Abstract
As a rich source of therapeutic agents, peptide natural products usually adopt a cyclic or multicyclic scaffold that minimizes structural flexibility to favor target binding. Inspired by nature, chemists have been interested in developing synthetic cyclic and multicyclic peptides that serve as biological probes and potential therapeutics. Herein we describe a novel strategy for peptide cyclization, in which intramolecular iminoboronate formation allows spontaneous cyclization under physiologic conditions to yield monocyclic and bicyclic peptides. Importantly the iminoboronate-based cyclization can be rapidly reversed in response to multiple stimuli, including pH, oxidation and small molecules. This highly versatile strategy for peptide cyclization should find applications in many areas of chemical biology.
TOC image
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.
Cyclic peptides are considered as privileged scaffolds because cyclization reduces conformational entropy and preorganizes key functional groups for target binding. This is nicely illustrated by the large number of peptide natural products that serve as therapeutic agents, including the immunosuppressant cyclosporine and the antibiotic of last resort daptomycin.1,2 Recently much attention has been paid to synthetic cyclic peptides designed to resemble the structure and function of their natural counterparts.3,4 A number of strategies have been introduced for the synthesis of cyclic and bicyclic peptides, including thioether formation,5,6 azide-alkyne click chemistry,7,8 olefin metathesis,9,10 KAHA ligation,11 C-H activation stapling,12 as well as formation of disulfide bonds and macrolactams.13 These strategies have enabled the preparation of cyclic and bicyclic peptide libraries, screening of which has yielded an impressive array of biological probes and potential therapeutics.3,13,14
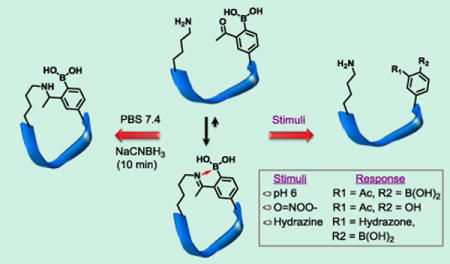
“Smart” peptides that can turn on or off their activity in response to biological stimuli are desirable for a wide range of applications.15,16 For example, they may serve as reporters of local environment or enable targeted drug delivery. Except the disulfide chemistry,17 the peptide cyclization strategies known to date forge permanent linkages, hence unable to sense and respond to physiologic stimuli. Herein, we describe responsive peptide cyclization via spontaneous and reversible formation of intramolecular iminoboronates. Iminoboronates refer to structures where an imine nitrogen forms a dative bond with an adjacent boronic acid (Fig. 1a, b). The dative bond provides thermodynamic stabilization to the imine product. Recent publications from several groups including our own have shown that intermolecular iminoboronate formation is rapidly reversible under physiologic conditions with dissociation constants in the low mM range.18–22 We postulated that intramolecular iminoboronate formation should allow spontaneous and highly efficient cyclization of peptides. Importantly, cyclic peptides with iminoboronate linkages should be able to linearize in response to physiologic stimuli, therefore allowing facile control of their functions.
Fig.1.
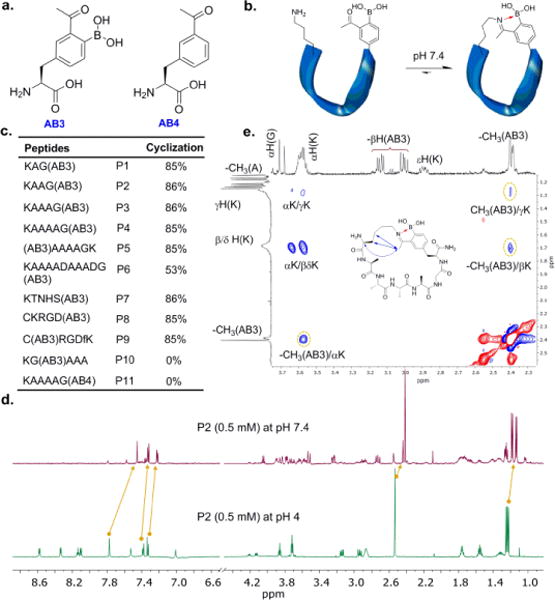
Iminoboronate-mediated peptide cyclization. a) Structure of the unnatural amino acids designed for iminoboronate chemistry; b) illustration of the iminoboronate-based peptide cyclization; c) peptide sequences and their cyclization efficiency at pH 7.4; % cyclization was determined by 1H-NMR with peptides at 0.5 mM, which is significantly lower than the Kd values of intermolecular iminoboronate formation (Figure S2). d) 1H-NMR spectra of P2 illustrating the pH-dependent cyclization; e) ROESY spectrum of P4 showing NOEs between AB3 and Lys.
