Abstract
Inhibitor of DNA binding 2 (ID2) is a helix-loop-helix transcriptional repressor rhythmically expressed in many adult tissues. Our previous studies have demonstrated that Id2 null mice have sex-specific elevated glucose uptake in brown adipose tissue (BAT). Here we further explored the role of Id2 in the regulation of core body temperature over the circadian cycle and the impact of Id2 deficiency on genes involved in insulin signaling and adipogenesis in BAT. We discovered a reduced core body temperature in Id2−/− mice. Moreover, in Id2−/− BAT, 30 genes including Irs1, PPARs, and PGC-1s were identified as differentially expressed in a sex-specific pattern. These data provide valuable insights into the impact of Id2 deficiency on energy homeostasis of mice in a sex-specific manner.
1. Introduction
The circadian clock is an autoregulatory network that regulates behavioral and metabolic programing in the context of a 24 h light-dark (LD) cycle [1]. Body temperature is one of the representing benchmarks of circadian patterning, which peaks in animals while awake and troughs while asleep [2]. Brown adipose tissue (BAT) is a major site for rodent thermogenesis, due to its involvement in controlling circadian thermogenic rhythms and influencing adaptability to environmental temperature challenges [3]. A previous study has revealed rhythmic expression patterns of over 5,000 genes in murine BAT, including genes associated with the circadian clock, adipose function, and metabolism [4]. Moreover, glucose uptake in BAT exhibits a diurnal rhythm [5].
The Inhibitor of DNA binding 2 (Id2) gene encodes a helix-loop-helix (HLH) transcriptional regulator, which is rhythmically expressed in many mammalian tissues and involved in the input pathway, core clock function, and output pathways of the circadian clock [6–9]. Our previous studies have shown that Id2−/− mice exhibit lower levels of locomotor activity, extended nighttime activity patterns of feeding and locomotor activity, and sex- and age-dependent enhanced glucose tolerance and insulin sensitivity [10]. Moreover, an energy-rich diet is able to rescue the disturbances to metabolic homeostasis and survival in the Id2−/− mice sex-specifically [11]. Importantly, Id2−/− mice show a sex-dependent elevated glucose uptake in interscapular BAT (iBAT) [10]. Id2 also plays a role in white adipose tissue (WAT) adipogenesis [10–12]. However, the role of Id2 on temperature homeostasis regulation and its influence on BAT physiology remain unknown. Therefore, we investigated the function of Id2 in the regulation of temperature rhythms under normal and thermoneutral conditions in a sex-specific manner and also profiled the expression of genes involved in insulin signaling and adipogenesis in BAT of Id2−/− mice, sex-specifically.
2. Materials and Methods
2.1. Animals
The generation of Id2−/− mice and genotype determination were performed as described previously [7, 10, 11]. Mice were maintained on a regular chow diet and sterile water containing antibiotic ad libitum [7, 10, 11]. All mice were housed in laboratory cages at normal temperature (21°C ± 1°C) and humidity of 50–65% under a 12 : 12 light : dark (LD) cycle with lights on at Zeitgeber time (ZT) 0 and lights off at ZT12. Controls were age- and sex-matched WT littermate mice. Animal experiments were approved by the University of Notre Dame Animal Care and Use Committee (Protocol number 14-02-1559) and performed in accordance with NIH Guidelines for the Care and Use of Laboratory Animals.
2.2. Temperature Measurement
Temperature measurements were carried out on 2-month–1.5-year- (5.5-month median) old male and female Id2−/− mice and WT littermates, housed individually in a climate-controlled room set to either normal (21°C ± 1°C) or thermoneutral (30°C ± 1°C) temperature. Body temperature sampling was conducted at 3 h intervals over the 24 h LD cycle. For thermoneutral conditions measurement, all WT and Id2−/− mice used in the studies were allowed to acclimate to thermoneutral temperature for 1 week before temperature measurement. Core body temperature was measured using subcutaneously surgically implanted telemetric transmitters positioned proximal to the iBAT (IPTT 300 transponders, Bio Medic Data Systems, Seaford, DE) following isoflurane anesthetization [3]. After a week of recuperation, core temperatures were recorded over a 24 h period.
