia sp. LCN16 is available in the European Nucleotide Archive (ENA) under the accession ERP013273.
Genome annotation was performed using PROKKA [63], and manually reviewed in ARTEMIS [64] (ENA accession ERS1015427). The circular genome image was plotted in DNA PLOTTER [65]. Protein annotation (InterPro and Gene Ontology) was further supported by BLAST2GO [66] and KAAS (KEGG Automatic Annotation Server) [67]. Genomic islands were annotated using online-tool Island Viewer 3.0 [31]. Genome to genome alignments of Serratia sp. LCN16, Serratia proteamaculans 568 (CP000826.1) and Serratia liquefaciens ATCC 27592 (CP006252.1) were conducted using MAUVE software [68]. The 16S rRNA and four housekeeping genes (atpD, dnaJ, gyrB and rpoB) were used to infer the phylogenetic relationship between Serratia sp. LCN16 and the following Serratia-type strains: S. liquefaciens ATCC 27592, S. marcescens subsp. marcescens Db11 (NZ_HG326223), S. marcescens WW4 (CP003959.1), S. plymuthica 4RX13 (CP006250.1), S. proteamaculans 568, and S. symbiotica “Cinara cedri” (CP0022951.1). Bacillus subtilis XF1 (CP004091.1) was used as out-group. Housekeeping sequence genes were concatenated using Seaview 4.0 [69]. All phylogenetic analyses were conducted in MEGA6 [70]. Phylogenetic robustness was inferred by bootstrap analysis using 1,000 iterations.
Functional analysis of oxidative stress resistance of Serratia sp. LCN16
Strains, plasmids, media and growth
All bacteria and plasmids used in this study are listed in Table 4. All bacteria were grown in LB at 28 °C and 200 rpm. The antibiotics used in this study were gentamicin (10 μg/ml and 30 μg/ml), chloramphenicol (50 μg/ml), kanamycin (50 μg/ml), and ampicillin (100 μg/ml).
Table 4.
List of strains, plasmids and primers used in the present study
| Strain, plasmid and primers | Genotype or phenotype |
|---|---|
| Serratia sp. | |
| LCN16 WT | WT resistant to ampicillin and erythromycin. |
| LCN16 oxyR::int(614) | Knockout mutant of oxyR with group II intron inserted in the position 614; Resistant to gentamicin and kanamycin. |
| LCN16 katA::int(808) | Knockout mutant of katA with group II intron inserted in the position 808; Resistant to gentamicin and kanamycin. |
| Escherichia coli | |
| DH5α | Competent cells (Sigma-Albrich) |
| Plasmids | |
| pAR1219 | TargeTron vector with chloramphenicol resistant |
| pAR1219-ΩGm | Constructed vector with gentamicin resistant gene aacC |
| pACD4K-C | TargeTron vector; kanamicin RAM marker (for chromosomal insertion) and chloramphenicol resistance (plasmid propagation) |
| pACD4K-C_oxyR | TargeTron vector with intron RNA retarget for oxyR gene |
| pACD4K-C_kat | TargeTron vector with intron RNA retarget for kat gene |
| pBK-miniTn7-ΩGm | Tn7 plasmid constructed with gentamicin resistant gene aacC |
| PCR Primers (5′-3′) | |
| aacC1_IfFor | CATACTCTTCCTTTTTCAATATTATTG |
| aacC1_IfRev | TAACTGTCAGACCAAGTTTACTC |
| oxyR_614|615 s-IBS | AAAAAAGCTTATAATTATCCTTAGGAAGCTGGTCAGTGCGCCCAGATAGGGTG |
| oxyR_614|615 s-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCTGGTCACTTAACTTACCTTTCTTTGT |
| oxyR_614|615 s-EBS2 | TGAACGCAAGTTTCTAATTTCGATTCTTCCTCGATAGAGGAAAGTGTCT |
| oxyR_KO_chkFor | CGTGGTCTGGAGGGAAACAA |
| oxyR_KO_chkRev | CATAACGACTGCGCAATGGG |
| katA_808|809a -IBS | AAAAAAGCTTATAATTATCCTTATAATCCGGATTTGTGCGCCCAGATAGGGTG |
| katA_808|809a EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGGATTTGCTAACTTACCTTTCTTTGT |
| katA_808|809a -EBS2 | TGAACGCAAGTTTCTAATTTCGGTTGATTATCGATAGAGGAAAGTGTCT |