To test our hypothesis, we have developed an efficient synthesis for the unnatural amino acid AB3 (Fig. 1a, Fig. S1, SI), which shows improved propensity for lysine conjugation than AB1, a similar amino acid previously reported by our group (Fig. S2, SI). We envisioned that peptides incorporating an AB3-Lys pair should readily cyclize via formation of an intramolecular iminoboronate (Fig. 1b). Towards this end, we have synthesized a panel of peptides that display an AB3-Lys pair in various contexts (Fig. 1c). Characterization by UV-vis, NMR spectroscopy and mass spectrometry clearly demonstrated formation of intramolecular iminoboronates for peptides P1–P9 (Fig. S3–13, SI). Taking P2 as an example, at pH 7.4, the sodium adduct of the cyclic peptide was clearly seen in mass-spec analysis, while only the linear precursor was observed at pH 4.0 (Fig. S6, SI). In 1H-NMR, with pH increase from 4.0 to 7.4, the acetyl –CH3 peak shifted from 2.6 to 2.4 ppm and the aromatic protons of AB3 displayed a more clustered pattern (Fig. 1d). Similar peak shifts were observed for intermolecular iminoboronate formation between AB3 and lysine (Fig. S4, SI). Further evidence of peptide cyclization was obtained from the 2D 1H-NMR data of P4 (Fig. 1e, Fig. S18–21, SI). At pH 7.8, the ROESY spectrum revealed a number of inter-residue cross peaks including Lys-α(H)↔Ac-CH3 (strong), Lys-β(H)↔Ac-CH3(medium) and Lys-α(H)↔Ac-CH3(weak), which disappeared upon acidification.
We note that cyclization of P2 afforded two new singlets (5:1 in ratio) at ~2.4 ppm, indicating a pair of stable conformational isomers of the iminoboronate product. Quantification with NMR showed that P2 cyclization proceeded to ~85% completion at pH 7.4. Interestingly, the extent of cyclization appeared insensitive to the AB3-to-Lys distance up to five amino acids (P1–5, Fig. 1c) and dropped to 53% for P6, in which AB3 and Lys are separated by nine amino acids. In addition to the Ala-rich peptides, we have also synthesized P7-9 that incorporate a variety of polar residues. All three peptides showed high cyclization efficiency suggesting the iminoboronate-based cyclization can be applied to various peptide sequences. Interestingly, no cyclization was observed for P10 (Fig. S14, SI), where AB3 and lysine are positioned at i and i+2 positions. This lack of cyclization is presumably due to the ring strain of cyclic product. Similarly, no cyclization was observed when an AB1 residue was placed adjacent to a lysine in peptide sequences.22 Finally, the negative control peptide P11 (Fig. S15, SI), which presents a deborylated residue (AB4, Fig. 1a), showed no cyclization as expected due to the unfavorable imine formation in absence of the stabilizing boronic acid.
We envisioned that installing two AB3-Lys pairs could elicit double cyclization to give bicyclic peptides. To avoid potential regioisomers of crosslinking, we intentionally placed an AB3 and a lysine at i and i+2 positions (Fig. 2a), which should prevent crosslinking of these two residues according to what we learned with P10. The peptides P12 and P13 were characterized by using 1H-NMR, 11B-NMR and mass spectrometry, which collectively support double cyclization as predicted. Specifically, mass-spec analysis of P12 revealed the molecular ion and the sodiated adduct of the bicyclic product at pH 7.4 (Fig. 2b). In contrast, only the linear precursor was observed at pH 4.0 (Fig. S16, SI). A series of peaks observed in our mass-spec data correspond to the bicyclic peptide losing one or more water molecules, which is commonly seen for compounds presenting boronic acids.21 Similar results were obtained for P13 in mass-spec analysis (Fig. S17, SI). Further, the 11B-NMR spectrum of P12 in neutral buffer exhibits a peak at ~8 ppm, as expected for the partly anioic boron in iminoboronates (Fig. 2d). In contrast, an unconjugated AB3 residue displays 11B resonances at ~30 ppm (Fig. 2c). The disappearance of the 30 ppm peak at neutral pH indicates complete bicyclization of the peptide. Although less straightforward to analyze, the 1H-NMR data (Fig. S16–17, SI) did show clustering of the AB3 aromatic peaks upon pH change from 4.0 to 7.4, which is a signature of iminoboronate formation of AB3 as shown in Fig. 1d.