2.3. iBAT PCR Array Preparation and Analysis
iBAT tissue was harvested at ZT16 (Id2 mRNA circadian rhythm in iBAT has a broad peak phase between ZT8 and ZT16) [4]. Id2−/− and WT male (WT = 8, Id2−/− = 6) and female (WT = 6, Id2−/− = 4) mice from 3–9 months (6.1-month median) were sacrificed and iBAT tissue was frozen in liquid nitrogen and stored at −80°C until analyzed. RNA extraction was performed as described previously [7, 13]. We also measured iBAT weight of these and additional mice (3–10-month-old Id2−/− mice and WT littermates; 6.3-month median; male, WT = 15, Id2−/− = 7; female, WT = 10, Id2−/− = 9) as described previously [10, 11]. Total RNA was purified following a Trizol extraction and sodium acetate/ethanol treatment. RNA integrity was assessed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA was subjected to a DNASe I treatment, and cDNA was synthesized by RT2 First Strand Kit (SABiosciences). Relative mRNA expression of 168 genes involved in insulin signaling and adipogenesis pathways was determined by using the mouse PCR arrays (PAMM-030ZC-24 and PAMM-049ZC-24, SABiosciences). Quantitative real-time PCR was performed using an Applied Biosystems 7500 system with RT2 SYBR green ROX qPCR master mix reagent (Qiagen). PCR array data were calculated by the comparative cycle threshold method and analyzed by Web-based free PCR array data analysis software provided by SABiosciences. Normalization of expression was to housekeeping genes provided on each array (Actb, B2m, Gapdh, Gusb, and Hsp90ab1). Clock controlled genes (CCGs) were identified from the CIRCA database of Mouse 1.OST Brown Adipose (Affymetrix) (http://bioinf.itmat.upenn.edu/circa/) where we defined CCGs as a JTK_CYCLE algorithm determined q < 0.1 value and a period length of 20–28 h as described previously [13–15]. Circadian phase was determined from the Lomb-Scargle phase values within CIRCA.
2.4. Statistics
Data were analyzed using Sigma Plot 12.0 software to run two-factor ANOVA. Where necessary, data were ranks transformed to correct for nonnormal distributions. The linear regression of iBAT temperature-body weight relationship was generated and analyzed using Prism 5.0 Graphpad software. PCR array data were analyzed using the Web-based free PCR array data analysis software provided by SABiosciences (Student's t-test).
3. Results
3.1. Loss of Id2 Results in a Reduced Core Body Temperature in Male and Female Mice
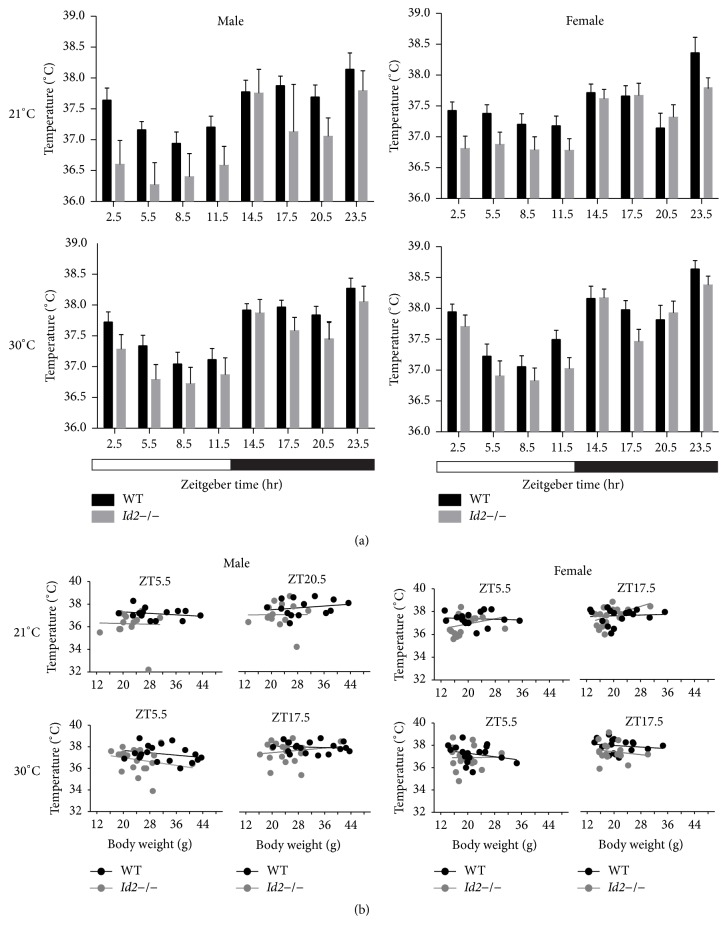
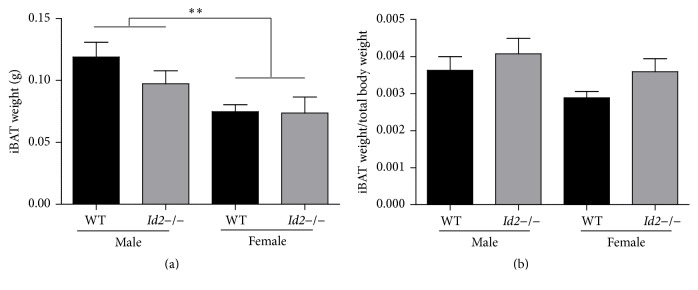
The discovery of a diurnal rhythm of glucose uptake in mice iBAT and a sex-dependent elevated glucose uptake in iBAT of Id2−/− mice prompted us to investigate whether Id2 contributes to thermoregulation [5, 10]. At normal ambient temperature conditions (21°C), ablation of Id2 reduced core body temperature across the 24 h day, in both male and female mice (Figure 1(a)) (males, wild types (WTs) = 14, Id2−/− = 14, ANOVA, time (T), p < 0.001, genotype (G) p < 0.001, interaction (I), n.s.; females, WTs = 18, Id2−/− = 17, ANOVA, T, p < 0.001, G, p < 0.05, I, n.s.). Considering the possibility of any confounding genetic background contribution and partial stimulation of BAT activity occurring under normal temperature conditions, Id2−/− mice core body temperature