| katA_KO_chkFor | GGTGAAGTTCCATTTCCGCTGC |
| katA_KO_chkRev | GGGTTCACCGCTACCTGTTCAAC |
| RT-qPCR primers (5′-3′) | |
| gyrA_For | TTATCTCCCTGATTGTGCCA |
| gyrA_Rev | CATTACGCTCGCTCACCTTA |
| oxyR_For | TTTAGAGTACCTGGTCGCCTTG |
| oxyF_Rev | ATCACACCCAGTTCGTCTTCC |
| katA_For | CCAGATTATGCCTGAACACG |
| katA_Rev | TGCAGTTCGAAGAAACCAAC |
| katG_For | AGCGGTAAGCCAAATACACC |
| katG_Rev | AATCGAAGTCAGGGTCCATC |
Gene knockout of oxyR and kat
The TargeTron® Gene Knockout System from Sigma-Aldrich (St. Loius, MO) was used to obtain complete gene knockouts of transcription factor OxyR and catalase KatA from Serratia sp. LCN16. The mobile group II intron sites for oxyR (LCN16_04688) and katA (LCN16_03339) were predicted using online TargeTron Design site (Sigma-Aldrich, St. Louis, MO). Intron PCR template was retargeted for both genes using the primers designed in the online tool and listed in Table 4. For oxyR gene, the following four primers were used: EBS universal primer, oxyR_614|615 s-IBS, oxyR_614|615 s-EBS1d, and oxyR_614|615 s-EBS2. For katA, the four primers used were: EBS universal primer, katA_808|809a-IBS, katA_808|809a-EBS1d, and katA_808|809a-EBS2. The amplified 350-bp DNA fragment was double digested with Hind III and BsrG I and ligated into the linear vector pACD4K-C (pACD4K-C_oxyR and pACD4K-C_katA, Table 4). Both plasmids were cloned in E. coli DH5α competent cells, following the manufacture’s instructions (Invitrogen). Since Serratia sp. LCN16 WT (wild type) is resistant to ampicillin, the plasmid pAR1219-ΩGm was constructed using pAR1219 linear without marker, previously amplified from the pAR1219 (Sigma-Aldrich, St. Louis, MO) using pAR1219_LFor and pAR1219_LRev, with gentamicin resistance used as an alternative antibiotic marker. Briefly, aaaC1 (gentamicin cassette) was amplified from pBK-miniTn7-ΩGm using aacC1_IfFor and aacC1_IfRev primers (Table 4). The PCR product was ligated into the pAR1219 linear with In-fusion® HD cloning system (Clontech Laboratories). The resultant plasmid was also cloned into E. coli DH5α competent cells, following the manufacture’s instructions (Invitrogen). TargeTron plasmids isolated from the correct clones were transformed sequentially into electrocompetent Serratia sp. LCN16 through electroporation. For each transformation, 50 μl of electrocompetent Serratia sp. LCN16 suspension was mixed with 5 μl of plasmid DNA and then added into a 0.2-cm precooled electroporation cuvette and incubated 2 min. Electroporation was carried out using the following conditions: 2,500 V of voltage, 25 μF of captaincy and 200 Ω of resistance. Afterwards, cells were incubated in LB for 1 h at 30 °C, and plated on selective medium (pACD4K-C_oxyR and pACD4K-C_kat in LB supplemented with 50 μg/ml of chloramphenicol and 30 μg/ml gentamicin; pAR1219-ΩGm in LB with 30 μg/ml gentamicin). Induction of the gene disruption was conducted in Serratia sp. LCN16 WT containing both plasmids (pACD4K-C_oxyR/kat and pAR1219-ΩGm) using 100 mM IPTG during 30 min at 25 °C. Induced cells were then pellet at high speed, suspended in LB, incubated at 25 °C for 1 h, and plated in LB supplemented with 50 μg/ml of kanamycin and 30 μg/ml gentamicin. Kanamycin-resistant colonies were picked for colony PCR to detect intron insertions, using the primers oxyR_KO_chkFor/oxyR_KO_chkRev; and katA_KO_chkFor/katA_KO_chkRev (Table 4). Successful mutants were named Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int (808).
