Abstract
Both the invasion of red blood cells (RBCs) by Plasmodium falciparum parasites and the sequestration of parasite-infected RBCs in the microvasculature are mediated in part by complement receptor one (CR1). RBC surface CR1 level can vary between individuals by more than 20-fold and may be associated with the risk of severe malaria. The factors that influence RBC CR1 level variation are poorly understood, particularly in African populations. We studied 3535 child residents of a malaria-endemic region of coastal Kenya and report, for the first time, that the CR1 Knops blood group alleles Sl2 and McCb, and homozygous HbSS are positively associated with RBC CR1 level. Sickle cell trait and ABO blood group did not influence RBC CR1 level. We also confirm the previous observation that α+thalassaemia is associated with reduced RBC CR1 level, possibly due to small RBC volume, and that age-related changes in RBC CR1 expression occur throughout childhood. RBC CR1 level in malaria-endemic African populations is a complex phenotype influenced by multiple factors that should be taken into account in the design and interpretation of future studies on CR1 and malaria susceptibility.
Introduction
Plasmodium falciparum is the most common cause of malaria world wide and is responsible for the vast majority of malaria deaths.1 P. falciparum exploits multiple host receptors during the process of red blood cell (RBC) invasion and in the subsequent sequestration of P. falciparum parasitised RBCs in the microvasculature.2 Understanding these host–parasite receptor–ligand interactions may help in the development of new methods for preventing and treating malaria.2 Complement receptor one (CR1) is a single-chain trans-membrane glycoprotein that is expressed on the surface of RBCs and leukocytes. The normal function of RBC CR1 is to regulate complement activation and to transfer immune complexes to phagocytic cells in the liver and spleen.3 In malaria, CR1 acts as a receptor on RBCs for both P. falciparum invasion4, 5, 6, 7 and rosette formation, in which two or more uninfected RBCs bind to a P. falciparum parasitised RBC in vitro.8, 9 Rosetting has been associated with severe malaria in multiple African study sites (reviewed in Rowe et al.10), potentially because it leads to enhanced parasitised RBC sequestration and decreased blood flow in post-capillary venules.11, 12 CR1 acts as a receptor for P. falciparum RBC invasion via its interaction with the parasite ligand P. falciparum reticulocyte-binding-like homologue protein 4.4, 5, 6, 7
The mean number of CR1 molecules expressed on the RBC surface varies between individuals, typically between 50 and 1500 CR1 molecules/RBC. RBC with <200 CR1 molecules per cell show reduced rosetting8 and negative or weak reactions with CR1 antibodies,13 being most marked at <100 CR1 molecules per cell. Low RBC CR1 level has also been associated with reduced P. falciparum RBC invasion in vitro.5, 6 Individuals with low RBC CR1 are common in some malaria-endemic regions,14, 15 with up to 80% of the population in coastal Papua New Guinea having fewer than 200 CR1 molecules per RBC.14 Variation in RBC CR1 level may contribute to individual differences in susceptibility to severe malaria. Low RBC CR1 might protect against cerebral malaria through the mechanism of reduced rosetting,8, 9, 14 whereas, in contrast, increasing the risk of immune-complex-mediated tissue damage and severe malarial anaemia.16, 17
Factors determining the variation in RBC CR1 expression are not fully understood, but are proposed to be both genetic and acquired.18 In Caucasian and Asian populations, variation in RBC CR1 level is genetically determined and is associated with a HindIII restriction fragment length polymorphism that is in tight linkage disequilibrium with several other single-nucleotide polymorphisms (SNPs) in exons 19, 22 and 33 of the CR1 gene.19, 20, 21, 22 However, in African populations, although RBC CR1 levels vary over the same range, there is no association between CR1 level and any of the above SNPs, and the causal mutations that determine RBC CR1 levels are unknown.21, 23 CR1 is also the site for SNPs giving rise to the Knops blood group system of antigens that include the allelic pairs Swain-Langley 1 and 2 (Sl1 and Sl2), McCoy a and b (McCa and McCb) and Knops a and b (Kna and Knb).24, 25, 26, 27 Sl2 and McCb occur at higher frequencies in African populations than non-African populations.28 To the best of our knowledge, the effect of the Knops Sl and McC genotypes on RBC CR1 level has not been studied previously.
One study in Papua New Guinea found a dose-dependent decrease in RBC CR1 level with α+thalassaemia,14 an association that might exist in African populations where α+thalassaemia is also common. It is unknown whether other high-frequency African polymorphisms associated with malaria resistance such as HbS and blood group O might influence RBC CR1 level. Two previous studies have shown that RBC CR1 expression level varies with age in both malaria-endemic and non-endemic populations, being lowest during early childhood.16, 29
In the current study, we investigated RBC CR1 level and the factors that influence it in children living within a malaria-endemic region on the Kenyan coast.
Results
RBC CR1 level in the Kilifi population varies with age during childhood
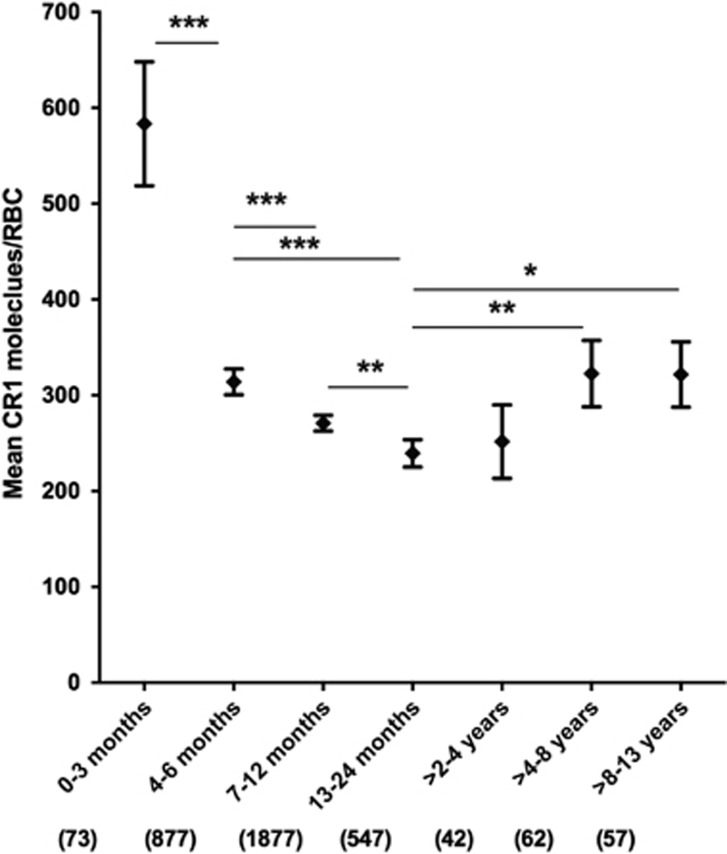
We measured RBC CR1 level by flow cytometry in a total of 3535 children 0–13 years of age in Kilifi County on the coast of Kenya. Previous work in Western Kenya suggested that RBC CR1 levels change with age during childhood,16, 29 and this relationship was confirmed in the Kilifi population (Figure 1). We analysed the data by analysis of variance using the same age categories as previous studies,16, 29 and found that mean RBC CR1 level was higher in newborns (0–3 months) than in other age groups, and significantly declined during each age interval in the first two years of life (Figure 1). CR1 level then increased and stabilized in older children (>4 years). We also analysed the relationship between RBC CR1 level and age as a continuous variable, using piecewise-linear regression analysis, which allows for adjustment for known confounding variables such as α+thalassaemia.14 This analysis assumes a linear effect within defined age ranges, using the same age categories as Figure 1. This confirmed the significant decline in RBC CR1 levels during each age category up to 2 years (shown by negative regression coefficients in Table 1). Then between 2 and 4 years of age, the RBC CR1 level significantly increased with age (shown by a positive regression coefficient, Table 1). There was no statistically significant change in CR1 level in the >4–8 years and >8–13 years age intervals, showing that CR1 levels plateau by about 4 years of age, as suggested previously.16, 29 When analysed across all samples, the regression analysis gave a value of r2=0.05, indicating that although there are significant changes in CR1 level within defined age groups as indicated above, overall, age explains only a minor part of the variation in RBC CR1 level, and other factors must be playing a role.
