Abstract
The detection of double-stranded (ds) DNA by SYBR Green I (SG) is important in many molecular biology methods including gel electrophoresis, dsDNA quantification in solution and real-time PCR. Biophysical studies at defined dye/base pair ratios (dbprs) were used to determine the structure–property relationships that affect methods applying SG. These studies revealed the occurrence of intercalation, followed by surface binding at dbprs above ∼0.15. Only the latter led to a significant increase in fluorescence. Studies with poly(dA) · poly(dT) and poly(dG) · poly(dC) homopolymers showed sequence-specific binding of SG. Also, salts had a marked impact on SG fluorescence. We also noted binding of SG to single-stranded (ss) DNA, although SG/ssDNA fluorescence was at least ∼11-fold lower than with dsDNA. To perform these studies, we determined the structure of SG by mass spectrometry and NMR analysis to be [2-[N-(3-dimethylaminopropyl)-N-propylamino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]. For comparison, the structure of PicoGreen (PG) was also determined and is [2-[N-bis-(3-dimethylaminopropyl)-amino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+. These structure–property relationships help in the design of methods that use SG, in particular dsDNA quantification in solution and real-time PCR.
INTRODUCTION
The detection of nucleic acids by fluorescent dyes such as SYBR Green I (SG) has become increasingly important for a variety of analytic and diagnostic applications (1). Since its introduction in the early 1990s (2), SG has been applied successfully in the detection of nucleic acids in gels (3–6), in solution (7,8), in the determination of DNase or telomerase activities (9,10), in fluorescence imaging techniques (11), in flow cytometry (12,13), in real-time PCR (14–17), in biochip applications (18), and more recently, in the demanding quantification of dsDNA in crude extracts of environmental samples that is usually hampered by a variety of quenching processes (19,20).
The wide spectrum of applications of SG and its overall convincing performance are due to its excellent properties. These include favorable photophysical properties, temperature stability, selectivity for dsDNA and high sensitivity (1,2). Furthermore, SG assays for the quantification of dsDNA in solution can be optimized to result in easy, robust and reliable one-step procedures that display a dynamic linear range of up to four orders of magnitude using a single dye solution (7,19).
Despite the excellent performance and significant impact of SG in analytic and diagnostic applications, studies on the structure–property relationships have not yet been performed in detail. The alterations in fluorescence emission of free and dsDNA-bound SG and the underlying mechanisms depend on the structure and binding mode of SG. Initially, the biphasic dependence of increasing guanidinium hydrochloride on SG/dsDNA complex fluorescence intensities (7) led to the notion that binding of SG to dsDNA may be described by at least two binding modes, intercalation and external binding (1), which was confirmed by subsequent studies (16,19). But this notion has not been proven yet. The dependence of the binding events on the dye/base pair ratios (dbpr) in conjunction with other parameters is also not known. However, this can significantly influence the performance of nucleic acids testing and the design of new methods (21). The structure of SG and its concentration in the commercial reagent have not been previously published, although some information may be deduced from the literature. According to the United States patent 5658751 (22) the core structure of SG is based on a monomeric unsymmetrical cyanine dye comprising an N-alkylated benzothiazolium or benzoxazolium ring system, which is linked by a monomethine bridge to a pyridinium or quinolinium ring system that carries a substituent with a heteroatom (1).
We have therefore determined the structure of SG, and its concentration within the commercially available reagent. With this knowledge we measured the binding of SG to different nucleic acids at defined dbprs. Also, absorption and fluorescence studies, gel electrophoresis and hydrodynamic measurements were performed. As initial studies indicated that SG and PG display comparable photophysical and DNA binding properties (7), the structure of PG was also resolved and fluorimetric titration experiments were performed to compare the results for both stains. Finally, the implications of the results on the use of SG are discussed.
MATERIALS AND METHODS
Materials
Ethidium bromide (EtBr), Hoechst 33258 (H 33258), PG and SG were purchased from Molecular Probes (Leiden, Netherlands), highly polymerized calf thymus (ct)DNA from Sigma-Aldrich (Deisenhofen, Germany) and the homopolymers poly(dA) · poly(dT) (dA · dT) and poly(dG) · poly(dC) (dG · dC) were from Amersham Pharmacia Biotech (Freiburg, Germany). DNA from bacteriophage lambda cI857 Sam 7 (λDNA) was obtained from Roche Diagnostics (Mannheim, Germany). The 10 bp DNA ladder was from Life Technologies (Karlsruhe, Germany). Miscellaneous chemicals and reagents were purchased from Merck (Darmstadt, Germany), Carl Roth (Karlsruhe, Germany) and Life Technologies (Karlsruhe, Germany) and were of the highest analytical grade available.
Methods
Commercially available SG reagent is dissolved in dimethyl sulfoxide (DMSO) (2). For NMR and mass spectrometry-fast atom bombardment (MS-FAB) experiments DMSO was removed by freeze drying. NMR experiments were carried out on a Bruker AC-250 spectrometer from Bruker Daltonik GmbH (Bremen, Germany) after dissolving the freeze-dried SG reagent in methanol-d4 with tetramethylsilane as internal standard. For determination of the accurate molecular mass of SG, the lyophilized SG reagent was dissolved in m-nitrobenzylalcohole and MS-FAB measurements were performed on a Finnigan MAT 95 mass spectrometer. To investigate fragmentation of the SG molecule and basic reactions of SG in aquatic solution the SG reagent was diluted 50-fold in water. MS and MS/MS spectra were recorded on Finnigan LCQ Deca mass spectrometer from Thermoquest (Egelsbach, Germany) with electrospray ionization. For structure analysis of PG, the PG reagent was diluted in DMSO and analyzed by MS/MS studies. The sum of anions and cations within the commercially available SG reagent was determined by using the ion-chromatograph DX100 from Dionex (Idstein, Germany) equipped with the column AS14 or AS11 (Dionex).
Solutions of dsDNA were prepared with 10 mM tris(hydroxymethyl)-aminomethane (Tris), pH 7.5, containing 1 mM ethylenediaminetetraacetic acid (EDTA) (TE buffer). dA · dT and dG · dC stock solutions were prepared in buffer containing 50 mM Tris, pH 7.5, 1 mM EDTA and 100 mM NaCl (TES buffer). For fluorescence measurements, dA · dT and dG · dC stock solutions were diluted in TE buffer. The concentration and purity of the stock solutions were determined by measuring the absorbance at 260 and 280 nm (23). ctDNA was solubilized by mild sonication (19). Analysis by agarose gel electrophoresis showed no profuse fragmentation.