Characterization of Serratia sp. LCN16
Unless otherwise specified, Serratia sp. LCN16 WT, Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808) were grown overnight at 28 °C from a single colony in LB with respective antibiotics, and adjusted to OD600 of 0.8.
Growth curves
The growth rates of all bacteria were determined in 5 ml of LB incubated at 30 °C with 200 r.p.m (initial OD600 0.05). Triplicate 100 μl aliquots were removed at various time points, and the culture turbidity (OD600) was determined using a multi-spectrophotometer 96-well plate reader (Bio-Tek, Synergy H1 microplate reader; Gen5 version 2.05). This experiment was repeated two times in independent days. Generation time (h) was determined in the exponential phase of bacterial growth.
Tolerance to H2O2
Serratia sp. LCN16 WT, Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808) were tested for their tolerance to H2O2, bacteria-only and in association with B. xylophilus Ka4. For the bacteria-only test, 100 μl of LB supplemented with H2O2 (final concentrations of 0, 50 and 100 mM) and 10 μl of overnight bacterial culture were incubated in 96-well plate for 24 h at 30 °C. Bacterial growth was read in a multi-spectrophotometer 96-well plate reader (Bio-Tek, Synergy H1 microplate reader; Gen5 version 2.05). Three independent biological replicates with three technical replicas per experiment were used for each treatment.
To test nematode-bacteria association in H2O2 conditions (final concentration 50 mM), firstly nematodes were surface-sterilized as described by Takemoto [71], and the concentration was adjusted to 150 nematodes per 50 μl of ddH2O. Secondly, nematode-bacteria association was performed by 1 h contact between surface cleaned nematodes and 1 ml of bacteria suspension (prepared as referred above) following the Han et al. [18] procedure. After contact, nematodes were washed and re-suspended in ddH2O. A 96-well plate was prepared as follows: each well received 50 μl of H2O2 suspension and 50 μl of each treatment (nematode-bacteria association and nematode alone). Control treatment of B. xylophilus Ka4 with ddH2O was also prepared. Three independent biological replicates with two technical replicas per experiment were used for each treatment. Nematode mortality was scored after 24 h. Nematodes were considered dead, if no movements were observed after mechanical stimulation.
H2O2 inhibition, swimming and swarming assays, and biofilm production
H2O2 inhibition and swimming/swarming assays were tested according to [56] and biofilm production adapted from [72]. For these experiments, three biological replicates with three technical replicas were performed for each bacterium.
H2O2 inhibition was tested by disk diffusion assays. Overnight bacteria culture was spread on LB medium and a sterile 6-mm paper disk was placed in the center of the plate, to which 10 μl of 30 % H2O2 (w/v, 9.79 M) was added. Plates were incubated overnight at 28 °C. H2O2 inhibition was determined by measuring the diameter of the inhibition halo surrounding the H2O2 disk. Swimming and swarming were tested in LB medium, respectively, with 0.3 %(w/v) and 0.6 %(w/v) agar. Bacteria was inoculated (10 μl) into semi-solid LB and incubated overnight at 28 °C. The ability to swim or swarm was inferred by visualization of colony expansion or shape.
For quantitative evaluation of biofilm production [72], 10 μl overnight bacteria culture (OD600 0.02) was inoculated into 150 μl LB in individual wells of a 96-well plate, and incubated for 48 h at 28 °C. Following incubation, the plate was gently washed with ddH2O, dried at 30 °C for 30 min, and wells filled with 150 μl of 0.1 % crystal violet stain. After 1 h incubation at room temperature, wells were gently rinsed with ddH2O, filled with 180 μl of ETOH (96 %, v/v), and incubated for 20 min at room temperature. The 96-well plate was read in a multi-spectrophotometer 96-well plate reader (Bio-Tek, Synergy H1 microplate reader; Gen5 version 2.05) at 590 nm.