was also measured under thermoneutral conditions (30°C) [3, 16]. Consistently, at thermoneutrality, Id2−/− mice displayed a reduced core body temperature (Figure 1(a)) (males, WTs = 19, Id2−/− = 20, ANOVA, T, p < 0.001, G, p < 0.01, I, n.s.; females, WTs = 18, Id2−/− = 17, ANOVA, T, p < 0.001, G, p < 0.01, I, n.s.). Under both conditions and in both sexes, no interaction between time and genotype was discovered, suggesting a generalized effect of the null mutation on core body temperature rather than a time-of-day specific contribution of the gene deletion. Regression analysis of time-of-day representative core body temperatures (day or night) revealed no significant relationships between temperature and body mass for either Id2−/− or WT mice. However, Id2−/− mice of both sexes showed consistently lower y-intercept lines compared to WT mice when examined during either the daytime (ZT5.5) or nighttime (ZT17.5 or ZT20.5), thus confirming the consistently lower temperature of the Id2 null mice (Figure 1(b); Supplementary Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/6785948). Lastly, Id2−/− mice exhibited no statistically significant difference in iBAT weight and iBAT to body weight ratio compared to WT controls (Figures 2(a) and 2(b)) (two-factor ANOVAs, iBAT weight, G, n.s., sex (S), p < 0.01, I, n.s.; iBAT/body weight ratio, G, n.s., S, n.s., I, n.s.). The mean and SEM body mass of WT and Id2−/− mice for both iBAT weight and body temperature experiments are shown in Supplementary Table 2: note that both male and female Id2−/− mice had on average a lower body mass compared to WT counterparts (two-factor ANOVA, G, p < 0.001, S, p < 0.001, I, n.s.).
Figure 1.
Sex-specific regulation of body temperature in Id2−/− mice. (a) Body temperature measurements of WT and Id2−/− male (left) and female (right) mice under normal temperature (upper panel) or thermoneutral temperature (lower panel) over 24 hrs. Values are mean ± SEM. Two-factor ANOVA was performed. ANOVAs revealed significantly lower body temperatures for both male and female Id2−/− mice compared to WT mice. The genotypic effect was independent of the prevailing ambient temperature. (b) Upper: regression analysis of body weight to body temperature of WT and Id2−/− mice under normal ambient temperature (left: male at ZT5.5 and ZT20.5; right: female at ZT5.5 and ZT17.5). Lower: regression analysis of body weight to body temperature of WT and Id2−/− mice under thermoneutral temperature conditions (left: male at ZT5.5 and ZT17.5; right: female at ZT5.5 and ZT17.5). Values are individual animal body temperatures and their respective measures of body mass. Note that no linear regression was found to be significant (n.s.), indicating that body mass does not predict body temperature for any group analyzed, examined under either 21°C or 30°C environmental temperatures.
Figure 2.
Brown Adipose tissue weight in Id2−/− mice. (a) Interscapular brown adipose tissue (iBAT) mass (g) from WT and Id2−/− mice. (b) Ratio of weight of iBAT tissue to total body mass from WT and Id2−/− mice. Values are mean ± SEM. Two-factor ANOVAs were performed followed by Tukey's post hoc tests, ∗∗ p < 0.01. No significant differences were observed between groups in the iBAT mass/body mass analysis.