Escherichia coli DH5α containing the plasmid pACYC184 (New England BioLabs, Beverly, USA) was cultured according to Sambrook et al. (23) and used as a pACYC184 source. The plasmid was purified by applying the Qiaprep Spin Miniprep kit from Qiagen (Hilden, Germany). Linearization of pACYC184 was performed by EcoRI restriction endonuclease (Promega, Mannheim, Germany) digestion according to the supplier's protocol and purified by applying the QIAquick PCR purification kit from Qiagen. For the preparation of ssDNA, an aliquot of EcoRI-digested pACYC184 was diluted with 20 mM Tris, pH 9.0, and denatured by heating (95°C, 3 min) according to Porter et al. (24). The sample was transferred into an ice/water bath after incubation. Complete denaturation was checked by gel electrophoresis. Another aliquot of EcoRI-digested pACYC184 that was not heat-denatured was used in the same buffer as control. Human embryonal kidney 293 T cells (25) were grown overnight in serum-derived medium RPMI-1640 from Invitrogen Life Technologies (Karlsruhe, Germany) and used as a source for total RNA. Isolation of total RNA was performed with the E.Z.N.A. total RNA kit from Peqlab (Erlangen, Germany).
The double-beam spectrophotometer Specord 200 from Analytik Jena (Jena, Germany), the Lambda2 spectrophotometer from Perkin Elmer (Überlingen, Germany; now Applied Biosystems) and the SpectraMax Plus384 plate reader from Molecular Devices (Ismaning, Germany) were used for spectrophotometric measurements. UV/VIS absorption spectra were carried out using plastic cuvettes from Brand (Wertheim, Germany) and UV microplates from Greiner Labortechnik (Frickenhausen, Germany). Interaction studies were performed by differential spectrophotometry as described in (26,27).
Spectrofluorimetric investigations were carried out with the Fluoromax 3 from Jobin Yvon (Grasbrunn, Germany), the Spectrafluor Plus from Tecan (Männedorf, Germany) and the Spectramax Gemini XS from Molecular Devices. Unless stated otherwise, excitation and emission wavelengths of 485 and 524 nm, respectively, were used for experiments with SG and PG (7,28). H 33258 was excited at 360 nm and fluorescence emission was registered at 465 nm (20).
In the present paper, the units dye/base pair ratio or dye/base ratio (dbr) are defined, respectively, as moles of dye per mole of DNA base pair or base. Investigations were carried out at room temperature with the solutions protected from light and incubated for ∼10 min before measurements were started (7).
Rod-like dsDNA for use in viscometry experiments was prepared by sonication of ctDNA in TE buffer pH 7.5. To avoid the presence of ssDNA due to the sonication procedure, the sample was heated to 95°C and renatured by a slowly decreasing temperature gradient. Fragmentation of the ctDNA was confirmed by gel electrophoresis. ctDNA fragment size was between 100 and 600 bp. Viscosity measurements were performed using an Ostwald-type viscometer (0.35 mm diameter of the capillar) at 21°C (29) and applying 0.5 mM rod-like ctDNA in TE buffer pH 7.5 with or without dye, i.e. EtBr, H 33258 or SG. dbprs of 0.1 were used.
Agarose (1%, w/v) gel electrophoresis was performed in 20 mM Tris–acetate, 2 mM EDTA, pH 8.2 (TAE). Experiments were carried out under light protection. DNA samples were incubated for ∼10 min with SG before loading the gels. When necessary, gels were stained with EtBr (0.5 μg/ml TAE buffer) for 0.5 h and destained for 1 h in TAE buffer to check whether alterations in SG fluorescence intensities were due to inadequate loading of the gel or based on the dbprs applied. Imaging was carried out with the IDA Gel Documentation System and the AIDA version 2.0 software from Raytest Isotopengerätebau (Straubenhardt, Germany). Fluorescence excitation was performed at 312 nm. Fluorescence emission was detected using an EtBr or a SG gel stain photographic filter purchased from Molecular Probes.
RESULTS
Determination of the structure of SG and PG
To permit investigations on the binding mode of SG to dsDNA and its implications for methodological applications, it was essential to analyze the structure of SG by MS, MS/MS and NMR, and determine the concentration of SG within the commercially available SG reagent by ion chromatography. MS-FAB analysis of the lyophilized SG reagent revealed a parent peak (M+•) with a mass/charge (m/z) ratio of 509.273 ± 0.005 a.m.u. (data not shown), which corresponds to the molecular formula C32H37N4S (calculated molecular mass: 509.2739 g/mol). Other peaks, correlated to the putative structure of SG, showed only minor intensities, e.g. m/z ratios of 424.2 and 255.1. The structure was further analyzed by additional MS and MS/MS studies after diluting the SG reagent 50-fold in water. Here, protolytic reactions, displayed by signals at m/z ratios of 255.3 and 424.3, were observed. The following m/z ratios were detected in several MS experiments:
MS (full scan, detected m/z range = 100 − 600): 509.3 (100%; M+•), 424.4 (65%), 409.4 (16%), 366.4 (15%), 255.3 (30%);
MS/MS of m/z ratio = 509.3 (detected m/z range = 200 − 520): 509.3 (100%; M+•), 367.4 (54%);
MS/MS of m/z ratio = 424.3 (detected m/z range = 115 − 520): 424.3 (25%), 409.4 (100%), 276.1 (51%), 233.3 (9%);
MS/MS of m/z ratio = 255.3 (detected m/z range = 70 − 520): 424.3 (100%), 184.2 (34%), 86.1 (15%).
In addition, the structure of SG was investigated by NMR experiments, which gave the following 1H and 13C spectra (chemical shift values given in p.p.m.):
1H: 8.55–7.16 (m, 14H), 6.83 (s, 1H), 3.98 (s, 3H), 3.30 (t, 2H), 3.24 (t, 2H), 2.31 (t, 2H), 2.26 (s, 6H), 1.59 (q, 2H), 1.39 (q, 2H), 0.85 (t, 3H);
13C: 160.79 (C), 160.03 (C), 151.48 (C), 142.36 (C), 142.25 (C), 140.65 (C), 133.68 (CH), 2x 131.97 (CH), 131.53 (CH), 2x 131.01 (CH), 129.45 (CH), 127.15 (CH), 125.97 (CH), 125.56 (CH), 125.31 (C), 123.81 (CH), 123.50 (C), 120.17 (CH), 113.37 (CH), 104.06 (CH), 88.35 (CH), 57.74 (CH2), 56.14 (CH2), 51.89 (CH2), 2x 45.35 (CH3), 33.93 (CH3), 25.93 (CH2), 21.45 (CH2), 11.76 (CH3).
Based on these data, the structure of SG was determined to be [2-[N-(3-dimethylaminopropyl)-N-propylamino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+ as shown in Table 1. As preliminary studies indicated that SG and PG show similar photophysical and DNA binding properties (7), the structure of PG was elucidated for comparison. PG reagent was diluted 100-fold in DMSO and analyzed by MS/MS. The following m/z ratios were detected:
Table 1. Chemical structures of monomeric cyanine dyes based on the same core structure.
The structure of SG [2-[N-(3-dimethylaminopropyl)-N-propylamino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+ as determined by MS and NMR studies. The structure of PG [2-[N-bis-(3-dimethylaminopropyl)-amino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+ was evaluated by MS/MS experiments.
MS (full scan, detected m/z range = 100 − 1000): 552.5 (100%; M+•), 467,5 (4%), 410.5 (7%), 367.5 (8%);
MS/MS of m/z ratio = 552.5 (detected m/z range = 150 − 600): 552.5 (100%; M+•), 367.5 (47%).