Figure 1.
RBC CR1 level varies with age. The relationship between RBC CR1 level and age was examined in a total of 3535 individuals aged between 0 months and 13 years by one-way analysis of variance with Tukey's post-hoc test to correct for multiple comparisons. Age was presented as a categorical variable defined into seven categories: 0–3 months, 4–6 months, 7–12 months, 1–2 years, >2–4 years, >4–8 years and >8–13 years. Error bars represent 95% confidence intervals of the mean RBC CR1 levels. Numbers in parenthesis reflect total number of samples tested for each group. Statistically significant differences in RBC CR1 level between groups are asterisked, *P<0.05, **P<0.01 and ***P<0.001. All age groups differed significantly from the 0–3 months age group (P <0.001), but these are not all asterisked on the graph in order to enhance readability of the figure.
Table 1. Regression coefficients for the relationship between age and RBC CR1 level over different age ranges in Kilifi, Kenya.
| Age intervals | Regression coefficients | 95% CI | P value | N |
|---|---|---|---|---|
| 0–3 Months | −31.24 | −64.83–2.35 | 0.068 | 73 |
| 4–6 Months | −48.08 | −62.08–−34.07 | <0.001 | 877 |
| 7–12 Months | −6.22 | −9.65–−2.79 | <0.001 | 1877 |
| 13–24 Months | −4.13 | −7.16–−1.09 | 0.008 | 547 |
| >2–4 Years | 4.25 | 0.73–7.78 | 0.018 | 42 |
| >4–8 Years | −0.33 | −2.66–1.99 | 0.778 | 62 |
| >8–13 Years | 0.94 | −1.31–3.19 | 0.411 | 57 |
Abbreviation: CI, confidence interval.
Differences in RBC CR1 levels by age as a continuous variable were analysed by piecewise-linear regression analysis with adjustment for confounding by α+thalassaemia and HbS. Age was classified into seven different ranges represented as 0–3 months, 4–6 months, 7–12 months, 1–2 years, >2–4 years, >4–8 years and >8–13 years.
RBC CR1 level is negatively associated with α+thalassaemia genotype
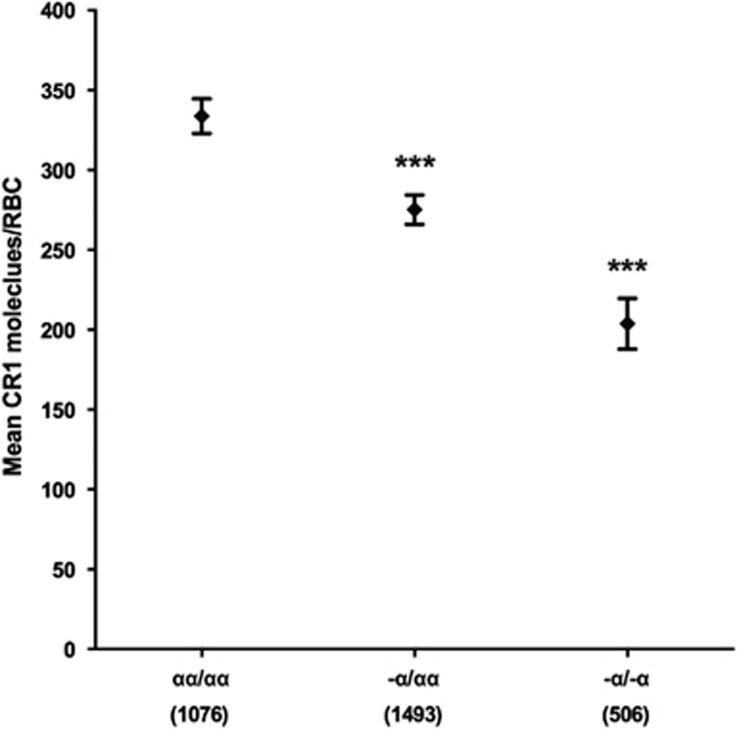
We investigated whether the association between α+thalassaemia and RBC CR1 level previously reported in Papua New Guinea14 might also hold in the Kilifi population. Compared with normal cells (mean CR1 molecules per RBC 334; 95% confidence interval (CI)=323–345), both heterozygous and homozygous α+thalassaemia were associated with significantly lower RBC CR1 levels: (275; 266–284; P<0.001) and (204; 188–220; P<0.001), respectively (Figure 2).
Figure 2.
RBC CR1 level is negatively associated with α+thalassaemia genotype. RBC CR1 level was examined in a total of 3535 children with known α+thalassaemia genotypes. Differences in RBC CR1 level by α+thalassaemia genotype were compared by multivariate linear regression analysis with adjustment for age, HbS, Sl and McC genotypes. The normal globin genotype (αα/αα) was used as the reference genotype. Error bars represent 95% confidence intervals of the mean RBC CR1 level. Numbers in parenthesis reflect total number of samples tested for each genotype group. Statistically significant differences in RBC CR1 level from the reference genotype are asterisked, ***P<0.001.