3.2. Sex-Specific Differential Gene Expression Associated with Insulin Signaling and Adipogenesis in iBAT of Id2−/− Mice
Our previous results showed sex-dependent enhanced insulin sensitivity and glucose uptake in iBAT of Id2−/− mice [10]. In the current study we observed a decreased core body temperature in Id2−/− mice as described above. To fully evaluate the impact of ablation of Id2 on BAT gene-regulation, we performed a gene expression analysis using RT2 Profiler PCR Arrays of BAT derived from Id2−/− mice and their WT littermates collected at the same time of the 24 h day (specifically ZT16). Deferentially regulated genes involved in insulin signaling and adipogenesis are shown in Tables 1 and 2, respectively. Thirty of 168 genes examined were identified as differentially expressed when analyzed as a cohort or as individual sex-specific groups. Using the CIRCA database as a resource [14], six genes were identified as clock controlled genes (CCGs), of which four oscillate in proximal phase with the rhythm of Peroxisome proliferator activated receptor alpha (Pparα), peaking during the middle of the day (~circadian time (CT) 6; CT12 = onset of night in prior LD cycle). Of importance for insulin signaling, glucose-6-phosphatase, catalytic (G6pc), was upregulated in Id2−/− females and the related G6pc family member G6pc2 downregulated in Id2−/− males (p = 0.079, approaching significance) compared to WTs. Insulin receptor substrate 1 (Irs1) was upregulated in both male and female Id2−/− mice. Protein Kinase C, iota (Prkcι), was downregulated in Id2−/− males. Insulin-like growth factor 2 (Igf2) was downregulated in female Id2−/− mice, while Fbp1, a rate-limiting enzyme in gluconeogenesis, and Shc1, a component in the IGF-1-regulated pathway, were upregulated. For adipogenesis, bone morphogenetic protein 4 (Bmp4) was elevated 1.7-fold (n.s.) in male and 1.6-fold in female Id2−/− mice. Consistent with the insulin signaling array, Irs1 was elevated in Id2−/− mice. Nuclear receptor coactivator 2 (Ncoa2), PR domain containing 16 (Prdm16), Pparα, and twist homolog 1 (twist1) were downregulated, in grouped analysis of male and female Id2−/− mice. Fatty acid synthase (Fasn), lipase, hormone sensitive (Lipe), and Peroxisome proliferative activated receptor, gamma, coactivator 1 beta (Ppargc1β/PGC-1β) were all downregulated in male Id2−/− mice. Female Id2−/− mice displayed a downregulation of proliferative activated receptor, gamma, coactivator 1 alpha (Ppargc1α/PGC-1α). A small 1.2-fold downregulation of Peroxisome proliferator activated receptor gamma (Pparγ) was detected in Id2−/− males, where the p value was approaching significance (p = 0.061). Note that the thermogenic protein, uncoupling protein 1 (ucp1), was present on both the insulin signaling and adipogenesis arrays, but its levels of expression were not significantly altered in the Id2−/− mice.
Table 1.
Differentially expressed genes from insulin signaling pathway of Id2−/− mice. Genes with significant differences (p < 0.05) are shown in bold. Peak phase value (determined by Lomb-Scargle phase values within CIRCA) in circadian time (CT) is provided where gene was identified as a clock control gene. Rhythmic genes identified from CIRCA database of Mouse 1.OST Brown Adipose (Affymetrix).
| Insulin signaling array | Symbol | Refseq | All | Male | Female | Clock control gene-peak phase | |||
|---|---|---|---|---|---|---|---|---|---|
| Fold change | p value | Fold change | p value | Fold change | p value | ||||
| Gene name | |||||||||
| AE binding protein 1 | Aebp1 | NM_009636 | 1.9 | 0.09 | 1.6 | 0.03 | — | ||
| Complement factor D (adipsin) | Cfd | NM_013459 | 2.4 | 0.01 | 3.6 | 0.03 | — | ||
| Fructose bisphosphatase 1 | Fbp1 | NM_019395 | 1.3 | 0.22 | 1.6 | 0.05 | — | ||
| Glucose-6-phosphatase, catalytic | G6pc | NM_008061 | 1.5 | 0.28 | 1.5 | 0.33 | 1.6 | 0.02 | — |
| Growth factor receptor bound protein 10 | Grb10 | NM_010345 | 1.5 | 0.08 | — | ||||
| Insulin receptor substrate 1 | Irs1 | NM_010570 | 1.4 | 0.01 | 1.3 | 0.07 | 1.4 | 0.05 | — |
| Protein tyrosine phosphatase, receptor type, F | Ptprf | NM_011213 | 1.7 | 0.05 | 2.6 | 0.05 | — | ||
| Scr homology 2 domain containing transforming protein C1 | Shc1 | NM_011368 | 1.25 | 0.01 | — | ||||
|
| |||||||||
| Glucose-6-phosphatase, catalytic, 2 | G6pc2 | NM_021331 | −3.2 | 0.08 | — | ||||
| Insulin-like growth factor 2 | Igf2 | NM_010514 | −1.2 | 0.21 | −1.4 | 0.04 | — | ||
| V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | Kras | NM_021284 | −1.1 | 0.09 | −1.25 | 0.03 | — | ||
| Protein kinase C, iota | Prkci | NM_008857 | −1.2 | 0.02 | −1.3 | 0.02 | CT0 | ||
| Thyroglobulin | Tg | NM_009375 | −1.8 | 0.29 | −3.5 | 0.02 | — | ||
Table 2.