As indicated by the MS/MS experiments, the parent peaks of SG and PG showed the same fragment ions with a m/z ratio of 367.5. For SG, this fragment was assigned to [[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+. Based on these MS data, it became evident that SG and PG share a common core structure. Further analysis of the detected parent peak of PG and its fragmentation revealed that PG is [2-[N-bis-(3-dimethylaminopropyl)-amino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+ (Table 1).
Recording of the MS-FAB spectrum of SG involved a lyophilization step to remove DMSO and dissolving the remaining compound in m-nitrobenzylalcohole. The resulting spectrum showed a single characteristic peak, the parent peak of SG. For the MS fragmentation studies, the SG reagent was diluted in water. Several peaks were recorded in the spectra and could be assigned to the structure of SG. The peak at the m/z ratio of 255.3 with a relative intensity of 30% (see above) corresponded to the double-charged form of SG. On the contrary, in the MS-FAB spectrum a peak of minor intensity below ∼2% was determined at the m/z ratio of 255.3.
Determination of the SG concentration and the molar absorption coefficient
As the data of the MS-FAB spectrum showed that SG was dominantly present in the single-charged form within the SG reagent, the molar concentration of SG within the reagent was determined by analyzing the sum of anions and cations by ion chromatography. Calculated as charge equivalents, the analysis revealed ∼14 mM chloride, ∼1.4 mM nitrate and ∼3.4 mN sulfate. Other anions, in particular typical dye counter anions like iodide and perchlorate, could not be detected. As for the cations, only trace amounts of sodium ions, 0.3 mM, were found. In sum, this led to a net normality of negative ions of at least 18.5 mN, which corresponded to a concentration of SG within the reagent of at least 9.4 mg/ml. Because of manufacturing reasons, one can easily conclude that the actual concentration is very likely to be ∼10 mg/ml, a common concentration of stock solutions of various dyes, including EtBr, H 33258, H 33342 and acridine orange (2). Thus, the SG reagent is likely to be a DMSO stock solution of 10 mg/ml SG, which corresponds to a molar concentration of ∼19.6 mM. Accordingly, the molar absorption coefficient of SG at the absorption maximum at 494 nm was determined to be ∼73 000 M−1 cm−1 in TE buffer pH 7.5.
Spectrophotometric investigations of SG in complex with dsDNA
Based on these results, absorption spectra of dsDNA-bound SG were recorded at defined dbprs to investigate the binding mode of SG to dsDNA (data not shown). TE buffer pH 7.5 containing 5.6 μM SG was incubated with increasing λDNA concentrations up to 22.4 μM bp. The overlay of the spectra revealed an isosbestic point at ∼408 nm. The visible absorption band of unbound SG at ∼494 nm was influenced in position, magnitude and shape upon binding to dsDNA (batho- and hypsochromic shifts depending on the dbpr). With respect to the absorption maximum of the free dye, binding of SG to dsDNA resulted in a slight increase in absorption maxima of ∼0.5% at low dbprs of ∼0.1 and 0.4. In contrast, higher dbprs of 1 and 4 revealed significant decreases in intensity of ∼4 and ∼16%, respectively.
As the interaction between SG and dsDNA influenced particularly the visible absorption band of SG, the interaction was analyzed in detail by differential absorption spectroscopy as described elsewhere (26,27). For differential spectroscopic studies at 494 nm equal volumes of SG solutions were added successively to a reference containing only TE buffer pH 7.5 and to the sample cuvette containing 15 μM bp of λDNA in TE buffer pH 7.5.
The studies displayed a dependence with three distinct areas (data not shown). First, differential absorption values increased with rising dbprs up to a maximum between dbprs of 0.1 and 0.15. Second, the titration curve decreased and reached a minimum at a dbpr of ∼2.5. Third, a further increase in the dbpr resulted in rising differential absorption values that started to approach a maximum at a dbpr of ∼10. Precipitation of SG/dsDNA complexes, probably due to high dye and DNA concentrations, prevented investigations at dbprs >12. In the range of these high dbprs, a fourth area of the dependence, the saturation area, began to show up (data not shown).
Fluorescence intensity of SG in complex with dsDNA
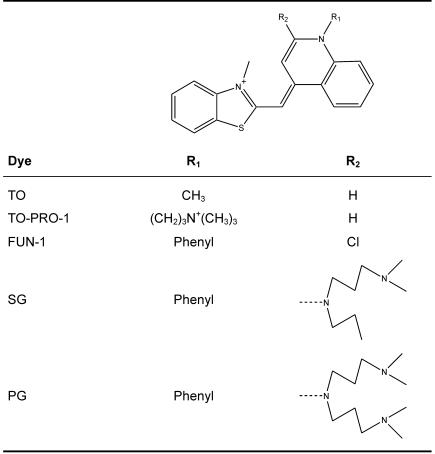
The dependence of the fluorescence intensities mirrored the dependence of the absorption values to a certain extent. Three distinct areas are displayed in Figure 1. First, up to a dbpr of ∼0.15 only a minor increase in fluorescence was observed. Second, between dbprs of ∼0.15 and ∼2 an abrupt increase in fluorescence occurred. Third, around a dbpr of 2.5 the steep increase in fluorescence intensities of the second phase entered a less steep increase until above dbprs of 3. A maximum was approached at a dbpr of ∼10. Then fluorescence intensities were more or less constant up to dbprs of ∼100. When a dbpr of 1000 was applied, fluorescence intensity decreased to ∼24% of that at a dbpr of 10 (latter data not shown). In contrast to differential absorption spectroscopy, higher dbprs could be investigated without precipitation of SG/dsDNA complexes due to much lower concentrations of SG and DNA applied (see experimental settings in Figure 1).
Figure 1.
Dependence of the relative fluorescence intensity on the dbpr of the SG/dsDNA complex. Samples containing 0.15 or 0.015 μM base pairs of λDNA were incubated with increasing dye concentrations ranging from 0 to 2.1 μM or from 0.15 to 1.52 μM, respectively.
The fluorescence intensities of λDNA, ctDNA, EcoRI-digested pACYC184, 10 bp DNA ladder at dbprs >0.15 were significantly different (data not shown). Setting the fluorescence intensity of the SG/λDNA complex at a dbpr of 10–100%, its fluorescence intensity at a dbpr of 4 was 93%. Fluorescence intensities of SG/EcoRI-digested pACYC184 complexes were 88 and 99% at dbprs of 4 and 10, respectively. Fluorescence intensities of SG/ctDNA complexes were 85 and 84%, respectively and for the 10 bp DNA ladder fluorescence intensities of 45 and 48% at dbprs of 4 and 10 were observed, respectively. For SG in complex with total RNA, fluorescence intensities were only 14 and 16% at the dbprs of 4 and 10, respectively.
Despite the discrepancies between different dsDNAs with respect to the dependences of the fluorescence intensities on the dbprs, fluorescence emission spectra were similar (data not shown). Fluorescence emission spectra recorded at dbprs of 0.4, 1.0 and 4.0 displayed the characteristic fluorescence emission band of the SG/dsDNA complex with an emission maximum at ∼524 nm. The emission maximum was not shifted batho- or hypsochromically within dbprs of 0.4 and 10 to a significant extent (data not shown).