RNA extraction and Real-time PCR
To understand the gene regulation under oxidative stress conditions, oxyR, katA and katG were selected for gene expression analysis. Predictions about general topology, domain/family and retrieved best matches were made using the online tools (Translate, ScanProsite, ProtParam) at Expasy WWW pages (http://www.expasy.org/).
All bacteria (WT and mutants) were grown in M9 medium supplemented with sucrose (2 % v/v) till mid-log phase (OD600 0.100–0.200, BioTek Synergy H1 multichannel spectrophotometer) and shocked with 50 mM H2O2 for 5 min. One-ml of each treatment was stabilized in RNA protect (Qiagen) and total RNA was extracted with RNeasy ® Minikit (Qiagen), following the manufacturer’s instructions. The concentration and quality of extracted RNA was measured using NanoVue plus spectrophotometer (GE Healthcare Life Science, USA). Total RNA was, firstly, treated with DNase I (Takara Bio Inc., Japan), adjusted to a final concentration of 500 ng/μl and reverse transcribed using random hexamers primers and PrimeScript RT enzyme from PrimeScript™ RT reagent kit (Perfect Real Time) (Takara Bio Inc., Japan). Quantitative RT-PCR was performed using CFX96™ Real-Time (Bio-Rad), and SYBR Premix Ex TaqTM II (Tli RNAse H Plus) kit (Takara Bio Inc., Japan). The housekeeping gyrA gene (LCN16_03331) was used as an internal control gene for the calculation of relative expression levels of each selected gene. Primers were designed using Primer3 software [73] (Table 4) and tested for specificity prior to qPCR. Two independent biological replicates with two technical replicas per experiment were used for each qPCR test. No template (NTC) and RNA controls were prepared for each qPCR run. Thermal cycling conditions were: initial denaturation at 95 °C for 30 s; 39 cycles of denaturation at 95 °C for sec, annealing and extension at 60 °C for 30 s; followed by the melting curve. Relative gene expression of each gene were analyzed using ΔΔCT method [74]. Data were analyzed with CT values in normal and stress conditions and using Eq. 2
| 2 |
The fold change of oxyR, katA and katG were normalized to gyrA and relative to the expression at normal conditions.
Statistical analyses
Statistical differences at 95–99 % confidence level between mutants (Serratia sp. LCN16 oxyR::int(614) and Serratia sp. LCN16 katA::int(808)) and Serratia sp. LCN16 WT were calculated using Student’s T-Test in Excel version 15.14.
Ethics and consent to participate
Not Applicable.
Availability of Data and Materials
The datasets supporting the conclusions of this article are available in the TreeBase repository (http://purl.org/phylo/treebase/phylows/study/TB2:S19116), in the European Nucleotide Archive (ENA) under the accession ERP013273, and included within the article and its additional files.
Acknowledgements
The authors would like to thank Prof. John Jones (The James Hutton Institute) for advice on an earlier draft of this manuscript; Sonia Humphris, Jenny A. Morris, and Pete Hedley (The James Hutton Institute) for all the support given in Serratia sp. LCN16 sequencing.
Funding
This work was supported by the JSPS KAKENHI Grant numbers P14394 (to CSLV) and 26450204 (to KH); the European Project REPHRAME - Development of improved methods for detection, control and eradication of pine wood nematode in support of EU Plant Health policy, European Union Seventh Framework Programme FP7-KBBE-2010-4; and FEDER Funds through the Operational Programme for Competitiveness Factors - COMPETE and National Funds through FCT - Foundation for Science and Technology under the Strategic Project PEst-C/AGR/UI0115/2011. The James Hutton Institute received funding from the Scottish Government.