Fig.2.
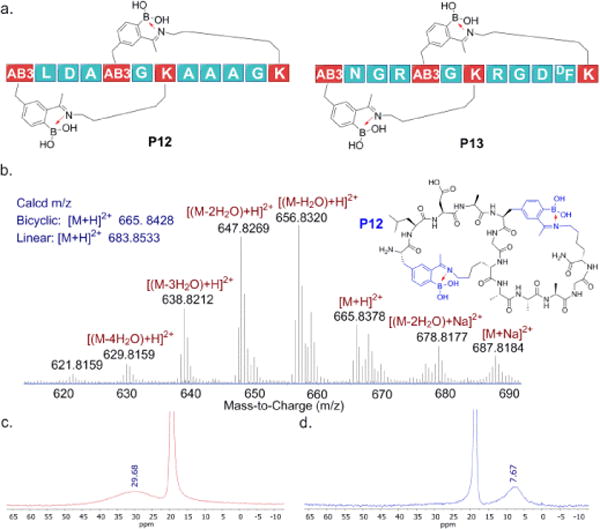
Iminoboronate-mediated peptide bicyclization. a) Peptide sequences designed for bicyclization; b) mass spectrum of P12 at pH 7.4; c) 11B-NMR spectrum of P12 at pH 4.0; d) 11B-NMR spectrum of P12 at pH 7.4. The truncated peaks at 20 ppm are from boric acid used as internal reference. The characterization data of P13 are included in Fig. S17, SI.
To assess the potential of the iminoboronate-cyclized peptides for biological applications, we first investigated the stability of the cyclic peptides in the presence of various biomolecules. Interestingly, we found that the peptide cyclization was not affected by small molecules like lysine (30 mM), glucose (5 mM) and glutathione (GSH, 5 mM) (Fig. S22, SI). Nor was it affected by model proteins or simply blood serum (Fig. S23–24, SI). The robust stability presumably originates from the intramolecularity of the peptide cyclization, which enjoys great thermodynamic advantage.
While stable at neutral conditions, the iminoboronate-based peptide cyclization is very sensitive to pH. To better understand the pH dependence, we performed a pH titration experiment with P4 cyclization quantified by 1H-NMR. The peptide was found to be >98% cyclized at pHs above 8 and the cyclization efficiency drops rapidly with acidification (Fig. 3a, Fig. S25, SI). Plotting the cyclization percentage against pH gives a sigmoidal curve with a mid-point of transition at pH 6.8. This makes the iminoboronate chemistry promising for applications in solid tumors, where mild acidification often occurs due to the high rate of metabolism.23,24
Fig. 3.
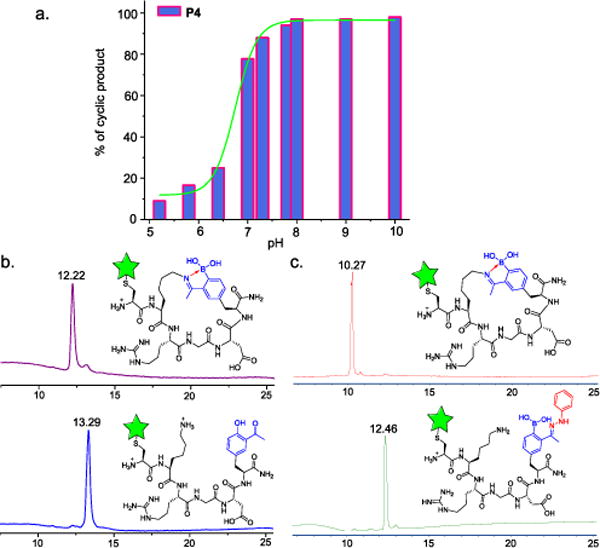
Iminoboronate-cyclized peptides responding to various stimuli. a) pH titration curves showcasing the sharp pH sensitivity of the iminoboronate-mediated peptide cyclization; b) LC chromatograms demonstrating the quick oxidation of AF488-P8 by peroxynitrite; c) LC traces showing linearization of AF488-P8 by phenylhydrazine.