Electrophoretic mobility of the SG/dsDNA complex
Electrophoretic gel mobility shift assays were performed to characterize the binding of SG to dsDNA in more detail, because they provide information on the electrophoretic stability, mobility and fluorescence intensities of the SG/dsDNA complexes at once (19).
A reduction of the electrophoretic mobility of dsDNA (EcoRI-digested pACYC184) was observed upon an increase of the dbpr of pre-stained dsDNA samples up to a dbpr of ∼1 (data not shown). Of note, at low dbprs the bands were sharp, but started to become diffuse and less intense at dbprs above ∼0.5, which may indicate partially denatured or degraded DNA, eventually by SG, undetectable sample components at low dbprs and/or an inhomogeneous distribution of SG molecules bound to dsDNA (data not shown).
DNA degradation may be based on photocleavage (30). Photocleavage of DNA by the cyanine dyes YO, YOYO and TOTO has been shown to mainly occur via single-strand cleavages (30), which is rather unlikely to lead to the observed diffuse bands. On the other hand, photocleavage mechanisms have been reported to be dependent on the dbpr, and single-strand breaks may accumulate to eventually produce double-strand cleavages (30) that may account for diffuse bands. However, as we performed electrophoretic mobility studies under light protection, one may hypothesize that DNA degradation may rather have occurred via backbone cleavage due to a nucleophilic attack of basic residues of SG, e.g. by SG's exocyclic secondary amine group.
In addition to DNA degradation, inhomogeneous distribution of SG molecules bound to dsDNA may account for the observation too. This scenario could explain why the mobility of diffuse bands at dbprs between 1.0 and 10 revealed the same front line. A closer look at the effects at low dbprs, showed that a significant increase in fluorescence intensity was observed when the dbpr reached a value between 0.13 and 0.17 (data not shown). Further studies that are beyond the scope of this work need to be performed to clarify the mechanisms underlying the observed diffuse bands and their intensity decrease at dbprs above ∼0.5. Of course, the intensity decrease may also be attributed to quenching processes that are particularly prominent at high dye concentrations (19).
Hydrodynamic studies of SG/dsDNA solutions
To determine whether intercalation occurred, hydrodynamic studies were performed. Intercalation of an agent into dsDNA leads to lengthening of the double helix. The increase in length of rod-like dsDNA (L) can be detected by hydrodynamic methods, e.g. viscosity measurements (31). In such measurements, viscosity is proportional to L3, and the relative viscosities in the presence or absence of an agent are calculated according to
![]()
where t0 is the measured flow time of the buffer and t is the observed flow time of the DNA solution (31). Generally, viscosity values are displayed as (η/η0)1/3. η0 and η indicate the relative viscosities of the dsDNA solution in the absence and presence of the dye, respectively. Hydrodynamic studies performed with 0.5 mM bp of rod-like ctDNA, revealed (η/η0)1/3values of 1 ± 0.03, 0.96 ± 0.07, 1.15 ± 0.08 and 1.13 ± 0.06, respectively, for uncomplexed rod-like ctDNA and at dbprs of 0.1 for the minor groove binding dye H 33258, the intercalator EtBr and for SG. As expected, the minor groove binding dye H 33258 did not increase relative viscosity. However, a significant increase of relative viscosity was observed when the intercalator EtBr was applied. The same was true for SG. This did clearly indicate an intercalative binding mode of SG at low dbprs.
Investigations on the surface binding of SG to dsDNA
After the hydrodynamic studies revealed intercalation of SG into dsDNA at low dbprs the binding mode of SG at higher dbprs was investigated in more detail. As agents which interact non-covalently with dsDNA mainly bind through intercalation or groove binding, in particular minor groove binding (32), and as monomeric cyanine dyes like TO (Table 1) and YO with structure homologies to SG were shown to express DNA surface binding at high dbprs (33,34), it appeared likely that SG binding at high dbpr was based on minor groove binding as well.
To determine whether minor groove binding may occur, differential absorption and fluorimetric studies were performed with dA · dT and dG · dC homopolymers and compared to λDNA (Figure 2). These homopolymers were chosen because their minor grooves differ in the electrostatic potential and because of their ability to form hydrogen bonds with a bound ligand and the sterical aspects that influence the binding of a dye (32). Therefore, surface binders, in particular minor groove binders, generally express a marked sequence specificity.
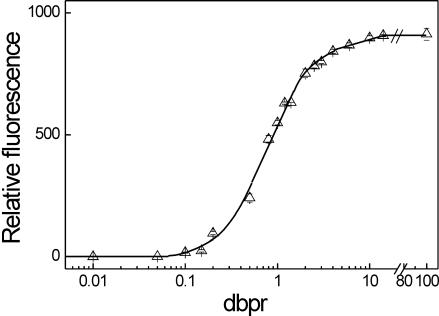
Figure 2.
Dependences of differential absorption values (A) and fluorescence intensities (B) of SG bound to λDNA (triangle), dA · dT (circle) and dG · dC (square). TES buffer pH 7.5 containing 15 μM base pairs of a nucleic acid type was used for differential absorption measurements. Fluorescence intensity studies were performed in TE buffer pH 7.5 containing 0.15 μM base pairs of a nucleic acids polymer.
The interaction analysis of SG with λDNA, dA · dT and dG · dC homopolymers performed by differential absorption spectroscopy at 494 nm revealed similar dependences at dbprs up to ∼0.1 (Figure 2A). Marked changes became obvious at higher dbprs (Figure 2A). In the experimental settings, TES buffer, pH 7.5, was applied. In contrast to the outcome with TE buffer, pH 7.5, comparable alterations of differential absorption values occurred at higher dbprs. This spread of data facilitated the analysis of changes. The data are spread, because salts (TES buffer contains 100 mM NaCl) reduced the affinity of SG to dsDNA (see below, Figure 3). Also, the application of TES buffer was recommended by the supplier of the homopolymers.
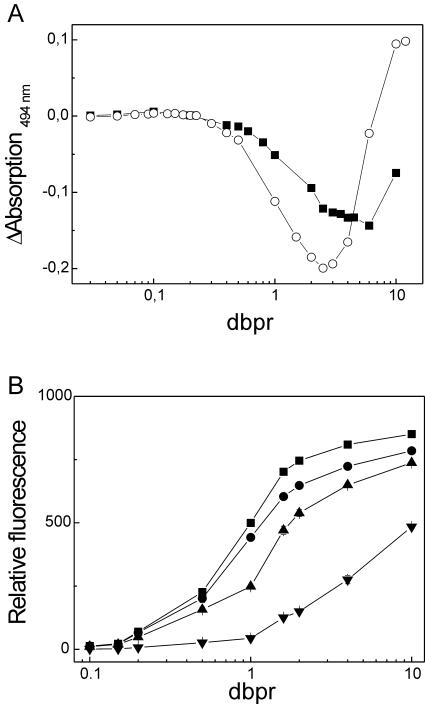
Figure 3.