Abbreviations
- CDS
coding sequences
- CFU
colony forming unit
- CRISPR
clustered regularly interspaced short palindromic repeats
- GI
genomic islands
- GO
gene ontology
- GPX
glutathione peroxidase
- GST
gluthathione S-transferase
- H2O2
hydrogen peroxide
- IAA
indole acetic acid
- KAT
catalase
- MGE
mobile genomic elements
- PRX
peroxiredoxin
- PWD
pine wilt disease
- PWN
pine wood nematode
- ROS
reactive oxygen species
- SOD
superoxidase dismutase
- TPX
thiol peroxidase
- TRX
thioredoxin
- WT
wild type
Additional files
Gene ontology of Serratia sp. LCN16 according to BLAST2GO [57]. (PDF 20 kb)
Comparison of Serratia sp. LCN16 genome and the closest Serratia representatives, S. proteamaculans 568 (Spro568) and S. liquefaciens ATCC 27592. Genome to genome alignment conducted using MAUVE software [64]. S1A figure shows the syntenic regions (locally collinear blocks, LCD) between genomes. In each LCD, the height of the similarity profile indicates the level of conservation in that genome region. White areas indicate specific sequences of the genome. The genome rearrangements are indicated as different colored lines. S1B figure presents the similarities between genomes are indicate as follows: purple indicates conserved regions in all genomes; green indicates Serratia sp. LCN16 and ATCC 27592 genomes (highlighted with red arrow); reds indicate Serratia sp. LCN16 and Spro568 genomes; and orange, conservation between Spro568 and ATCC 27592. (PDF 3065 kb)
List of genes predicted in Serratia sp. LCN16 genomic islands (GI). GIs at least one of the two methods SIGI-HMM or IslandPath-DIMOB in IslandViewer server [25]. (PDF 263 kb)
List of genes predicted to be involved in a plant-associated life-style of Serratia sp. LCN16. (PDF 113 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors read and approved the final manuscript. Conceived and designed the experiments: CSLV and KH. Performed the experiments: CSLV and YI. Analyzed the data: CSLV, FXN, PC, KH. Contributed reagents/materials/analysis tools: CSLV, PJAC, MM, KH. Wrote the paper: CSLV and KH.
Authors’ information
CSLV has a PhD in Soil Microbiology from Universidad Pablo de Olavide (Seville, Spain) and six-years postdoctoral experience working in pine wilt disease, mainly investigating the playing-role of nematode-associated bacteria on the development of this disease. Currently, CSLV is working in the Department of Environmental Biology (College of Bioscience and Biotechnology) of Chubu University under the supervision of KH, while maintaining collaboration with MM in NemaLab, ICAAM-University of Évora as a eligible member of this institute.
KH was graduated from Kyoto University (Graduate School of Agriculture) and obtained his PhD in Agriculture in Kyoto University. KH is specialized in Applied Entomology, Nematology and Genetics, and his main research areas involve the evolution of symbiosis and the mechanisms of animal response against environmental stress. KH is currently Associate Professor in the Department of Environmental Biology (College of Bioscience and Biotechnology) of Chubu University.
References
- 1.Vicente C, Espada M, Vieira P, Mota M. Pine Wilt Disease: a threat to European forestry. Eur J Plant Pathol. 2012;133:89–99. doi: 10.1007/s10658-011-9924-x. [DOI] [Google Scholar]
- 2.Futai K. Pine Wood Nematode, Bursaphelenchus xylophilus. Annu Rev Phytopathol. 2013;51:61–83. doi: 10.1146/annurev-phyto-081211-172910. [DOI] [PubMed] [Google Scholar]
- 3.Kiyohara T, Tokushige Y. Inoculation experiments of a nematode, Bursaphelenchus sp., onto pine trees. J Jpn For Soc. 1971;53:210–8. [Google Scholar]
- 4.Zhao BG, Futai K, Sutherland JR, Takeuchi Y. Pine Wilt Disease. 2008. [Google Scholar]