MCV largely accounts for the association between α+thalassaemia and RBC CR1 level
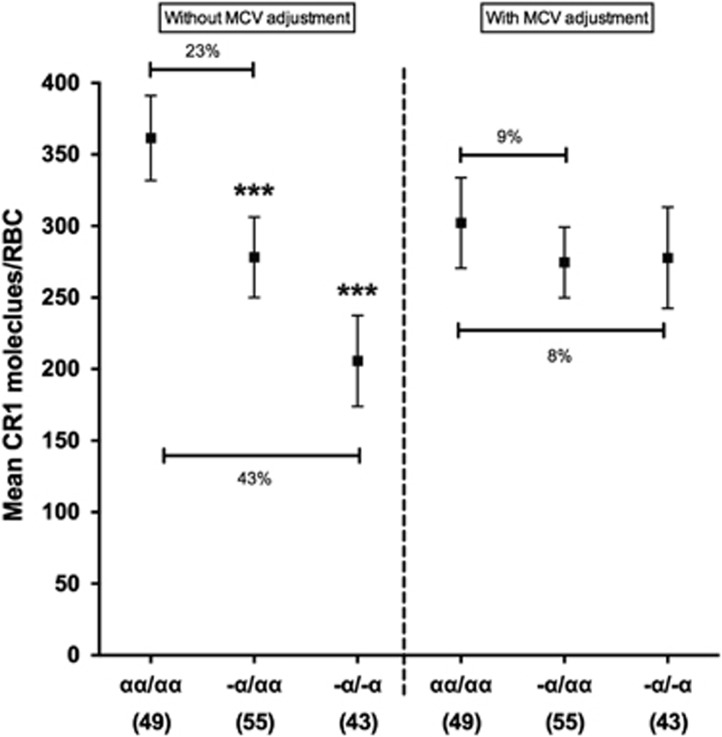
The mechanism leading to reduced RBC CR1 level with α+thalassaemia has not been explored previously. α+Thalassaemia RBCs are microcytic, with significantly lower mean cell volumes (MCVs) compared with normal cells. We therefore examined whether reduced MCV could explain the lower CR1 levels seen with α+thalassaemia. One hundred and forty-seven RBC samples from the Mild Disease Cohort included in the above analyses had MCV measurements available. As expected, α+thalassaemia was associated with reduced MCV in both the heterozygous (MCV=73.5; 95% CI=72.0–74.9; P<0.001) and homozygous state (65.1; 63.5–66.8; P<0.001) when compared with normal RBCs (79.6; 78.0–81.1). As seen in the larger data set, unadjusted for MCV, RBC CR1 levels declined with α+thalassaemia in the 147 samples: CR1 level in heterozygous (278; 250–306, P<0.001) and homozygous (206; 174–237; P<0.001) α+thalassaemic RBCs compared with normal RBCs (361; 332–391; Figure 3, left side). After adjusting for MCV, the difference in CR1 level between normal RBCs (302; 271–334) and heterozygous (275; 250–299, P=0.172) and homozygous α+thalassaemic RBCs (278; 242–313, P=0.382) was no longer statistically significant (Figure 3, right side).
Figure 3.
MCV largely accounts for the association between α+thalassaemia and RBC CR1 level. Association between RBC CR1 level and α+thalassaemia in 147 samples without adjustment for MCV (left side), or with adjustment for MCV (right side). Differences in CR1 level by α+thalassaemia were first analysed by multivariate linear regression analysis with adjustment for age, HbAS and Sl and McC genotypes and compared with a separate model with the additional adjustment for MCV. The normal globin genotype (αα/αα) was used as the reference genotype. Error bars represent 95% confidence intervals of the mean RBC CR1 level. The percentages shown represent the relative difference in mean CR1 level between the respective genotypes. Numbers in parenthesis reflect total number of samples tested for each genotype group. Statistically significant differences in RBC CR1 level from the reference genotype are asterisked, ***P<0.001.
RBC CR1 level is affected by homozygous sickle cell disease (HbSS) but not by heterozygous sickle cell trait (HbAS) or ABO blood group
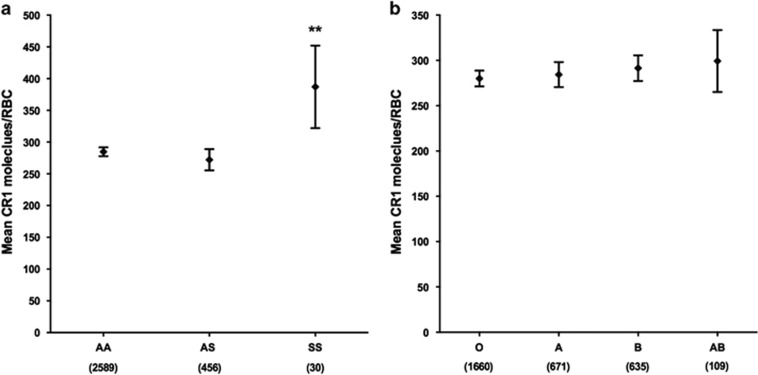
We also investigated the relationship between RBC CR1 level and other common polymorphisms in the Kilifi population, including the sickle cell mutation (HbS)30 and blood group O of the ABO blood group system31 in 3075 individuals for whom full genotype data were available. We found no significant association between RBC CR1 level and either sickle cell trait (HbAS; Figure 4a) or ABO blood group (Figure 4b). Although the number of subjects studied was small (n=30), homozygous sickle cell disease (HbSS) was associated with significantly elevated RBC CR1 level (387; 322–452; P=0.002) in comparison to normal HbAA RBCs (285; 278–292; Figure 4a).
Figure 4.
RBC CR1 level increases with homozygous HbS (HbSS) but is not affected by sickle cell trait (HbAS) or ABO blood group. (a) RBC CR1 level by HbS genotype. (b) RBC CR1 level by ABO blood group. Differences in RBC CR1 level were examined in a total of 3075 individuals by multivariate regression analysis with adjustment for age, α+thalassaemia, Sl and McC genotypes. The normal globin genotype (HbAA) and blood group O were used as the reference groups, respectively. Error bars represent 95% confidence intervals of the mean RBC CR1 level. Numbers in parenthesis reflect total number of samples tested for each genotype group. Statistically significant differences in RBC CR1 level from the reference groups are asterisked, **P<0.01.