It is well documented that aryl boronic acids can be oxidized by biologically generated oxidants (O=NOO−, O2•, H2O2).25,26 We assessed the response of an iminoboronate-cyclized peptide (AF488-P8) to oxidation. Surprisingly, incubation of AF488-P8 with H2O2 at biologically relevant concentrations (200 μM) resulted in no oxidation even after 24 hours (Fig. S26, SI). In contrast, unsubstituted phenyl boronic acid was oxidized in a few minutes. The much reduced propensity of the iminoboronate for oxidation is presumably due to the B-N dative bond, which protects the boronic acid against oxidation. Interestingly, the iminoboronate linkage in AF488-P8 was found to be very sensitive to peroxynitrite, which at 20 μM concentration converted the boronic acid moiety into a phenol in less than 10 min. Due to loss of the dative bond, the iminoboronate oxidation resulted in rapid linearization of the cyclic peptide as confirmed by LC-MS analysis (Fig. 3b, Fig. S27, SI).
In addition to pH and oxidation, the iminoboronate-cyclized peptides show rapid response to α-nucleophiles, which we recently found to react with 2-acetylphenylboronic acid (the side chain of AB3) at neutral pH to form oximes and hydrazones.27 The oxime/hydrazone formation is reversible and exhibits more favorable thermodynamic equilibrium with Kd values in the low micromolar to nanomolar range. We envisioned that iminoboronate-cyclized pepitdes, although stable to endogenous nucleophiles like amines and thiols, could be readily linearized by the addition of α-nucleophiles. Indeed, treating AF488-P7 at neutral pH with phenylhydrazine (11 μM) resulted in complete linearization of the cyclic peptide in less than 10 min according to LC-MS analysis (Fig. 3c, Fig. S28, SI). Similar results were observed when a cyclic peptide was mixed with aminoxy hexanoic acid (AOHA, Fig. S22, SI).
To further probe the potential of the iminoboronate-cyclized peptides for biological applications, we tested an iminoboronate-cyclized RGD peptide against SKOV3 cells, an ovarian cancer cell line known to overexpress the αvβ3 integrin (Fig. 4).28 Specifically we synthesized the peptide P9 that presents an RGDf (f: D-Phe) motif flanked by an AB3-Lys pair. The RGDf motif elicits potent and selective binding to the αvβ3 integrin when placed in an appropriate cyclic scaffold.28 According to 1H-NMR and mass-spec analyses (Fig. S13, SI), the peptide P9 cyclizes to 85% at neutral pH and is completely linear at pH 4.0. We postulated that P9 in cyclic forms should be able to bind SKOV3 cells, and the cell binding should be readily controllable with pH modulation or with exogenous small molecule modulators.
Fig. 4.
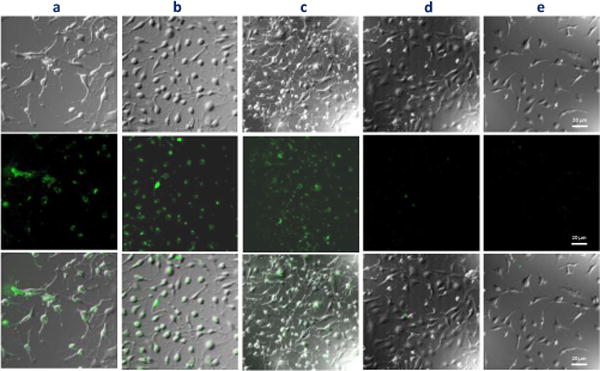
Integrin binding of an iminoboronate-cyclized peptide. The images shown are phase contrast (top), fluorescence (middle) and overlay (bottom) respectively. a) AF488-P14 at pH 7.4; b) AF488-P9 at pH 7.4; c) AF488-P14 at pH 6.0; d) AF488-P9 at pH 6.0; e) AF488-P9L, a phenylhydrazine conjugate of AF488-P9 at pH 7.4. All peptides were used at 10 μM concentration for the cell binding studies. Scale bar: 20 μm.
A commercially available, head–to-tail cyclized peptide P14 (cyclo-(RGDfC)) was used as a positive control. The SKOV3 cells were incubated with the AF488-labeled P14 and P9, at pH 7.4 and pH 6.0 respectively. As expected, the positive control P14 elicited strong staining of the SKOV3 cells at both pH (Fig. 4a, c). Interestingly, P9 at pH 7.4 afforded equally strong, if not stronger fluorescence staining of the cells (Fig. 4b), suggesting the iminoboronate-based cyclization fully supports the integrin binding of the RGDf motif. In contrast, at pH 6.0, the AF488-labeled P9 yielded little staining of the cells, presumably due to the acid-triggered linearization of the peptide. Consistently, linearization of AF488-P9 with phenylhydrazine abolished the peptide’s propensity to stain SKOV3 cells as well. These results nicely demonstrate the biological applicability of the iminoboronate-based cyclic peptides, as well as their versatility in responding to both endogenous and exogenous stimuli.