The influence of NaCl on the dbpr dependence of SG/dsDNA differential absorption values (A) and fluorescence (B). (A) Differential absorption values at 494 nm of SG/λDNA complexes at increasing dbprs in TE buffer pH 7.5 that did not contain NaCl (circle) and in TES buffer pH 7.5 that contained 100 mM NaCl (solid square). (B) Samples containing 0.15 μM bp of λDNA were titrated with increasing amounts of SG ranging from 0 to 1.5 μM in the presence of 0 mM (solid squares), 25 mM (solid circles), 50 mM (solid triangles, up) and 300 mM (solid triangles, down) NaCl.
In agreement with the photometric studies, the overall characteristics of the dependences of fluorescence intensities of SG in complex with λDNA, dA · dT and dG · dC homopolymers with increasing dbprs were similar, but differences in the magnitudes were observed (Figure 2B). Dependences were similar up to dbprs of ∼0.2 (Figure 2B). Significant changes were observed at dbprs above ∼0.5. At a dbpr of 4, dA · dT- and dG·dC-bound SG reached ∼90 and ∼40% of the fluorescence of the SG/λDNA complex, respectively. Of note, at dbprs above ∼2, the fluorescence intensities of the SG/dG·dC complexes decreased slightly with increasing dbprs (Figure 2B), which may be due to collisional quenching (19). Maximal fluorescence intensity was detected at a dbpr of 2. Fluorescence intensities were 0.9-fold lower at a dbpr of 10.
The data show that SG exhibits sequence-specific binding, in particular at dbprs above ∼0.2. This may indicate minor groove binding. Of note, the differences between fluorescence intensities of PG/dA · dT complexes and PG/dG · dC complexes compared to PG/λDNA complexes were more pronounced than those of the respective SG/DNA complexes. However, the differences between the dye/dA · dT complexes and dye/dG · dC complexes of PG were lower than those of SG. Fluorescence intensities of PG/dA · dT complexes and PG/dG · dC complexes revealed ∼45 and ∼30% of the fluorescence intensity of the PG/λDNA complex at dbprs of 4.
Effects of mono- and divalent cations on the fluorescence of SG/dsDNA complexes
Another profound influence on SG assays can be expected from salts, because the affinity of cationic dyes to dsDNA can be sensitive to the ionic strength of the solution (35). Therefore, the impact of cations on differential absorption values and the fluorescence of SG/dsDNA complexes depending on the dbpr were assessed. The dependence of differential absorption values on the dbprs was significantly altered at dbprs above ∼0.3 in the presence of 100 mM NaCl (Figure 3A). More detailed fluorescence studies confirmed the impact of salts. Here, it should be noted that increasing concentrations of neither NaCl nor MgCl2 led to changes in the shape of the corresponding emission and excitation spectra of the SG/dsDNA complexes (data not shown). However, the dependence of the fluorescence intensities of SG/dsDNA complexes on the dbpr appeared to be significantly influenced by the NaCl concentration (Figure 3B). The most significant impact of NaCl was observed at dbprs of ∼1. The same was true for MgCl2 (data not shown). The influence of MgCl2 was more pronounced than that of NaCl.
To quantitatively correlate fluorescence quenching to the concentration of the quencher, i.e. the salts, data were transformed into Stern–Volmer plots according to the Stern–Volmer equation (36):
![]()
where F0 and F are the fluorescence intensities in the absence and presence of the quencher, respectively. KSV is the Stern–Volmer constant and Q is the concentration of the quencher. KSV−1 represents the quencher concentration where only 50% of the quencher-free fluorescence intensity is detected. Consequently, KSV−1 provides quantitative information on fluorescence quenching. In this case, it may be used to indicate the extent of change in the affinity of the dye to dsDNA with increasing quencher, i.e. salt concentrations.
At dbprs of 0.1, 0.15, 0.2, 0.5, 1.0, 1.6 and 2.0, the Stern–Volmer plots were characterized by a non-linear behaviour with upward curvatures, i.e. concave toward the y-axis (data not shown). This showed that the impact of salts was not based on a single quenching mechanism. However, with increasing dbprs Stern–Volmer plots converted into linear dependences. This was probably due to the increasing dominance of one quenching mechanism. Based on the linear dependences for dbprs of 4 and 10, KSV−1 values of 155 and 408 mM for NaCl, and 12 and 47 mM for MgCl2 were determined by linear regression, respectively. Correlation coefficients were 0.9901 and 0.9973, respectively, for the experiments with NaCl, and 0.9990 and 0.9646, respectively, for the studies with MgCl2.
The KSV−1 values showed that MgCl2 reduced the affinity of SG to dsDNA to a much greater extent than NaCl. As the magnitude of the difference between the KSV−1 values of NaCl and MgCl2 may not only be described by the different ionic forces, the influence of MgCl2 on the structure of dsDNA could play a role as well.
Binding of SG to ssDNA
SGs selectivity for dsDNA is important for many of its applications, but surely crucial in SG real-time PCR and dsDNA melting curve analysis (16,37,38). Therefore, the binding behaviour of SG to ssDNA was investigated. As absorption measurements displayed rather weak signals, fluorimetric and electrophoretic mobility studies were chosen for the investigation.
When compared to SG/λDNA complexes, SG/ssDNA complexes exhibited a different dependence of fluorescence intensities on the dbr (Figure 4A). Maximal fluorescence was observed at a dbr of ∼0.25. An increase of the dbr resulted in a decrease of fluorescence intensities, which was possibly due to quenching (19). At dbrs of 0.25, 1 and 10, the fluorescence intensities of the SG/ssDNA complex were only 8, 6 and 4%, respectively, when compared to the SG/dsDNA complex at dbprs of 10.
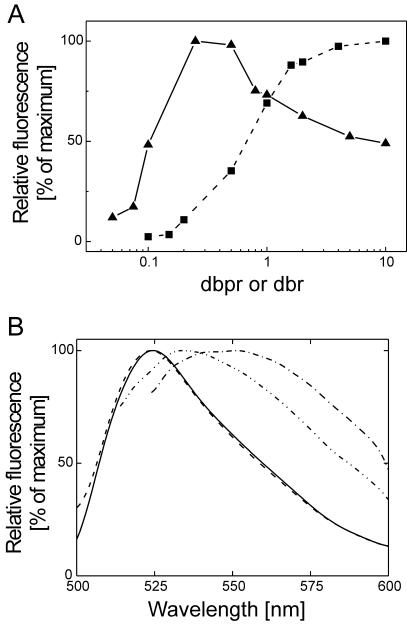
Figure 4.
Comparison of fluorescence of SG/ssDNA complexes and SG/dsDNA complexes. (A) Dependence of fluorescence intensities of SG/ssDNA complexes on dbr in comparison with the corresponding SG/dsDNA complexes. Samples containing 0.15 μM (in bp) EcoRI-digested pACYC184 plasmid (solid squares) or 0.3 μM (in bases) heat-denatured, EcoRI-digested pACYC184 plasmid (solid triangles) were incubated with increasing concentrations of SG (0–1.5 or 0–3 μM, respectively). For reasons of comparison, the fluorescence intensities of the SG/dsDNA complex and of the SG/ssDNA complex were set to 100% at a dbpr of 10 and 0.25, respectively. (B) Fluorescence spectra of SG in complex with dsDNA (EcoRI-digested pACYC184) at dbprs of 1 (dashed line) and 10 (solid line) and with ssDNA (heat-denatured EcoRI-digested pACYC184) at dbr of 1 (dash dot dot line) and 10 (dash dot line).