Differentially expressed genes from adipogenesis pathway of Id2−/− mice. Genes with significant differences (p < 0.05) are shown in bold.
| Adipogenesis array | Symbol | Refseq | All | Male | Female | Clock control gene-peak phase | |||
|---|---|---|---|---|---|---|---|---|---|
| Fold change | p value | Fold change | p value | Fold change | p value | ||||
| Gene name | |||||||||
| Bone morphogenetic protein 4 | Bmp4 | NM_007554 | 1.6 | 0.04 | 1.7 | 0.12 | 1.6 | 0.01 | — |
| Complement factor D (adipsin) | Cfd | NM_013459 | 2.5 | 0.02 | 4.3 | 0.03 | — | ||
| Insulin receptor substrate 1 | Irs1 | NM_010570 | 1.2 | 0.03 | 1.1 | 0.29 | 1.4 | 0.02 | — |
| TSC22 domain family, member 3 | Tsc22d3 | NM_010286 | 1.6 | 0.04 | 1.9 | 0.09 | CT16 | ||
|
| |||||||||
| Axin 1 | Axin1 | NM_009733 | −1.2 | 0.07 | −1.3 | 0.05 | — | ||
| Cyclin-dependent kinase 4 | Cdk4 | NM_009870 | −1.4 | 0.003 | −1.7 | 0.001 | CT6 | ||
| Delta-like 1 homolog (Drosophila) | Dlk1 | NM_010052 | −1.9 | 0.01 | −2.2 | 0.01 | — | ||
| Fatty acid synthase | Fasn | NM_007988 | −1.2 | 0.06 | −1.4 | 0.02 | — | ||
| Lipase, hormone sensitive | Lipe | NM_010719 | −1.2 | 0.06 | −1.5 | 0.02 | — | ||
| Nuclear receptor coactivator 2 | Ncoa2 | NM_008678 | −1.2 | 0.07 | −1.3 | 0.07 | — | ||
| Peroxisome proliferator activated receptor alpha | Ppara | NM_011144 | −2.1 | 0.01 | −2.3 | 0.06 | −1.7 | 0.09 | CT6 |
| Peroxisome proliferator activated receptor gamma | Pparg | NM_011146 | −1.2 | 0.06 | |||||
| Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
Ppargc1a
(PGC-1 α ) |
NM_008904 | −1.2 | 0.09 | −1.5 | 0.004 | CT8 | ||
| Peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
Ppargc1b
(PGC-1 β ) |
NM_133249 | −1.4 | 0.01 | −1.4 | 0.04 | CT5 | ||
| PR domain containing 16 | Prdm16 | NM_027504 | −1.3 | 0.03 | −1.5 | 0.03 | — | ||
| Twist homolog 1 (Drosophila) | Twist1 | NM_011658 | −1.8 | 0.01 | −2.0 | 0.06 | −1.5 | 0.04 | — |
| Actin, beta | Actb | NM_007393 | −1.2 | 0.02 | −1.4 | 0.004 | — | ||
4. Discussion
In the present study, we discovered a reduced core body temperature in Id2−/− mice, and this effect was not found to be dependent upon the time-of-day. Moreover, from the iBAT of Id2−/− mice, genes involved in insulin signaling and adipogenesis were differentially regulated in a sex-dependent manner. These results reveal a role of Id2 in the regulation of thermogenesis and BAT metabolic functions.
Our previous study revealed that Id2−/− mice exhibit less activity as demonstrated by daily counts of general activity and the wheel running activity, which could partially explain the reduced core body temperature, since less physical activity would generate less heat [10]. Moreover, Id2−/− mice show a reduced body mass and less gonadal adipose deposits [6, 10, 11]. As the subcutaneous and intradermal fat functions as thermal insulation for mice to preserve heat loss, Id2−/− mice with low fat content might tend to lose heat more readily than WT mice. Furthermore, the reduced body temperature associated with lower fat content might contribute to the high death rate observed previously (mice housed under normal temperature), which was rescued by a high fat diet that resulted in increased total body fat [11]. Specifically in male Id2−/− mice iBAT we observed increased glucose uptake and reduced TG accumulation [10], suggesting alterations in its metabolic programing. Additionally, both male and female Id2−/− mice exhibited an increased activated iBAT volume [10]. Interestingly, our results suggest that the role of Id2 in thermoregulation is opposite to the function of another member of this HLH family, Id1, whose deficiency results in higher thermogenesis and an elevated BAT expression of thermogenic proteins [17]. Notably, Id1 has a distinct and opposite function in WAT adipogenesis compared to Id2, despite both Id1 and Id2 null mice exhibiting reduced adiposity [10–12, 17]. Lastly, we examined the relationship between body mass and body temperature in Id2−/− mice by regression analysis and revealed a limited relationship between the two variables [18]. No significant relationship was observed between body mass and body temperature at any time of the day or in the two sexes. However, as can be seen with the y-intercept of the regression lines, both Id2−/− male and female mice expressed a consistently lower temperature compared to WT controls, irrespective of body mass, and this feature was observed during both the day and night phases of the LD cycle. These results suggest a role for Id2 in the regulation of core body temperature.