As described above, intramolecular iminoboronate formation allows peptide cyclization in a reversible manner. Finally we show that the iminoboronate linkage could be trapped via reduction of the imine to give permanently cyclized product (Fig. 5). Indeed, when a fluorophore-labeled peptide AF488-P8 was treated with sodium cyanoborohydride (NaCNBH3), a clean conversion (>95%) was obtained within 20 min according to LC-MS analysis (Fig. 5b, Fig. S29, SI). The clean reduction was further confirmed by 1H-NMR (Fig. S30, SI), which showed a dramatic shift of the acetyl –CH3 peak. Further, two sets of peaks were observed for the easily identifiable protons, indicating that the reduced product exists as a pair of diastereomers. The reduction strategy applies to bicyclic peptides as well: treating AF488-P13 resulted in a clean product with both iminoboronate linkages reduced (Fig. 5c, Fig. S29, SI).
Fig. 5.
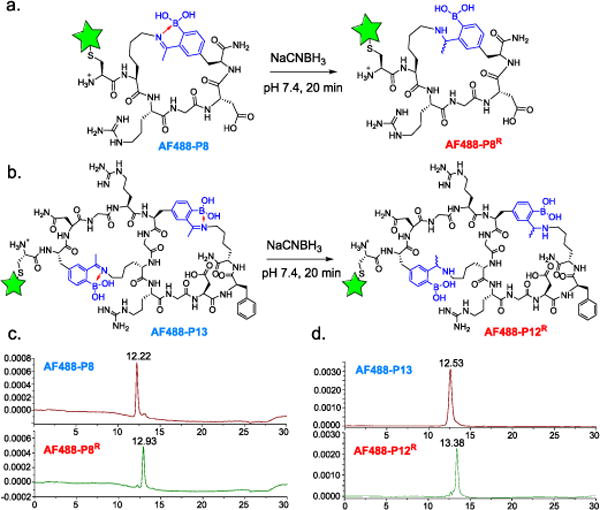
Iminoboronate reduction yielding permanently cyclized peptides. a) Schematic illustration of the iminoboronate reduction to give irreversible peptide cyclization; b) LC chromatograms of AF488-P8 before (top) and after (bottom) reduction; c) LC chromatograms of AF488-P13 before (top) and after reduction (bottom) reduction.
To summarize, this contribution describes the use of iminoboronate chemistry for peptide cyclization. Specifically we have developed a synthetic amino acid AB3, which, when incorporated into a peptide, readily conjugates an adjacent lysine to give cyclic and bicyclic peptides with iminoboronate linkages. We note that several papers exist in literature documenting peptide cyclization via formation of intramolecular imines.29–31 This unusual mechanism of peptide cyclization originates from the conformational rigidity of the linear precursor and is extremely sensitive to the amino acid composition of the peptides.30 This is perhaps not surprising given the unfavorable thermodynamic equilibrium of imine formation in aqueous media. In contrast to simple imines, iminoboronates enjoy much greater thermodynamic stability due to stabilization by the dative bond. At neutral pH, the iminoboronate-cyclized peptides display surprisingly robust stability against commonly seen biomolecules. Excitingly, despite the robustness in biological milieu, the iminoboronate-mediated cyclization can be readily reversed with acidification, oxidation, and addition of exogenous small molecule modulators. We believe the quick response to multiple stimuli will make the iminoboronate cyclization strategy useful for a wide range of applications in biotechnology.
Supplementary Material
Acknowledgments
The authors acknowledge the financial support from the National Institutes of Health via the grant GM102735.