- 5.Jones JT, Moens M, Mota M, Li H, Kikuchi T. Bursaphelenchus xylophilus: opportunities in comparative genomics and molecular host–parasite interactions. Mol Plant Pathol. 2008;9:357–68. doi: 10.1111/j.1364-3703.2007.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuchi T, Jones JT, Aikawa T, Kosaka H, Ogura N. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004;572:201–5. doi: 10.1016/j.febslet.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi T, Shibuya H, Aikawa T, Jones JT. Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol Plant Microbe Interact. 2006;19:280–7. doi: 10.1094/MPMI-19-0280. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi T, Aikawa T, Kosaka H, Pritchard L, Ogura N, Jones JT. Expressed sequence tag (EST) analysis of the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus. Mol Biochem Parasitol. 2007;155:9–17. doi: 10.1016/j.molbiopara.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Shinya R, Takeuchi Y, Futai K. A technique for separating the developmental stages of the propagative form of the pine wood nematode, Bursaphelenchus xylophilus. Nematol. 2009;11:305–7. doi: 10.1163/156854108X399164. [DOI] [Google Scholar]
- 10.Shinya R, Morisaka H, Takeuchi Y, Ueda M, Futai K. Comparison of the surface coat proteins of the pine wood nematode appeared during host pine infection and in vitro culture by a proteomic approach. Phytopathol. 2010;100:1289–97. doi: 10.1094/PHYTO-04-10-0109. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi T, Cotton JA, Dalzell JJ, Hasegawa K, Kanzaki N, McVeigh P, et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinya R, Morisaka H, Kikuchi T, Takeuchi Y, Ueda M, Futai K. Secretome Analysis of the Pine Wood Nematode Bursaphelenchus xylophilus Reveals the Tangled Roots of Parasitism and Its Potential for Molecular Mimicry. PLoS One. 2013;8:e67377. doi: 10.1371/journal.pone.0067377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicente CSL, Ikuyo Y, Shinya R, Mota M, Hasegawa K. Catalases Induction in High Virulence Pinewood Nematode Bursaphelenchus xylophilus under Hydrogen Peroxide-Induced Stress. PLoS One. 2015;10:e0123839. doi: 10.1371/journal.pone.0123839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espada M, Silva AC, Eves van den Akker S, Cock PJ a., Mota M, Jones JT. Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy. Mol Plant Pathol. 2015. doi: 10.1111/mpp.12280. [DOI] [PMC free article] [PubMed]
- 15.Nascimento FX, Hasegawa K, Mota M, Vicente CSL. Bacterial role in pine wilt disease development - review and future perspectives. Environ Microbiol Rep. 2015;7:51–63. doi: 10.1111/1758-2229.12202. [DOI] [PubMed] [Google Scholar]
- 16.Vicente CSL, Nascimento F, Espada M, Barbosa P, Mota M, Glick BR, et al. Characterization of Bacteria Associated with Pinewood Nematode Bursaphelenchus xylophilus. PLoS One. 2012;7:e46661. doi: 10.1371/journal.pone.0046661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawazu K, Zhang H, Yamashita H, Kanzaki H. Relationship between the pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus, and phenylacetic acid production. Biosci Biotechnol Biochem. 1996;60:1413–5. doi: 10.1271/bbb.60.1413. [DOI] [PubMed] [Google Scholar]
- 18.Han ZM, Hong YD, Zhao BG. A Study on Pathogenicity of Bacteria Carried by Pine Wood Nematodes. J Phytopathol. 2003;151:683–9. doi: 10.1046/j.1439-0434.2003.00790.x. [DOI] [Google Scholar]
- 19.Zhao BG, Lin F. Mutualistic symbiosis between Bursaphelenchus xylophilus and bacteria of the genus Pseudomonas. For Pathol. 2005;35:339–45. [Google Scholar]
- 20.Paiva G, Proença DN, Francisco R, Verissimo P, Santos SS, Fonseca L, et al. Nematicidal Bacteria Associated to Pinewood Nematode Produce Extracellular Proteases. PLoS One. 2013;8:e79705. doi: 10.1371/journal.pone.0079705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng XY, Tian XL, Wang YS, Lin RM, Mao ZC, Chen N, et al. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci Rep. 2013;3:1–10. doi: 10.1016/0167-5729(83)90005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vicente CSL, Ikuyo Y, Mota M, Hasegawa K. Pinewood nematode-associated bacteria contribute to oxidative stress resistance of Bursaphelenchus xylophilus. BMC Microbiol. 2013;13:299. doi: 10.1186/1471-2180-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta KJ, Igamberdiev AU. Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants. In: K. J. Gupta AUI, Editor. Reactive Oxygen and Nitrogen Species. 2015. pp. 1–14. doi:10.1007/978-3-319-10079-1.