In this study we also measured iBAT mass and iBAT/body mass ratio. While there was a tendency for higher iBAT/body mass in both Id2−/− male and female mice, this was not determined to be a significant difference. Note that the average body mass of Id2−/− mice used for both the iBAT weight and body temperature experiments was found to be significantly lower, consistent with our previous studies [7, 10]. Important is the fact that a lower body mass, found for some of the Id2−/− mice and for males in particular, does not correlate with a lower body temperature, and body mass in this situation is therefore an independent factor when predicting core body temperature.
It is important to note that while the objective of examining body temperature using the implanted thermometers was to record “core” body temperature, the position of the implants may not give an exact measure of true core body temperature. However, in a comparable study of mouse body temperatures, temperature measurements were similar whether derived from similarly subcutaneously implanted thermometers in the interscapular region of WT and Rev-erbα mutant mice or as determined using dataloggers that were implanted within the abdomen [3].
Id2 is rhythmically expressed in BAT [4, 15] amongst other tissues [6, 7]. ID2 protein has also been observed to be rhythmic in its abundance over the 24 hr diurnal/circadian cycle within the liver and heart [6] (Ward, Fernando, Hou, and Duffield, unpublished data). A role for ID2 has been established as a mediator of circadian clock output and control of expression patterns of clock controlled genes (CCGs) within the liver [6]. CCGs encompass ~10% transcriptome in individual tissues [19]. It is for this reason that we examined whether any of the genes identified as differentially expressed in iBAT were in fact known CCGs. Using the CIRCA database [14, 15], 5 of the 17 differentially genes associated with adipogenesis were found to be CCGs (e.g., Pparα and PGC-1α), and so a possible role for ID2 is in mediating circadian regulatory effects on these genes within BAT. However, further investigation would be required to test this hypothesis. The observation that few of the differentially regulated genes involved in insulin signaling are CCGs (1 out of 13 genes) suggests that the contribution of ID2 to insulin signaling intrinsic to BAT is independent of the role of ID2 in mediating circadian clock output [6].
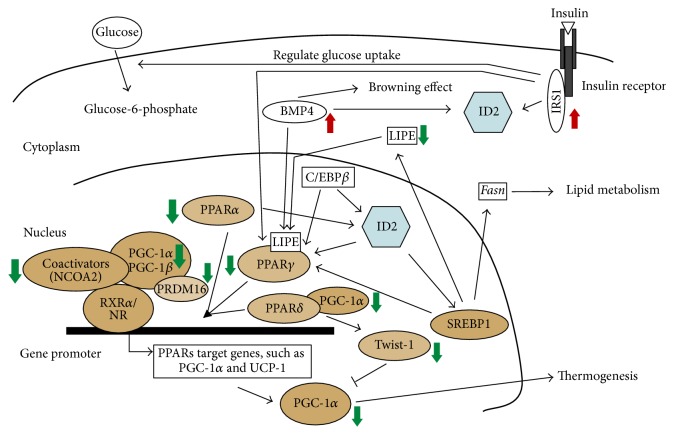
In order to explain how Id2 deficiency has an impact on BAT insulin signaling and adipogenesis, we propose a network model (Figure 3). The nuclear receptor PPARs are fundamentally important for energy homeostasis and Id2 plays a role in interfacing with the molecular pathways upstream or downstream of these transcriptional factors. Expression of two members of the PPAR subfamily of ligand-activated nuclear receptors, PPARα and PPARγ, was downregulated in our study. PPARα is highly expressed in BAT and considered a marker of BAT; it also plays an important role in the overall regulation of lipid metabolism; and its target genes are involved in mitochondrial and peroxisomal β-oxidation of fatty acids (FAs) [20–22]. Moreover, PPARα regulates the expression of uncoupling protein 1 (ucp1), which confers on BAT its thermogenic capacity [23]. PGC-1α (downregulated in our study) is a transcriptional coactivator involved in the control of energy metabolism and critical for BAT thermogenesis and enhancing overall mitochondrial oxidative activity [24]. PPARα can induce PGC-1α gene expression and contributes to the thermogenic activation of brown fat [25]. PRDM16 exhibits a brown fat selective expression pattern and regulates the thermogenic gene program in brown and beige adipocytes [26]. The observation of reduced Prdm16 expression in Id2−/− mice is consistent with the role of PRDM16 as a transcriptional regulator of PGC-1α [27]. Likewise, studies have demonstrated the linkage between Id2 and PPARα [28]. PPARγ is essential for adipocyte differentiation, and PPARγ alone generates a fat phenotype that is common to both WAT and BAT. The CCAAT enhancer binding protein beta (C/EBPβ) and PGC-1α are critical for controlling PPARγ expression in BAT and for determining BAT-specific programs [29, 30]. The PPARγ thermogenic effect in BAT is mediated by PGC-1α [24]. It has been observed that overexpression of Id2 associates with PPARγ expression, ID2 acts upstream of PPARγ, and C/EBPβ induces Id2 expression during the adipogenesis process [12, 31]. Cofactors such as NCoA2 (downregulated in our study) can interact directly with PPARγ to initiate its own transactivation [32]. Moreover, LIPE (downregulated in our study) could