Footnotes
Supporting Information
Supporting information includes details of synthesis and characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Baeriswyl V, Heinis C. ChemMedChem. 2013;8:377. doi: 10.1002/cmdc.201200513. [DOI] [PubMed] [Google Scholar]
- 2.Bock JE, Gavenonis J, Kritzer JA. ACS Chem Biol. 2013;8:488. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passioura T, Katoh T, Goto Y, Suga H. Annu Rev Biochem. 2014;83:727. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 4.Heinis C, Winter G. Curr Opin Chem Biol. 2015;26:89. doi: 10.1016/j.cbpa.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Goto Y, Ohta A, Sako Y, Yamagishi Y, Murakami H, Suga H. ACS Chem Biol. 2008;3:120. doi: 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- 6.Bionda N, Cryan AL, Fasan R. ACS Chem Biol. 2014;9:2008. doi: 10.1021/cb500311k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne WS, Olsen CA, Beierle JM, Montero A, Ghadiri MR. Angew Chem Int Ed. 2009;48:4718. doi: 10.1002/anie.200805900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wuo MG, Mahon AB, Arora PS. J Am Chem Soc. 2015;137:11618. doi: 10.1021/jacs.5b05525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell HE, Sadowsky JD, Howard RJ, Sampson JN, Chao JA, Steinmetz WE, O’Leary DJ, Grubbs RH. J Org Chem. 2001;66:5291. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- 10.Kim YW, Grossmann TN, Verdine GL. Nat Protoc. 2011;6:761. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 11.Rohrbacher F, Deniau G, Luther A, Bode JW. Chem Sci. 2015;6:4889. doi: 10.1039/c5sc01774b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendive-Tapia L, Preciado S, Garcia J, Ramon R, Kielland N, Albericio F, Lavilla R. Nat Commun. 2015;6:7160. doi: 10.1038/ncomms8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian W, Upadhyaya P, Rhodes CA, Liu Y, Pei D. J Am Chem Soc. 2013;135:11990. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinis C, Rutherford T, Freund S, Winter G. Nat Chem Biol. 2009;5:502. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 15.Branco MC, Sigano DM, Schneider JP. Curr Opin Chem Biol. 2011;15:427. doi: 10.1016/j.cbpa.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Xu B. Bioconjug Chem. 2015;26:987. doi: 10.1021/acs.bioconjchem.5b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowerman CJ, Nilsson BL. J Am Chem Soc. 2010;132:9526. doi: 10.1021/ja1025535. [DOI] [PubMed] [Google Scholar]
- 18.Hutin M, Bernardinelli G, Nitschke JR. Chem Eur J. 2008;14:4585. doi: 10.1002/chem.200800074. [DOI] [PubMed] [Google Scholar]
- 19.Galbraith E, Kelly AM, Fossey JS, Kociok-Kohn G, Davidson MG, Bull SD, James TD. New J Chem. 2009;33:181. [Google Scholar]
- 20.Gutierrez-Moreno NJ, Medrano F, Yatsimirsky AK. Org Biomol Chem. 2012;10:6960. doi: 10.1039/c2ob26290h. [DOI] [PubMed] [Google Scholar]
- 21.Cal PM, Vicente JB, Pires E, Coelho AV, Veiros LF, Cordeiro C, Gois PM. J Am Chem Soc. 2012;134:10299. doi: 10.1021/ja303436y. [DOI] [PubMed] [Google Scholar]
- 22.Bandyopadhyay A, McCarthy KA, Kelly MA, Gao J. Nat Commun. 2015;6:6561. doi: 10.1038/ncomms7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubbs M, McSheehy PM, Griffiths JR, Bashford CL. Mol Med Today. 2000;6:15. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 24.Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO, Engelman DM, Reshetnyak YK. Proc Natl Acad Sci U S A. 2007;104:7893. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippert AR, De Bittner GCV, Chang CJ. Acc Chem Res. 2011;44:793. doi: 10.1021/ar200126t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZJ, Ren W, Wright QE, Ai HW. J Am Chem Soc. 2013;135:14940. doi: 10.1021/ja408011q. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay A, Gao J. Chem Eur J. 2015;21:14748. doi: 10.1002/chem.201502077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mas-Moruno C, Rechenmacher F, Kessler H. Anticancer Agents Med Chem. 2010;10:753. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker JE, Moore RE, Moore BS. Gene. 2004;325:35. doi: 10.1016/j.gene.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Kopp F, Mahlert C, Grunewald J, Marahiel MA. J Am Chem Soc. 2006;128:16478. doi: 10.1021/ja0667458. [DOI] [PubMed] [Google Scholar]
- 31.Evans BS, Ntai I, Chen Y, Robinson SJ, Kelleher NL. J Am Chem Soc. 2011;133:7316. doi: 10.1021/ja2015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