The fluorescence emission bands of SG in complex with ssDNA were significantly broader and the emission maxima were found at longer wavelengths (Figure 4B). At dbrs of 2 and 10, the emission maxima were detected at 535 and 552 nm, respectively. By contrast, the fluorescence emission bands of SG in complex with dsDNA were not influenced at dbprs of 1 and 10.
Fluorescence of SG bound to ssDNA was low, but the results did support an interaction of SG with ssDNA. This notion was further analyzed by electrophoretic mobility shift assays (data not shown). EcoRI-digested pACYC184 was denatured by heating to obtain ssDNA, which shows an increased electrophoretic mobility compared to dsDNA (24). Aliquots of the same volume were taken as dsDNA controls before heat denaturation. Before electrophoresis, aliquots of the ssDNA solution containing 227 pmol bases were incubated with increasing amounts of SG up to 182 pmol. In contrast to the results obtained for dsDNA, interaction of SG with ssDNA increased the electrophoretic mobility of ssDNA. Relative to unstained ssDNA, binding of SG to ssDNA resulted in a continuous increase in fronting of the SG/ssDNA bands with increasing dbrs. At dbrs ∼0.5 fronting reached a maximum. An increase of the electrophoretic mobility of dye/ssDNA bands was also observed for M13mp18 ssDNA on binding to the unsymmetrical cyanine dye thiazole orange homodimer (TOTO) (39). Here, it was postulated that the extended, coil-like structure of ssDNA was altered by interaction with a cationic dye due to diminished coulombic repulsions between the negative charges of phosphate groups, resulting in a complex with decreased sphere-resistance during electrophoresis (39). The same could be true for the SG/ssDNA complexes.
DISCUSSION
Structure
We have determined the structure of SG (Table 1), its concentration within the SG reagent and have characterized its binding to dsDNA to provide insight into its structure–property relationships, as this knowledge affects the use of SG in nucleic acids testing. MS and NMR investigations showed that SG shares its core structure, 4-[[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-quinolinium]+. with dyes like TO, TO-PRO-1 and FUN-1 (Table 1) (2,40).
Determination of the structure of PG (Table 1) revealed significant structure homologies between SG and PG. The structure and properties observed for SG and PG were in alignment with those displayed in the US patent for Molecular Probes dyes 937 and 993, respectively. As expected from the structural homology, SG and PG possess similar absorption coefficients of ∼73.000 M−1 cm−1 at 494 nm (see above) and ∼70.000 M−1 cm−1 at 500 nm (28), respectively. For SG, it should be noted that the concentration of SG was determined indirectly, based on some assumptions, which may have introduced some errors. However, the agreement of the absorption coefficients of SG and PG suggest that the counter ion approach is reliable.
Both, SG and PG, carry a phenyl group at position one of the quinolinium ring, but SG has a N-(3-dimethylaminopropyl)-N-propylamino and PG a N-bis-(3-dimethylaminopropyl)-amino residue at position two of the quinolinium ring (Table 1). Therefore, we advocate that SG carries two positive charges under standard conditions, whereas PG carries three positive charges. One positive charge is based on the electron deficiency of the conjugated, mesomeric heteroaromatic systems linked by a methine bridge. One or two additional charges are contributed by the protonation of the amino group(s) of the 3-dimethylaminopropyl residue(s) of SG or PG, respectively.
The two positive charges of SG are likely to contribute to the high binding affinity for dsDNA. This may explain why SG/dsDNA complexes are stable under electrophoretic conditions. Benson et al. (41) showed that dye/dsDNA complexes, pre-stained with analogs of EtBr or TO bearing various polycationic substituents, displayed lower off-rates under electrophoretic conditions than dye/dsDNA complexes pre-stained with EtBr or TO. They concluded that the derivatives were bound more tightly to dsDNA due to the polycationic side chains. Also, the introduction of cationic side chains into the cyanine dye TO resulted in a series of derivatives, e.g. TO-PRO-1 (Table 1), which exhibited higher binding affinities for nucleic acid polymers than the parent compound (42).
dsDNA binding
SG was shown to intercalate at low dbprs by hydrodynamic studies that have been reported to reliably indicate intercalation (31). In agreement with most simple intercalators (32) and PG (28), SG did not display a marked sequence binding preference at low dbprs, i.e. intercalation, as shown by differential absorption spectroscopy or fluorimetric studies. A significant sequence dependence of SG binding was observed at high dbprs though (see below). Here, it should be noted that the reported property of PG, not to be sequence specific (28), does apply only to low dbpr, but not to high dbprs that are generally used for the quantification of DNA in solution by PG and SG. Fluorescence intensities of the SG/dsDNA complexes at dbprs below ∼0.15 were only marginal. Marked radiative relaxation of dsDNA-bound SG occurred at dbprs above ∼0.2. Therefore, other binding events or reorientation must have taken place.
Investigations on the interaction of cyanine dyes like TO, TO-PRO-1 and YO-PRO-1 (34), YO (33), SYTOX Orange (43) and PG (44) with dsDNA revealed a biphasic binding mode. External binding at high dbprs for YO and YOYO has been reported to be responsible for the biphasic binding mode (33). Also, BEBO (40,45) and DAPI (46) have been shown to intercalate or to groove bind depending on the dsDNA sequence. Actually, the unsymmetrical cyanine dyes BETO and BOXTO exhibit minor groove binding (40). Because of structure similarities, it appears likely that SG exhibits minor-groove binding at dbprs above ∼0.2. The observed sequence specificity of SG and the impact of ions support this notion (see below).
The arguments mentioned above favour the notion that the second binding mode of SG is an interaction of SG with the minor groove. Small molecules having aromatic rings connected by bonds with torsional freedom, and thus being able to fit into the helical curve of the minor groove, are reported to bind in the minor groove (32). The flexibility and torsional freedom of SG would allow for minor-groove binding. Also, the structure of SG should allow for electrostatic interaction of its cationic groups with the negative electrostatic potential in the minor groove as well as proximate van der Waals contacts within the boundaries of the minor groove. Now, the minor grooves of AT- and GC-rich sequences differ in some general aspects, which are important for the interaction with cationic molecules (32,47–50).
In accordance, for SG a significant AT preference was observed at high dbprs. Fluorescence intensities of SG/dA · dT complexes at high dbprs were higher compared to the corresponding SG/dG · dC complexes. Here, further investigations with different sequences are necessary, in order to elucidate binding preferences of SG (48,51).
Apart from the sequence, the influence of salts was also significant; as expected for a cationic dye (32,35,45). Stern–Volmer plots indicated the occurrence of several salt-dependent quenching mechanisms at low dbprs (36). This indicates the occurrence of combined static and dynamic quenching processes and eventually other processes like competitive binding (19). This is in agreement with the multiple effects that salts express on dye binding and the structure of dsDNA (32,52,53). KSV−1 values indicated an up to almost 13-fold more pronounced impact of MgCl2 on fluorescence compared to NaCl. An aspect that should be considered in particular when SG real-time PCR is performed. With respect to SG real-time PCR, the different fluorescence intensities of SG bound to dsDNA and ssDNA are of importance also. Electrophoretic mobility studies showed that SG binds efficiently to ssDNA, however, fluorescence intensities were rather low.