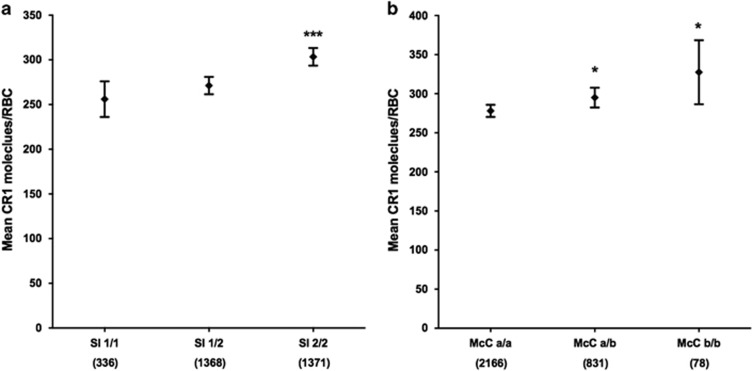
RBC CR1 level is positively associated with both the Sl2 and McC b CR1 Knops blood group genotypes
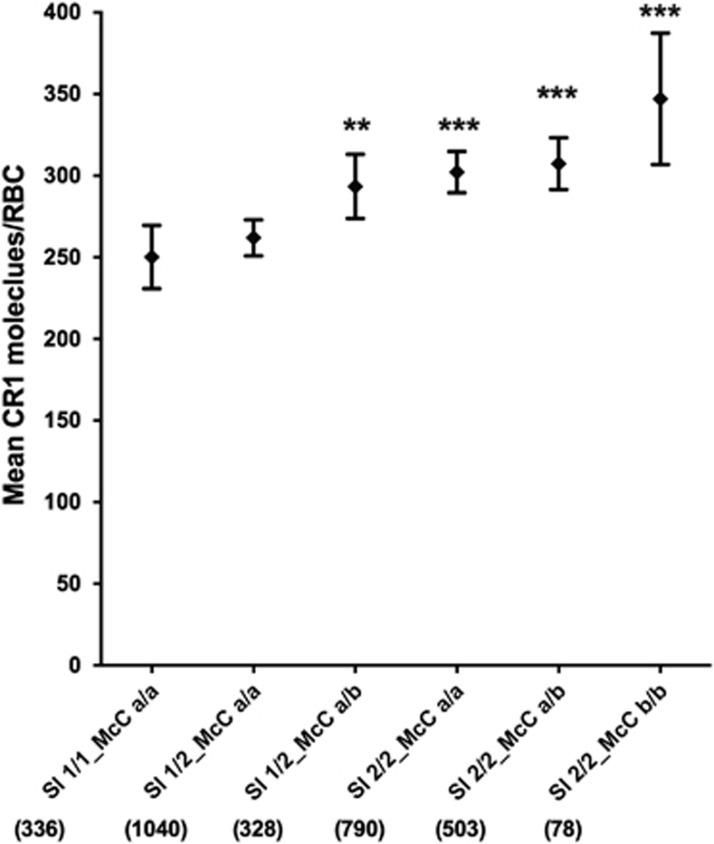
The CR1 Knops blood group Sl2 and McCb alleles are present at high frequencies in African populations,28 but it is unknown whether they influence RBC CR1 level. We found that homozygous Sl2 genotype was associated with an increased RBC CR1 level (303; 294–313; P<0.001 when compared with homozygous Sl1 256; 236–276; Figure 5a). Similarly, both heterozygous and homozygous McCb were associated with increased RBC CR1 level (295; 282–308; P=0.028) and (328; 287–369; P=0.021), respectively, in comparison to homozygous McCa (278; 270–286; Figure 5b). This association was even more marked when looking at Sl-McC genotype combinations, with the homozygous Sl2-McCb haplotype expressing the highest CR1 level (347; 307–387; P<0.001), when compared with the homozygous Sl1-McCa (250; 231–270; Figure 6).
Figure 5.
RBC CR1 level varies by Swain-Langley and McCoy genotypes. RBC CR1 level by CR1 Knops blood genotype. (a) Swain-Langley (Sl) and (b) McCoy (McC). Differences in RBC CR1 level by Sl and McC genotypes were tested in a total of 3075 individuals by multivariate linear regression analysis with adjustment for age, α+thalassaemia and HbS genotypes. Error bars represent 95% confidence intervals of the mean RBC CR1 level. Numbers in parenthesis reflect total number of samples tested for each genotype group. The Sl1/1 and McCa/a genotypes were used as the reference genotypes, respectively. Statistically significant differences in RBC CR1 level from the reference genotypes are asterisked, *P<0.05, ***P<0.001.
Figure 6.
RBC CR1 level by Swain-Langley and McCoy genotype combinations. Differences in RBC CR1 level by Sl and McC genotype combinations were compared by multivariate linear regression analysis in a total of 3075 individuals with adjustment for age, α+thalassaemia and HbS genotypes. Error bars represent 95% confidence intervals of the mean RBC CR1 level. Numbers in parenthesis reflect total number of samples tested for each genotype combination group. Statistically significant differences in RBC CR1 level from the reference Sl1/1_McCa/a genotype combination are asterisked, **P<0.01, ***P<0.001.
Discussion
An increasing body of evidence from in vitro studies supports a role for CR1 in the pathogenesis of severe malaria,4, 5, 6, 7, 8, 9 yet little is known about the determinants of RBC CR1 level variation in African populations. We and others have shown previously that the SNPs in the CR1 gene that are associated with RBC CR1 level in Caucasian and Asian populations do not influence RBC level in African populations.21, 23 In the current study, we have shown that the CR1 Knops blood group Sl2 and McCb genotypes are associated with increased RBC CR1 levels. In addition, we have confirmed age-associated changes in RBC CR1 during childhood, and that α+thalassaemia is associated with reduced RBC CR1 levels, an observation that could largely be explained by the microcytic nature of thalassaemic RBCs.
To the best of our knowledge, the association between RBC CR1 level and the CR1 Knops blood group antigens has not been investigated previously. We have shown that both Sl2 and McCb are associated with increased RBC CR1 level, even after adjusting for known confounders (Figures 5 and 6). The mechanisms involved, however, remain unclear. Sl2 and McCb alleles are the products of substitution of the basic amino acid arginine for the neutral glycine and substitution of the basic amino acid lysine for the acidic glutamic acid, respectively.27 CR1 is known to be cleaved from the RBC surface as cells age,32 although the specific proteases involved are unknown. Several proteases attack peptide bonds at the amino or carboxyl side of arginine or lysine residues, therefore one hypothesis is that the Sl2 and McCb substitutions cause loss of proteolytic cleavage sites and hence result in higher CR1 levels on RBC. Alternatively, the substitutions might cause conformational changes in CR1 that might render the molecule less susceptible to proteolytic cleavage. However, previous work using nuclear magnetic resonance was unable to demonstrate any structural differences between the different polymorphic forms of CR1.33
The mechanism by which α+thalassaemia results in lower RBC CR1 levels, as shown in this study (Figure 2) and reported previously,14 has not yet been defined. We found that RBC size is an important factor, as the difference between thalassaemic and normal cells was no longer statistically significant after adjusting for the reduced MCV of α+thalassaemic RBCs34 (Figure 3). Microcytic RBCs due to α+thalassaemia and other causes such as iron-deficiency have been shown to have reduced rosette frequencies with P. falciparum-infected RBC, which could influence susceptibility to severe malaria.35 It remains unclear whether microcytic RBC synthesise lower amounts of CR1 than normal cells, or whether some CR1 is lost, for example, through exocytosis of membrane vesicles during normal RBC ageing,36 as well as by proteolysis as described above. Increased vesiculation has been documented in α+thalassaemia RBCs,37, 38, 39 which might therefore provide a mechanism for CR1 loss.