modulate adipose metabolism by reducing the availability of ligands for PPARγ, since gene knockout of LIPE in mice attenuates activation of PPARγ [33]. LIPE is also able to hydrolyze stored TGs in adipose tissue and to mobilize free FA from adipose tissue [34]. Furthermore, PPARγ is a direct target of the transcription factor sterol response element binding protein 1 (SREBP1), whose transcriptional activity is modulated by ID2 and which regulates downstream lipid metabolism genes such as lipe and Fasn [35, 36]. Additionally, Irs1 (upregulated in our study) plays essential roles in the differentiation of brown adipocytes and expression of PPARγ [37, 38]. Previous studies have revealed IRS1-regulated Id2 gene expression [39], although in the current study it is unclear whether this is a direct effect or a feedback response. As for the mechanism by which IRS1 is elevated in Id2−/− iBAT, it is unclear and warrants further investigation. BMP4 (upregulated in our study) is able to induce the white to brown transition of adipose cells, which could indirectly regulate PPARγ activation [40, 41]. The elevated BMP4 expression in the context of reduced PPARs is surprising since BMP4 upregulation is associated with increased BAT adipogenesis and the WAT browning effect [42]. Interestingly, the Id2 gene promoter has BMP-response elements and has been shown to be a target of BMP signaling [43]. PPARδ plays an integral role in transcriptional network regulation of fat-burning genes and brown fat metabolism. PGC-1α/PPARδ could regulate brown fat metabolism through Twist-1 tuning [44]. Twist-1 (downregulation in our study) encodes a basic HLH transcription factor, and overexpression of Twist-1 is associated with Id2 expression [45].
Figure 3.
Id2 network model in insulin signaling, thermogenesis, and adipogenesis pathways. A schematized and partial view of the signaling pathway indicating its major downstream targets and factors susceptible to interfering with signaling. Function of ID2 is not necessarily limited to the nucleus [8]. ↑ or ↓, up- or downregulation of gene expression, respectively, as determined by PCR array analysis in the current study.
Note that the gene encoding the thermogenic protein, UCP1, was present on the PCR arrays, but no difference in its expression was observed between genotypes of either sex. Interestingly, in the Id1 null mouse, ucp1 gene expression is elevated in iBAT, and this is associated with an increased core body temperature phenotype [17]. Thus, it is surprising that in the Id2−/− mouse that exhibits a reduced body temperature phenotype we do not observe a reduction in ucp1 gene expression. Of course, this does not exclude, however unlikely, the possibility of an altered UCP1 protein abundance through a posttranscriptional/posttranslational process.
Hypoxia, while not a focus of the current study, is known to reduce body temperature in mammals and contribute to the thermogenic activity of BAT [46]. It is noteworthy that in a recent study of human glioblastoma cells/tissue, an important role was established for ID2 in modulating the cellular effects of hypoxia and its activation of the HIF2α pathway [47]. The Id2 gene is also a target for HIF1α and HIF2α [47, 48], making it part of a positive feedback loop mechanism, at least in models of brain tumor. It is plausible that the hypoxic effects on BAT function might also include a contribution from ID2, and this would be an important pathway to examine in future experiments in this context.
It is a somewhat contradictory finding that elements of the thermogenic pathway are reduced (e.g., PGC1-α) or unaltered (ucp1) in Id2−/− mice but that Id2−/− male iBAT exhibits increased glucose uptake (PET-FDG) coupled with reduced iBAT triglyceride levels and a systemwide enhanced insulin sensitivity [10, 11] and that core body temperature is consistently reduced in both Id2−/− male and female mice. Id2−/− mice also exhibit an altered 24 hr locomotor activity and feeding profile and an overall reduction in nocturnal locomotor activity [10], the latter of which suggests a reduction in energy expenditure/increased energy conservation. Clearly the BAT adipogenic program is altered in both male and female Id2−/− mice, as is WAT adipogenesis [10–12], and it is likely that a change in BAT function contributes to the reduced temperature phenotype. Due to the nature of whole body knockout of ID2, it is possible that the temperature phenotype and iBAT gene changes observed are secondary to whole body metabolic changes. Clearly additional experiments are required to elucidate the relative contributions of these and other potential components in generating the altered core body temperature phenotype.