- 24.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Biol. 1997;48:251–75. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 25.Torres MA. ROS in biotic interactions. Physiol Plant. 2010;138:414–29. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 26.Quan LJ, Zhang B, Shi WW, Li HY. Hydrogen Peroxide in Plants: a Versatile Molecule of the Reactive Oxygen Species Network. J Integr Plant Biol. 2008;50:2–18. doi: 10.1111/j.1744-7909.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- 27.Nanda AK, Andrio E, Marino D, Pauly N, Dunand C. Reactive Oxygen Species during Plant-microorganism Early Interactions. J Integr Plant Biol. 2010;52:195–204. doi: 10.1111/j.1744-7909.2010.00933.x. [DOI] [PubMed] [Google Scholar]
- 28.Torres MA, Jones JDG, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–8. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashelford KE, Fry JC, Bailey MJ, Day MJ. Characterization of Serratia isolates from soil, ecological implications and transfer of Serratia proteamaculans subsp. quinovora Grimont et al. 1983 to Serratia quinivorans corrig., sp. nov. Int J Syst Evol Microbiol. 2002;52:2281–9. doi: 10.1099/00207713-52-6-2281. [DOI] [PubMed] [Google Scholar]
- 30.Grimont F, Grimont P. The Genus Serratia. Prokaryotes. 2006;6:219–44. [Google Scholar]
- 31.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, et al. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 2015;43:W104–8. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobrindt U, Hochhut B, Hentschel U, Hacke J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004;2:414–24. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 33.Ali S, Duan J, Charles TC, Glick BR. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J Theor Biol. 2014;343:193–8. doi: 10.1016/j.jtbi.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Taghavi S, van der Lelie D, Hoffman A, Zhang Y-B, Walla MD, Vangronsveld J, et al. Genome Sequence of the Plant Growth Promoting Endophytic Bacterium Enterobacter sp. 638. PLoS Genet. 2010;6:e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitter B, Petric A, Shin MW, Chain PSG, Hauberg-Lotte L, Reinhold-Hurek B, et al. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci. 2013;4:1–15. doi: 10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank AC. Endophytes of Forest Trees. In: Pirttilä AMFAC, Editor. Endophytes of Forest Trees: Biology and Applications. Springer Science+Business Media; 2011. doi: 10.1007/978-94-007-1599-8.
- 37.Schellhorn HG. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol Letters. 1994;131:113–9. doi: 10.1111/j.1574-6968.1995.tb07764.x. [DOI] [PubMed] [Google Scholar]
- 38.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–82. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jong A. Genome2D webserver for analysis and visualization of bacterial genomes and transcriptome data. University of Groningen. http://pepper.molgenrug.nl Accessed 3 Feb 2016.
- 40.Izumi H, Anderson IC, Killham K, Moore ERB. Diversity of predominant endophytic bacteria in European deciduous and coniferous trees. Can J Microbiol. 2008;54:173–9. doi: 10.1139/W07-134. [DOI] [PubMed] [Google Scholar]
- 41.Izumi H. Diversity of Endophytic Bacteria in Forest Trees. In: Pirttilä AMFAC, Editor. Endophytes of Forest Trees: Biology and Applications. Springer Science+Business Media; 2011. doi: 10.1007/978-94-007-1599-8.