In agreement with two previous studies,16, 29 we found that RBC CR1 levels were influenced by host age in the Kilifi population (Figure 1). The mechanisms leading to these age-related changes are unknown. RBC MCV is known to vary with age, falling in early childhood up to about 15 months of age, and then steadily rising into adulthood.40 Changes in MCV with age were relatively minor in our data set of 147 samples (Supplementary Figure S1), and seem unlikely to account for the differences in CR1 levels between different age groups during early childhood. However, further studies of RBC CR1 level, MCV and age with larger sample sizes and over a wider age range will be needed to examine this relationship more fully.
Although factors affecting RBC CR1 level have been clearly demonstrated in this study, the work has some limitations. In addition to the Knops blood groups polymorphisms, CR1 also displays four allelic size variants: CR1-A (220 kDa), CR1-B (250 kDa), CR1-C (190 kDa) and CR1-D (280 kDa). These differences are a result of unequal crossing-over leading to duplications or deletions of a long homologous repeat (LHR) unit, with CR1-A, B, C and D consisting of 4, 5, 3 and 6 LHR's, respectively. CR1-A and B are the most common, occurring at frequencies of between 75–98% and 2–13%, respectively, whereas CR1-C and D are rare.41 The association between CR1 allelic variants and RBC CR1 level and Knops antigen type is unknown and was not examined in our study. Different anti-CR1 monoclonal antibodies are known to recognise epitopes on different LHRs;42, 43 for example, J3D3 binds epitopes on LHR-A, B and C.43 Therefore, depending on the overall frequencies of these different allelic variants in our population, and how they associate with the different genotypes tested, it is possible that we may have either under- or over-estimated CR1 levels using our current methods.44 Furthermore, the allelic types of the samples of known CR1 level used as standards in our CR1 flow cytometry assay are unknown, and again could influence the CR1 levels measured.
Another uncertainty is whether the variation in RBC CR1 level that we describe here has functional significance in terms of malaria host–parasite interactions. Previous work on CR1-mediated invasion of RBC by P. falciparum shows a significant positive correlation between RBC CR1 level and binding of the parasite invasion ligand P. falciparum reticulocyte-binding-like homologue protein 4(ref. 7) and parasite invasion rate,5 suggesting that relatively small increases in RBC CR1 level may have the potential to increase invasion efficiency and increase overall parasite burden. Previous work on P. falciparum rosetting suggests that very low levels of RBC CR1 (less than 100 molecules per cell) reduce rosetting,8, 9 but there is no evidence for a direct positive correlation between increasing amounts of CR1 and increasing rosetting. Instead, it seems more likely that there is a threshold effect, such that cells with less than 100–200 molecules per cell show reduced rosetting, whereas those with more than 100–200 molecules per cell show similar rosetting capacity over the full range of CR1 levels (200–1500 molecules per cell). The overall levels of CR1 measured in this study are relatively low, with mean values between 250 and 350 molecules per RBC for most age groups and genotypes (Figures 1,2,3,4,5,6), but they are higher than those measured in other parts of the world where low CR1 has been associated with protection against severe malaria, such as Papua New Guinea14 and India.15 Further work will be needed to study the relationship between RBC CR1 level, P. falciparum invasion and rosetting and severe malaria in African populations to determine the functional significance of the CR1 level variation described here.
One unexplained contradiction in our data is that low CR1 is thought to protect against severe malaria,14, 15 yet the Knops mutations are associated with increased CR1 levels (Figures 5 and 6). This appears inconsistent with suggestions that the Knops mutations have been selected to high frequencies in African populations because of a protective advantage against severe malaria.8, 45 Recent in vitro data using truncated recombinant proteins with the Knops polymorphisms found no effect of the Sl2 and McCb mutations on CR1 complement regulatory functions or P. falciparum rosetting or invasion.33 In addition, the SNPs determining Sl and McC blood group antigens are located within LHR-D of CR1,8, 27 whereas the sites implicated in rosetting and invasion are in LHR-A, -B and -C.8, 9, 6, 46 It is possible that the Knops polymorphisms might protect against severe malaria through mechanisms in which increased CR1 is advantageous. CR1 is involved in clearance of C3b/C4b-coated immune complexes and regulation of complement activation, and RBCs with low CR1 may be more susceptible to complement-mediated damage and increased clearance, which are mechanisms associated with severe malarial anaemia.29, 47, 48 Alternatively, Sl2 and McCb could be under selection through other diseases unrelated to malaria. CR1 is a receptor-mediating phagocytic uptake and cell invasion by a number of infectious agents including Mycobacterium tuberculosis, M. leprae and Leishmania major.49, 50, 51, 52, 53, 54, 55, 56, 57 Preliminary studies suggest that McCb is associated with protection against M. tuberculosis and M. leprae in West Africa.58, 59 Recent evidence from our group also points to both McCb and Sl2 protecting against common non-malaria-related illnesses among children living on the Coast of Kenya (Opi, et al., manuscript in preparation).
In summary, we have shown that a number of host factors including Knops blood group genotype, α+thalassaemia and age are associated with RBC CR1 level in Kenyan children. The exact mechanisms by which levels of RBC CR1 are controlled remain unknown, but variation in RBC CR1 might be partly explained by differences in haematological indices such as MCV or by other complex processes including susceptibility to proteases and RBC vesiculation that are potential areas for future study.
Materials and methods
Study populations
All samples used in this study were collected from residents of the area served by the Kilifi Health and Demographic Surveillance System (KHDSS) on the coast of Kenya. This area and the resident population have been described in detail previously.60 The study involved a total of 3535 individuals aged 0 months to 13 years of age recruited into three different study cohorts, each of which have been previously described: the Kilifi Genetic Birth Cohort (2928 individuals),61 Birth Cohort Study 2 (460 individuals)62, 63 and Mild Disease Cohort (147 individuals).64, 65 Following informed consent from parents or guardians, blood samples were collected by either venipuncture or by capillary sampling into sterile tubes containing EDTA (BD Vacutainer systems, Franklin Lakes, NJ, USA). Because CR1 levels are known to decline during malaria infections,16 samples were only collected from malaria negative individuals, as tested by a rapid diagnostic test (OptiMAL Diamed, Morat, Switzerland) and by light microscopy of thick and thin Giemsa-stained blood smears.