In conclusion, the data indicate that dbprs >0.2 should be applied to discriminate between ssDNA and dsDNA. For maximum dsDNA selectivity, also with respect to other compounds, e.g. RNA, proteins, etc. (7) the application of dbprs of at least 10 appears advantageous. This should also reduce the impact of salts, other quenchers (19) and dsDNA sequence specificity that can affect SG real-time PCR and melting curve analysis (16). However, inhibition of PCR has to be considered (54) as well as the potential degradation of dsDNA at least at high dbprs. This should also be taken into account for other applications, in particular for the determination of DNase or telomerase activities (9), in fluorescence imaging techniques (11) and for comet assays (55). However, in PCR the dbpr is not constant. It changes with the cycle number as more dsDNA is produced. Therefore, the number of cycles can influence melting curve analysis (16). Again, the application of sufficiently high dbprs, just below a value where significant inhibition is observed, appears advantageous. Also, for dsDNA quantification in solution the actual amount of dsDNA in a sample under investigation is unknown at the time of analysis. But as already reported (19,22) and according to the data, considerations on the dbpr applied are important. Finally, our findings on SG's structure–property relationships and the determined influences of the dbpr, salts and SG's nucleic acid specificity should help in the optimal utilization of this excellent dye in a broad variety of nucleic acids methods.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Joachim Opitz for performing MS-FAB measurements and the Institute for Applied Macromolecular Chemistry of the University of Stuttgart for giving us the opportunity to perform NMR measurements. We are grateful to Timm Eiseler and Gabriele Beck-Schwadorf for excellent technical assistance. We thank Michael Breuer, Bernhard Hauer, Iris Kräuter and Carmen Gruber for helpful discussions. We also thank Felicitas Vitzthum and Fritz Behrens for critical reading of the manuscript. We are grateful to the Peter and Traudl Engelhorn-Stiftung ‘zur Förderung der Biotechnolgie und Gentechnik’, the Fraunhofer Institute for Interfacial Engineering and Biotechnology, and the BASF Incorporation for supporting this work as part of a screening program for novel enzymes from microbial communities.
REFERENCES
- 1.Vitzthum F. and Bernhagen,J. (2002) SYBR Green I: an ultrasensitive fluorescent dye for double-standed DNA quantification in solution and other applications. Recent Res. Devel. Anal. Biochem., 2, 65–93. [Google Scholar]
- 2.Haugland R.P. (2001) Handbook of Fluorescent Probes and Research Chemicals, 8th edn. Molecular Probes, Inc., Eugene, OR. [Google Scholar]
- 3.Schneeberger C., Speiser,P., Kury,F. and Zeillinger,R. (1995) Quantitative detection of reverse transcriptase–PCR products by means of a novel and sensitive DNA stain. PCR Methods Appl., 4, 234–238. [DOI] [PubMed] [Google Scholar]
- 4.Jin X., Dong,F. and Singer,V.L. (1996) SYBR Green I nucleic acid gel stain provides a sensitive fluorescent method for detecting gel mobility shift products. FASEB J., 10, A1128. [Google Scholar]
- 5.Kiltie A.E. and Ryan,A.J. (1997) SYBR Green I staining of pulsed field agarose gels is a sensitive and inexpensive way of quantitating DNA double-strand breaks in mammalian cells. Nucleic Acids Res., 25, 2945–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diggle C.P., Bentley,J. and Kiltie,A.E. (2003) Development of a rapid, small-scale DNA repair assay for use on clinical samples. Nucleic Acids Res., 31, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitzthum F., Geiger,G., Bisswanger,H., Brunner,H. and Bernhagen,J. (1999) A quantitative fluorescence-based microplate assay for the determination of double-stranded DNA using SYBR Green I and a standard ultraviolet transilluminator gel imaging system. Anal. Biochem., 276, 59–64. [DOI] [PubMed] [Google Scholar]
- 8.Rengarajan K., Cristol,S.M., Mehta,M. and Nickerson,J.M. (2002) Quantifying DNA concentrations using fluorometry: a comparison of fluorophores. Mol. Vis., 8, 416–421. [PubMed] [Google Scholar]
- 9.Iida R., Yasuda,T., Tsubota,E., Nakashima,Y., Sawazaki,K., Aoyama,M., Matsuki,T. and Kishi,K. (1998) Detection of isozymes of deoxyribonucleases I and II on electrophoresed gels with picogram sensitivity using SYBR Green I. Electrophoresis, 19, 2416–2418. [DOI] [PubMed] [Google Scholar]
- 10.Wege H., Chui,M.S., Le,H.T., Tran,J.M. and Zern,M.A. (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res., 31, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwabuchi S., Muramatsu,H., Chiba,N., Kinjo,Y., Murakami,Y., Sakaguchi,T., Yokoyama,K. and Tamiya,E. (1997) Simultaneous detection of near-field topographic and fluorescence images of human chromosomes via scanning near-field optical/atomic-force microscopy (SNOAM). Nucleic Acids Res., 25, 1662–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbesti S., Citterio,S., Labra,M., Baroni,M.D., Neri,M.G. and Sgorbati,S. (2000) Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry, 40, 214–218. [PubMed] [Google Scholar]
- 13.Brussaard C.P., Marie,D. and Bratbak,G. (2000) Flow cytometric detection of viruses. J. Virol. Methods, 85, 175–182. [DOI] [PubMed] [Google Scholar]
- 14.Wittwer C.T., Ririe,K.M., Andrew,R.V., David,D.A., Gundry,R.A. and Balis,U.J. (1997) The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 15.Bengtsson M., Karlsson,H.J., Westman,G. and Kubista,M. (2003) A new minor groove binding asymmetric cyanine reporter dye for real-time PCR. Nucleic Acids Res., 31, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giglio S., Monis,P.T. and Saint,C.P. (2003) Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res., 31, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pattyn F., Speleman,F., De Paepe,A. and Vandesompele,J. (2003) RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res., 31, 122–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webster J.R., Burns,M.A., Burke,D.T. and Mastrangelo,C.H. (2001) Monolithic capillary electrophoresis device with integrated fluorescence detector. Anal. Chem., 73, 1622–1626. [DOI] [PubMed] [Google Scholar]
- 19.Zipper H., Buta,C., Lämmle,K., Brunner,H., Bernhagen,J. and Vitzthum,F. (2003) Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res., 31, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachoon D.S., Otero,E. and Hodson,R.E. (2001) Effects of humic substances on fluorometric DNA quantification and DNA hybridization. J. Microbiol. Methods, 47, 73–82. [DOI] [PubMed] [Google Scholar]
- 21.Dutton M.D., Varhol,R.J. and Dixon,D.G. (1995) Technical considerations for the use of ethidium bromide in the quantitative analysis of nucleic acids. Anal. Biochem., 230, 353–355. [DOI] [PubMed] [Google Scholar]
- 22.Yue S.T., Singer,V.L., Roth,B.L., Mozer,T.J., Millard,P.J., Jones,L.J., Jin,X. and Haugland,R.P. (1997) Patent US 5,658,751.