- 42.Nicholson WL, Leonard MT, Fajardo-Cavazos P, Panayotova N, Farmerie WG, Triplett EW, et al. Complete Genome Sequence of Serratia liquefaciens Strain ATCC 27592. Genome Announc. 2013;1:27592. doi: 10.1128/genomeA.00548-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mardanova A. Draft genome sequence of Serratia grimesii strain A2. Genome Annouc. 2014;2:7–8. doi: 10.1128/genomeA.00937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicente CSL, Nascimento F, Espada M, Mota M, Oliveira S. Bacteria associated with the pinewood nematode Bursaphelenchus xylophilus collected in Portugal. Ant Van Leeuwenhoek. 2011;100:477–81. [DOI] [PubMed]
- 45.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol. 2014;27:30–7. [DOI] [PMC free article] [PubMed]
- 46.Krause A, Ramakumar A, Bartels D, Battistoni F, Bekel T, Boch J, et al. Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat Biotechnol. 2006;24:1385–91. doi: 10.1038/nbt1243. [DOI] [PubMed] [Google Scholar]
- 47.Alavi P, Starcher MR, Thallinger GG, Zachow C, Müller H, Berg G. Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics. 2014;15:482. doi: 10.1186/1471-2164-15-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter A, Mittler R, Suzuki N. ROS as key players in plant stress signalling. J Exp Bot. 2014;65:1229–40. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Liu Y, Chew SC, Tay M, Salido MMS, Teo J, et al. Complete Genome Sequence and Transcriptomic Analysis of the Novel Pathogen Elizabethkingia anophelis in Response to Oxidative Stress. Genome Biol Evol. 2015;7:1676–85. doi: 10.1093/gbe/evv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, Yang SL, Chung KR, Diaz SA, Mooring EQ, Rens EG, et al. Resistance of oxidative stress via regulating siderophore-mediated iron acquisition by the citrus fungal pathogen Alternaria alternata. Microbiology. Microbiol. 2014;160:970–9. doi: 10.1099/mic.0.076182-0. [DOI] [PubMed] [Google Scholar]
- 51.Eisendle M, Schrettl M, Kragl C, Muller D, Illmer P, Haas H. The Intracellular Siderophore Ferricrocin Is Involved in Iron Storage, Oxidative-Stress Resistance, Germination, and Sexual Development in Aspergillus nidulans. Eukaryot Cell. 2006;5:1596–603. doi: 10.1128/EC.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lambowitz AM, Zirnmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Li X, Milne CB, Janssen H, Lin W, Phan G, Hu H, Jin YS, Price N, Blaschek HP. Development of a Gene Knockout System using Mobile Group II introns (Targetron) and Genetic Disruption of Acid Production Pathways in Clostridium beijerinckii. Applied Environ Microbiol. 2013;79:5853–63. doi: 10.1128/AEM.00971-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng M, Wang X, Templeton LJ, Dana R, Larossa RA, Storz G, et al. DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. J Bacteriol. 2001;183:4562. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubbs JM, Mongkolsuk S. Peroxide-Sensing Transcriptional Regulators in Bacteria. J Bacteriol. 2012;194:5495–503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shanks RMQ, Stella NA, Kalivoda EJ, Doe MR, O’Dee DM, Lathrop KL, et al. A Serratia marcescens OxyR Homolog Mediates Surface Attachment and Biofilm Formation. J Bacteriol. 2007;189:7262–72. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jamet A, Sigaud S, Van de Sype G, Puppo A, Herouart D. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol Plant Microbe Interact. 2003;16:217–25. doi: 10.1094/MPMI.2003.16.3.217. [DOI] [PubMed] [Google Scholar]
- 58.Eason MM, Fan X. The role and regulation of catalase in respiratory tract opportunistic bacterial pathogens. Microb Pathog. 2014;74:50–8. doi: 10.1016/j.micpath.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol Nature. 2013;11:443–54. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu XQ, Li LP, Shen PQ. Feedback regulation of an Agrobacterium catalase gene katA involved in Agrobacterium-plant interaction. Mol Microbiol. 2001;42:645–57. doi: 10.1046/j.1365-2958.2001.02653.x. [DOI] [PubMed] [Google Scholar]
- 61.Margulies M, Egholm M, Altman W, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chevreux B, Wetter T, Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. Comput Sci Biol: Proceedings of the German Conference on Bioinformatics (GCB) 1999;99:45–56. [Google Scholar]
- 63.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 64.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 65.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–20. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 67.Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 70.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takemoto S. Pine Wilt Disease. Tokyo: Springer; 2008. Population ecology of Bursaphelenchus xylophilus. [Google Scholar]
- 72.Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In Vitro Biofilm Formation of Commensal and Pathogenic Escherichia coli Strains: Impact of Environmental and Genetic Factors. J Bacteriol. 2006;188:3572–81. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer 3- new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the TreeBase repository (http://purl.org/phylo/treebase/phylows/study/TB2:S19116), in the European Nucleotide Archive (ENA) under the accession ERP013273, and included within the article and its additional files.