RBC surface CR1 quantification by flow cytometry
Whole blood was washed thrice in supplemented RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA; with 25 mM HEPES, 2 mM l-glutamine (Invitrogen), 25 μg ml−1 gentamicin, 20 mM d-glucose (Sigma) and 6 mM NaOH) by centrifugation and plasma and white blood cells removed from the RBC pellet by aspiration. RBC surface CR1 level was quantified by flow cytometry on formaldehyde-fixed RBC samples, as described in detail previously.66 Previous data indicated that CR1 levels determined on formaldehyde-fixed RBCs are equivalent to those determined using fresh RBCs, as long as the standards (with known CR1 levels) are fixed and stored in an identical way.66 Fixed cells were stored at 4 °C and CR1 quantified within 8 weeks. Briefly, fixed RBC pellets were stained in duplicate with 0.5 μg ml−1 of mouse anti-CR1 monoclonal antibody J3D3 (catalogue number COIM0195, Beckman-Coulter Inc., Fullerton, CA, USA) followed by a secondary staining with 5 μg ml−1 of Alexa Fluor 488-conjugated goat anti-mouse IgG (catalogue number A-11001, Molecular Probes, Leiden, the Netherlands) and the mean fluorescence intensity determined on a FC500 flow cytometer (Beckman-Coulter Inc.). CR1 levels of unknown samples were then extrapolated from a standard curve generated using three RBC samples of known CR1 levels (low ~350, intermediate ~650 and high CR1 levels ~900) included in each experiment, and prepared and stored in the same way as the test samples.66 Thirteen samples were collected but were not included in the study due aberrant forward/side scatter plots or readings that were >2 standard deviations above the highest point on the standard curve.
DNA extraction and genotyping
DNA was extracted either from fresh whole blood samples using the semi-automated ABI PRISM 6100 Nucleic acid prep station (Applied Biosystems, Foster City, CA, USA) or from EDTA blood samples, previously stored at −80 °C, using the QIAamp DNA and Blood Mini Kit (Qiagen, Valencia, CA, USA). Genotyping for the Sl and McC blood group genotypes was carried out as described previously using the SEQUENOM MassARRAY platform (Agena Biosciences, Hamburg, Germany) multiplex system following DNA amplification by whole-genome amplification.67 ABO blood groups were determined by either standard haemagglutination methods or were inferred from A, B and O SNP haplotypes determine by Sequenom typing as detailed previously.67 Genotyping for HbAS and α+thalassaemia was by PCR as described in detail elsewhere.68, 69
MCV determination
Full blood counts for the determination of RBC MCV were performed on fresh EDTA blood samples from the mild disease cohort using a Coulter MDII-18 machine (Beckman-Coulter Inc.).
Sample availability
A total of 3535 samples were successfully typed for RBC CR1 level, HbS and α+thalassaemia genotypes, whereas ABO and the Sl and McC Knops genotypes were available for all samples except the 460 samples from the Birth Cohort Study 2. MCV data were only available for the 147 samples collected from members of the Mild Disease Cohort Study.
Statistical analysis
Statistical analyses were conducted using STATA v11 (StataCorp LP, TX, USA) and Graph Pad Prism v5 (GraphPad Software Inc, San Diego, CA, USA) and all graphs were generated using GraphPad Prism. The relationship between RBC CR1 level and age as a categorical variable was assessed using one-way analysis of variance with Tukey's post-hoc test to correct for multiple comparisons. We also analysed the relationship between RBC CR1 level and age as a continuous variable using piecewise-linear regression analysis allowing for adjustment for confounding by α+thalassaemia and HbS. Unlike conventional linear models that assume linearity over the whole range of a continuous independent variable, this model assumes a linear effect within defined ranges of the continuous variable that are connected. The age categories and ranges chosen were 0–3 months, 4–6 months, 7–12 months, 13–24 months, >2–4 years, >4–8 years and >8–13 years, as used in previous studies.16, 29 The relationship between RBC CR1 level and α+thalassaemia, HbS, ABO blood group and the Sl and McC genotypes was tested by multivariate linear regression analysis. Each variable was first tested in a univariate regression model and all explanatory variables displaying a P value of <0.05 (all except ABO) were included in a final multivariate model. Age as a categorical variable was also included in the multivariate model, using the age intervals described above. ABO blood group and Sl and McC genotypes were not available for the 460 children from Birth Cohort Study 2, therefore the multivariate analyses are restricted to the 3075 samples with full genotype data. For ease of interpretation, predicted mean CR1 levels and 95% CIs estimated from the multivariate regression results are presented in figures, rather than regression coefficients, using the ‘margins' post-estimation command in Stata. A two-sided significance level of <0.05 was considered statistically significant for all analyses.
Ethical statement
Individual informed written consent to collect blood was obtained from all participants' parents or legal guardians. The study was approved by the Kenya Medical Research Institute National Ethical Review Committee in Nairobi and was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
We thank the children and their families who made these studies possible, and the medical, nursing and research staff of the KEMRI-Wellcome Trust Research Programme who assisted with these studies. We particularly thank Metrine Tendwa, Johnston Makale and Adan Mohammed for laboratory support, Oscar Kai for flow cytometry support and Gideon Nyutu and Caroline Ndila for help with data management. We are also grateful to the volunteers from KEMRI-Wellcome Trust Research Programme who donated blood used as controls in RBC CR1 level determination. This paper was published with permission from the Director of the KENYA Medical Research Institute (KEMRI). This work was funded by the Wellcome Trust through Senior Research Fellowships awarded to TNW (grant no 091758) and JAR (grant no 084226), through core support to the KEMRI-Wellcome Trust Programme (grant no 084535) and through a sub-grant from a Wellcome Trust Strategic Award (grant no 084538) to DHO. The research leading to these results also received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement 242095.