- 23.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Porter D. and Sanborn,B.M. (1992) Analysis of heat-denatured DNA using native agarose gel electrophoresis. Biotechniques, 13, 406–410. [PubMed] [Google Scholar]
- 25.Kleemann R., Hausser,A., Geiger,G., Mischke,R., Burger-Kentischer,A., Flieger,O., Johannes,F.J., Roger,T., Calandra,T., Kapurniotu,A. et al. (2000) Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature, 408, 211–216. [DOI] [PubMed] [Google Scholar]
- 26.Bisswanger H. (1994) Enzymkinetik: Theorie und Methoden, 2nd edn. VCH Verlagsgesellschaft mbH, Weinheim, Germany. [Google Scholar]
- 27.Geiger G., Bernhagen,J., Wagner,E., Bisswanger,H., Brunner,H. and Vitzthum,F. (2001) Standardized measurements and differential spectroscopy in microplates. Anal. Biochem., 296, 29–40. [DOI] [PubMed] [Google Scholar]
- 28.Singer V.L., Jones,L.J., Yue,S.T. and Haugland,R.P. (1997) Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal. Biochem., 249, 228–238. [DOI] [PubMed] [Google Scholar]
- 29.Atkins P.W. (1990) Physikalische Chemie, 2nd edn. VCH Verlagsgesellschaft mbH, Weinheim, Germany. [Google Scholar]
- 30.Akerman B. and Tuite,E. (1996) Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res., 24, 1080–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh D. and Chaires,J.B. (1995) Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem., 3, 723–728. [DOI] [PubMed] [Google Scholar]
- 32.Blackburn G.M. and Gait,M.J. (1996), Nucleic Acids in Chemistry and Biology. 2nd edn. Oxford University Press, Oxford, pp. 329–370. [Google Scholar]
- 33.Larsson A., Carlsson,C., Jonsson,M. and Albinsson,B. (1994) Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J. Am. Chem. Soc., 116, 8459–8465. [Google Scholar]
- 34.Petty J.T., Bordelon,J.A. and Robertson,M.E. (2000) Thermodynamic characterization of the association of cyanine dyes with DNA. J. Phys. Chem. B, 104, 7221–7227. [Google Scholar]
- 35.Nygren J., Svanvik,N. and Kubista,M. (1998) The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers, 46, 39–51. [DOI] [PubMed] [Google Scholar]
- 36.Lakowicz J.R. (1999) Principles of Fluorescence Spectroscopy, 2nd edn. Kluwer Academic/Plenum Publishers, NY. [Google Scholar]
- 37.Ririe K.M., Rasmussen,R.P. and Wittwer,C.T. (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem., 245, 154–160. [DOI] [PubMed] [Google Scholar]
- 38.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–131, 134–138. [DOI] [PubMed] [Google Scholar]
- 39.Rye H.S. and Glazer,A.N. (1995) Interaction of dimeric intercalating dyes with single-stranded DNA. Nucleic Acids Res., 23, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlsson H.J., Lincoln,P. and Westman,G. (2003) Synthesis and DNA binding studies of a new asymmetric cyanine dye binding in the minor groove of [poly(dA–dT)](2). Bioorg. Med. Chem., 11, 1035–1040. [DOI] [PubMed] [Google Scholar]
- 41.Benson S.C., Mathies,R.A. and Glazer,A.N. (1993) Heterodimeric DNA-binding dyes designed for energy transfer: stability and applications of the DNA complexes. Nucleic Acids Res., 21, 5720–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue S.T., Johnson,I.D., Huang,Z., Haugland,R.P. and Richard,P. (1992) Patent US 5,321,130.
- 43.Yan X., Habbersett,R.C., Cordek,J.M., Nolan,J.P., Yoshida,T.M., Jett,J.H. and Marrone,B.L. (2000) Development of a mechanism-based, DNA staining protocol using SYTOX orange nucleic acid stain and DNA fragment sizing flow cytometry. Anal. Biochem., 286, 138–148. [DOI] [PubMed] [Google Scholar]
- 44.Cosa G., Focsaneanu,K.S., McLean,J.R., McNamee,J.P. and Scaiano,J.C. (2001) Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution. Photochem. Photobiol., 73, 585–599. [DOI] [PubMed] [Google Scholar]
- 45.Eriksson M., Karlsson,H.J., Westman,G. and Akerman,B. (2003) Groove-binding unsymmetrical cyanine dyes for staining of DNA: dissociation rates in free solution and electrophoresis gels. Nucleic Acids Res., 31, 6235–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson W.D., Tanious,F.A., Barton,H.J., Jones,R.L., Fox,K., Wydra,R.L. and Strekowski,L. (1990) DNA sequence dependent binding modes of 4′,6-diamidino-2-phenylindole (DAPI). Biochemistry, 29, 8452–8461. [DOI] [PubMed] [Google Scholar]
- 47.Lavery R., Zakrzewska,K. and Pullman,B. (1986) Binding of non-intercalating antibiotics to B-DNA: a theoretical study taking into account nucleic acid flexibility. J. Biomol. Str. Dyn., 3, 1155–1170. [DOI] [PubMed] [Google Scholar]
- 48.Loontiens F.G., Regenfuss,P., Zechel,A., Dumortier,L. and Clegg,R.M. (1990) Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A–T)], and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry, 29, 9029–9039. [DOI] [PubMed] [Google Scholar]
- 49.Latt S.A. and Stetten,G. (1976) Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J. Histochem. Cytochem., 24, 24–33. [DOI] [PubMed] [Google Scholar]
- 50.Teng M.K., Usman,N., Frederick,C.A. and Wang,A.H. (1988) The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res., 16, 2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao R., Venezia,C.F. and Armitage,B.A. (2001) Investigation of DNA binding modes for a symmetrical cyanine dye trication: effect of DNA sequence and structure. J. Biomol. Struct. Dyn., 18, 844–856. [DOI] [PubMed] [Google Scholar]
- 52.Jerkovic B. and Bolton,P.H. (2001) Magnesium increases the curvature of duplex DNA that contains dA tracts. Biochemistry, 40, 9406–9411. [DOI] [PubMed] [Google Scholar]
- 53.Chiu T.K. and Dickerson,R.E. (2000) 1 A crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J. Mol. Biol., 301, 915–945. [DOI] [PubMed] [Google Scholar]
- 54.Nath K., Sarosy,J.W., Hahn,J. and Di Como,C.J. (2000) Effects of ethidium bromide and SYBR Green I on different polymerase chain reaction systems. J. Biochem. Biophys. Methods, 42, 15–29. [DOI] [PubMed] [Google Scholar]
- 55.Ward T.H. and Marples,B. (2000) Technical report: SYBR Green I and the improved sensitivity of the single-cell electrophoresis assay. Int. J. Radiat. Biol., 76, 61–65. [DOI] [PubMed] [Google Scholar]