Author contributions
DHO, SU, TNW and JAR conceived and designed the experiments. DHO, SU and EN performed the experiments. DHO, TNW and JAR analysed the data. DHO, TNW and JAR wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- WHO. World Malaria Report 2013. WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Miller LH, Ackerman HC, Su XZ, Wellems TE. Malaria biology and disease pathogenesis: insights for new treatments. Nat Med 2013; 19: 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CQ, Kennedy AT, Tham WH. More than just immune evasion: Hijacking complement by Plasmodium falciparum. Mol Immunol 2015; 67: 71–84. [DOI] [PubMed] [Google Scholar]
- Awandare GA, Spadafora C, Moch JK, Dutta S, Haynes JD, Stoute JA. Plasmodium falciparum field isolates use complement receptor 1 (CR1) as a receptor for invasion of erythrocytes. Mol Biochem Parasitol 2011; 177: 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C, Awandare GA, Kopydlowski KM, Czege J, Moch JK, Finberg RW et al. Complement receptor 1 is a sialic acid-independent erythrocyte receptor of Plasmodium falciparum. PLoS Pathog 2010; 6: e1000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Schmidt CQ, Hauhart RE, Guariento M, Tetteh-Quarcoo PB, Lopaticki S et al. Plasmodium falciparum uses a key functional site in complement receptor type-1 for invasion of human erythrocytes. Blood 2011; 118: 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Wilson DW, Lopaticki S, Schmidt CQ, Tetteh-Quarcoo PB, Barlow PN et al. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc Natl Acad Sci USA 2010; 107: 17327–17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature 1997; 388: 292–295. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Rogerson SJ, Raza A, Moulds JM, Kazatchkine MD, Marsh K et al. Mapping of the region of complement receptor (CR) 1 required for Plasmodium falciparum rosetting and demonstration of the importance of CR1 in rosetting in field isolates. J Immunol 2000; 165: 6341–6346. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Claessens A, Corrigan RA, Arman M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev Mol Med 2009; 11: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul DK, Roth EF Jr., Nagel RL, Howard RJ, Handunnetti SM. Rosetting of Plasmodium falciparum-infected red blood cells with uninfected red blood cells enhances microvascular obstruction under flow conditions. Blood 1991; 78: 812–819. [PubMed] [Google Scholar]
- Nash GB, Cooke BM, Carlson J, Wahlgren M. Rheological properties of rosettes formed by red blood cells parasitized by Plasmodium falciparum. Br J Haematol 1992; 82: 757–763. [DOI] [PubMed] [Google Scholar]
- Moulds JM, Moulds JJ, Brown M, Atkinson JP. Antiglobulin testing for CR1-related (Knops/McCoy/Swain-Langley/York) blood group antigens: negative and weak reactions are caused by variable expression of CR1. Vox Sang 1992; 62: 230–235. [DOI] [PubMed] [Google Scholar]
- Cockburn IA, Mackinnon MJ, O'Donnell A, Allen SJ, Moulds JM, Baisor M et al. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci USA 2004; 101: 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Jha GN, Anand P, Qidwai T, Pati SS, Mohanty S et al. CR1 levels and gene polymorphisms exhibit differential association with falciparum malaria in regions of varying disease endemicity. Hum Immunol 2009; 70: 244–250. [DOI] [PubMed] [Google Scholar]
- Waitumbi JN, Donvito B, Kisserli A, Cohen JH, Stoute JA. Age-related changes in red blood cell complement regulatory proteins and susceptibility to severe malaria. J Infect Dis 2004; 190: 1183–1191. [DOI] [PubMed] [Google Scholar]
- Stoute JA. Complement receptor 1 and malaria. Cell Microbiol 2011; 13: 1441–1450. [DOI] [PubMed] [Google Scholar]
- Khera R, Das N. Complement Receptor 1: disease associations and therapeutic implications. Mol Immunol 2009; 46: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JG, Murphy EE, Wong WW, Klickstein LB, Weis JH, Fearon DT. Identification of a restriction fragment length polymorphism by a CR1 cDNA that correlates with the number of CR1 on erythrocytes. J Exp Med 1986; 164: 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JG, Wong WW, Schur PH, Fearon DT. Mode of inheritance of decreased C3b receptors on erythrocytes of patients with systemic lupus erythematosus. N Engl J Med 1982; 307: 981–986. [DOI] [PubMed] [Google Scholar]
- Herrera AH, Xiang L, Martin SG, Lewis J, Wilson JG. Analysis of complement receptor type 1 (CR1) expression on erythrocytes and of CR1 allelic markers in Caucasian and African American populations. Clin Immunol immunopathol 1998; 87: 176–183. [DOI] [PubMed] [Google Scholar]
- Xiang L, Rundles JR, Hamilton DR, Wilson JG. Quantitative alleles of CR1: coding sequence analysis and comparison of haplotypes in two ethnic groups. J Immunol 1999; 163: 4939–4945. [PubMed] [Google Scholar]
- Rowe JA, Raza A, Diallo DA, Baby M, Poudiougo B, Coulibaly D et al. Erythrocyte CR1 expression level does not correlate with a HindIII restriction fragment length polymorphism in Africans; implications for studies on malaria susceptibility. Genes Immun 2002; 3: 497–500. [DOI] [PubMed] [Google Scholar]
- Moulds JM, Nickells MW, Moulds JJ, Brown MC, Atkinson JP. The C3b/C4b receptor is recognized by the Knops, McCoy, Swain-langley, and York blood group antisera. J Exp Med 1991; 173: 1159–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulds JM, Thomas BJ, Doumbo O, Diallo DA, Lyke KE, Plowe CV et al. Identification of the Kna/Knb polymorphism and a method for Knops genotyping. Transfusion 2004; 44: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulds JM, Zimmerman PA, Doumbo OK, Diallo DA, Atkinson JP, Krych-Goldberg M et al. Expansion of the Knops blood group system and subdivision of Sl(a). Transfusion 2002; 42: 251–256. [DOI] [PubMed] [Google Scholar]
- Moulds JM, Zimmerman PA, Doumbo OK, Kassambara L, Sagara I, Diallo DA et al. Molecular identification of Knops blood group polymorphisms found in long homologous region D of complement receptor 1. Blood 2001; 97: 2879–2885. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Opi DH, Williams TN. Blood groups and malaria: fresh insights into pathogenesis and identification of targets for intervention. Curr Opin Hematol 2009; 16: 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odhiambo CO, Otieno W, Adhiambo C, Odera MM, Stoute JA. Increased deposition of C3b on red cells with low CR1 and CD55 in a malaria-endemic region of western Kenya: implications for the development of severe anemia. BMC Med 2008; 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TN, Mwangi TW, Wambua S, Peto TE, Weatherall DJ, Gupta S et al. Negative epistasis between the malaria-protective effects of alpha+-thalassemia and the sickle cell trait. Nat Genet 2005; 37: 1253–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Handel IG, Thera MA, Deans AM, Lyke KE, Kone A et al. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci USA 2007; 104: 17471–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche J, Sim RB. Loss of complement receptor type 1 (CR1) on ageing of erythrocytes. Studies of proteolytic release of the receptor. Biochem J 1986; 235: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh-Quarcoo PB, Schmidt CQ, Tham WH, Hauhart R, Mertens HD, Rowe A et al. Lack of evidence from studies of soluble protein fragments that Knops blood group polymorphisms in complement receptor-type 1 are driven by malaria. PLoS One 2012; 7: e34820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TN, Maitland K, Ganczakowski M, Peto TE, Clegg JB, Weatherall DJ et al. Red blood cell phenotypes in the alpha+thalassaemias from early childhood to maturity. Br J Haematol 1996; 95: 266–272. [DOI] [PubMed] [Google Scholar]
- Carlson J, Nash GB, Gabutti V, al-Yaman F, Wahlgren M. Natural protection against severe Plasmodium falciparum malaria due to impaired rosette formation. Blood 1994; 84: 3909–3914. [PubMed] [Google Scholar]
- Dervillez X, Oudin S, Libyh MT, Tabary T, Reveil B, Philbert F et al. Catabolism of the human erythrocyte C3b/C4b receptor (CR1, CD35): vesiculation and/or proteolysis? Immunopharmacology 1997; 38: 129–140. [DOI] [PubMed] [Google Scholar]
- Pattanapanyasat K, Noulsri E, Fucharoen S, Lerdwana S, Lamchiagdhase P, Siritanaratkul N et al. Flow cytometric quantitation of red blood cell vesicles in thalassemia. Cytometry B Clin Cytom 2004; 57: 23–31. [DOI] [PubMed] [Google Scholar]
- Allan D, Limbrick AR, Thomas P, Westerman MP. Release of spectrin-free spicules on reoxygenation of sickled erythrocytes. Nature 1982; 295: 612–613. [DOI] [PubMed] [Google Scholar]
- Alaarg A, Schiffelers RM, van Solinge WW, van Wijk R. Red blood cell vesiculation in hereditary hemolytic anemia. Front Physiol 2013; 4: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serjeant GR, Grandison Y, Lowrie Y, Mason K, Phillips J, Serjeant BE et al. The development of haematological changes in homozygous sickle cell disease: a cohort study from birth to 6 years. Br J Haematol 1981; 48: 533–543. [DOI] [PubMed] [Google Scholar]
- Birmingham DJ, Hebert LA. CR1 and CR1-like: the primate immune adherence receptors. Immunol Rev 2001; 180: 100–111. [DOI] [PubMed] [Google Scholar]
- Cook J, Fischer E, Boucheix C, Mirsrahi M, Jouvin MH, Weiss L et al. Mouse monoclonal antibodies to the human C3b receptor. Mol Immunol 1985; 22: 531–539. [DOI] [PubMed] [Google Scholar]
- Nickells M, Hauhart R, Krych M, Subramanian VB, Geoghegan-Barek K, Marsh HC Jr. et al. Mapping epitopes for 20 monoclonal antibodies to CR1. Clin Exp Immunol 1998; 112: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri RA, Schifferli JA. Over-estimation of the number of complement receptor type 1 (CR1) on erythrocytes. Scand J Immunol 1992; 36: 125–130. [DOI] [PubMed] [Google Scholar]
- Thathy V, Moulds JM, Guyah B, Otieno W, Stoute JA. Complement receptor 1 polymorphisms associated with resistance to severe malaria in Kenya. Malar J 2005; 4: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Guariento M, Maciejewski M, Hauhart R, Tham WH, Cowman AF et al. Using mutagenesis and structural biology to map the binding site for the Plasmodium falciparum merozoite protein PfRh4 on the human immune adherence receptor. J Biol Chem 2014; 289: 450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owuor BO, Odhiambo CO, Otieno WO, Adhiambo C, Makawiti DW, Stoute JA. Reduced immune complex binding capacity and increased complement susceptibility of red cells from children with severe malaria-associated anemia. Mol Med 2008; 14: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoute JA, Odindo AO, Owuor BO, Mibei EK, Opollo MO, Waitumbi JN. Loss of red blood cell-complement regulatory proteins and increased levels of circulating immune complexes are associated with severe malarial anemia. J Infect Dis 2003; 187: 522–525. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Torano A. Immune adherence-mediated opsonophagocytosis: the mechanism of Leishmania infection. J Exp Med 1999; 189: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey JS, Carroll MC, Brown EJ. A macrophage invasion mechanism of pathogenic mycobacteria. Science 1997; 277: 1091–1093. [DOI] [PubMed] [Google Scholar]
- Hirsch CS, Ellner JJ, Russell DG, Rich EA. Complement receptor-mediated uptake and tumor necrosis factor-alpha-mediated growth inhibition of Mycobacterium tuberculosis by human alveolar macrophages. J Immunol 1994; 152: 743–753. [PubMed] [Google Scholar]
- Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol 1990; 144: 2771–2780. [PubMed] [Google Scholar]
- Schlesinger LS, Horwitz MA. Phagocytosis of leprosy bacilli is mediated by complement receptors CR1 and CR3 on human monocytes and complement component C3 in serum. J Clin Invest 1990; 85: 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva RP, Hall BF, Joiner KA, Sacks DL. CR1, the C3b receptor, mediates binding of infective Leishmania major metacyclic promastigotes to human macrophages. J Immunol 1989; 143: 617–622. [PubMed] [Google Scholar]
- Robledo S, Wozencraft A, Valencia AZ, Saravia N. Human monocyte infection by Leishmania (Viannia) panamensis. Role of complement receptors and correlation of susceptibility in vitro with clinical phenotype. J Immunol 1994; 152: 1265–1276. [PubMed] [Google Scholar]
- Payne NR, Horwitz MA. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J Exp Med 1987; 166: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogembo JG, Kannan L, Ghiran I, Nicholson-Weller A, Finberg RW, Tsokos GC et al. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep 2013; 3: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noumsi GT, Tounkara A, Diallo H, Billingsley K, Moulds JJ, Moulds JM. Knops blood group polymorphism and susceptibility to Mycobacterium tuberculosis infection. Transfusion 2011; 51: 2462–2469. [DOI] [PubMed] [Google Scholar]
- Fitness J, Floyd S, Warndorff DK, Sichali L, Mwaungulu L, Crampin AC et al. Large-scale candidate gene study of leprosy susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg 2004; 71: 330–340. [PubMed] [Google Scholar]
- Scott JA, Bauni E, Moisi JC, Ojal J, Gatakaa H, Nyundo C et al. Profile: The Kilifi Health and Demographic Surveillance System (KHDSS). Int J Epidemiol 2012; 41: 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TN, Uyoga S, Macharia A, Ndila C, McAuley CF, Opi DH et al. Bacteraemia in Kenyan children with sickle-cell anaemia: a retrospective cohort and case-control study. Lancet 2009; 374: 1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokes DJ, Okiro EA, Ngama M, Ochola R, White LJ, Scott PD et al. Respiratory syncytial virus infection and disease in infants and young children observed from birth in Kilifi District, Kenya. Clin Infect Dis 2008; 46: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English M, Muhoro A, Aluda M, Were S, Ross A, Peshu N. Outcome of delivery and cause-specific mortality and severe morbidity in early infancy: a Kenyan District Hospital birth cohort. Am J Trop Med Hyg 2003; 69: 228–232. [PubMed] [Google Scholar]
- Wambua S, Mwangi TW, Kortok M, Uyoga SM, Macharia AW, Mwacharo JK et al. The effect of alpha+-thalassaemia on the incidence of malaria and other diseases in children living on the coast of Kenya. PLoS Med 2006; 3: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TN, Mwangi TW, Wambua S, Alexander ND, Kortok M, Snow RW et al. Sickle cell trait and the risk of Plasmodium falciparum malaria and other childhood diseases. J Infect Dis 2005; 192: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Donvito B, Cohen JH, Rowe JA. A simple method for accurate quantification of complement receptor 1 on erythrocytes preserved by fixing or freezing. J Immunol Methods 2002; 271: 59–64. [DOI] [PubMed] [Google Scholar]
- Fry AE, Griffiths MJ, Auburn S, Diakite M, Forton JT, Green A et al. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum Mol Genet 2008; 17: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfall CM, Cobb BD. Single tube genotyping of sickle cell anaemia using PCR-based SNP analysis. Nucleic Acids Res 2001; 29: E119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SS, Boehm CD, Higgs DR, Cutting GR. Single-tube multiplex-PCR screen for common deletional determinants of alpha-thalassemia. Blood 2000; 95: 360–362. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.