Our previous study showed enhanced glucose uptake in iBAT of Id2−/− mice [10], and the present study reveals a reduced core body temperature in Id2−/− mice. This discrepancy could partially be explained by the differential regulation of Irs1, Lipe, PPARs, and PGC-1s. It has been reported that degradation of IRS1 leads to impaired glucose uptake in adipose tissue [49]. Therefore, upregulation of IRS1 might explain the increased glucose uptake we observed before [10], whereas downregulation of LIPE, PPARs, and PGC-1s might contribute to reduced FA oxidation, impaired adipogenesis, and a lower body temperature. Inactivation of PPARs is associated with insulin resistance [50, 51], yet paradoxically Id2−/− mice show enhanced insulin sensitivity with downregulated PPARs [10, 11]. It has been suggested that mice that lack one allele of the PPARγ gene are more sensitive to insulin, which could partially explain the enhanced insulin sensitivity we observe in Id2 null mice [10, 51]. Furthermore, the differential regulation of genes specifically in female mutant mice, such as Fbp1, a rate-limiting enzyme in gluconeogenesis, Shc1, a component in the IGF-1-regulated pathway, and Igf2, suggests a sex-specific physiological program for ID2 in BAT.
5. Conclusion
Inhibitor of DNA binding 2 is rhythmically expressed in BAT [4, 15], and the observation that few of the differentially regulated genes involved in insulin signaling are CCGs suggests that the contribution of ID2 to insulin signaling intrinsic to BAT is independent of the role of ID2 in mediating circadian clock output [6]. Overall, ID2 seems to be an important coordinator of energy homeostasis including insulin signaling, adipogenic programing, and thermoregulation. In conclusion, our finding that ID2 contributes to the regulation of body temperature and energy homeostasis presents the possibility that ID2 could be a potential therapeutic target for metabolic disease. Further, these data emphasize the influence of Id2 on BAT molecular signaling and physiology in a sex-specific manner.
Supplementary Material
Supplementary Table 1 summarizes the regression analysis of time-of-day representative core body temperatures (day and night).
Supplementary Table 2 shows the mean ± SEM body mass of wild type and Id2−/− mice.
Acknowledgments
The work was supported by grants to Giles E. Duffield from NIGMS (R01-GM087508) and AHA (10SDG4030011). The authors are grateful to N. J. Balmert, Y. Xi, D. Lee, J. H. Werner, A. D. Hummel, A. Nguyen, and R. Nabrzyski for assistance with genotyping; D. Acri for assistance with figure preparation; the Freimann Life Science Center for assistance with animal care and training; and Drs X. C. Dong and T. Hou for comments on the paper.
Additional Points
Supplementary Table 1 summarizes the regression analysis of time-of-day representative core body temperatures (day and night). Supplementary Table 2 shows the mean ± SEM body mass of wild type and Id2−/− mice.
Disclosure
The present address of Peng Zhou is Division of Endocrinology, Diabetes, and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA 02215.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Mohawk J. A., Green C. B., Takahashi J. S. Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buhr E. D., Yoo S.-H., Takahashi J. S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhart-Hines Z., Feng D., Emmett M. J., et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zvonic S., Ptitsyn A. A., Conrad S. A., et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 5.Van Der Veen D. R., Shao J., Chapman S., Leevy W. M., Duffield G. E. A diurnal rhythm in glucose uptake in brown adipose tissue revealed by in vivo PET-FDG imaging. Obesity. 2012;20(7):1527–1529. doi: 10.1038/oby.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou T. Y., Ward S. M., Murad J. M., Watson N. P., Israel M. A., Duffield G. E. ID2 (inhibitor of DNA binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver. The Journal of Biological Chemistry. 2009;284(46):31735–31745. doi: 10.1074/jbc.m109.013961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffield G. E., Watson N. P., Mantani A., et al. A role for Id2 in regulating photic entrainment of the mammalian circadian system. Current Biology. 2009;19(4):297–304. doi: 10.1016/j.cub.2008.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward S. M., Fernando S. J., Hou T. Y., Duffield G. E. The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression. Journal of Biological Chemistry. 2010;285(50):38987–39000. doi: 10.1074/jbc.M110.175182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalsbeek A., la Fleur S., Fliers E. Circadian control of glucose metabolism. Molecular Metabolism. 2014;3(4):372–383. doi: 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew D., Zhou P., Pywell C. M., et al. Ablation of the ID2 gene results in altered circadian feeding behavior, and sex-specific enhancement of insulin sensitivity and elevated glucose uptake in skeletal muscle and brown adipose tissue. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073064.e73064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P., Hummel A. D., Pywell C. M., Charlie Dong X., Duffield G. E. High fat diet rescues disturbances to metabolic homeostasis and survival in the Id2 null mouse in a sex-specific manner. Biochemical and Biophysical Research Communications. 2014;451(3):374–381. doi: 10.1016/j.bbrc.2014.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kye W. P., Waki H., Villanueva C. J., et al. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-γ expression and adipocyte differentiation. Molecular Endocrinology. 2008;22(9):2038–2048. doi: 10.1210/me.2007-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Ross R. A., Pywell C. M., Liangpunsakul S., Duffield G. E. Disturbances in the murine hepatic circadian clock in alcohol-induced hepatic steatosis. Scientific Reports. 2014;4, article 3725 doi: 10.1038/srep03725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pizarro A., Hayer K., Lahens N. F., Hogenesch J. B. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Research. 2013;41(1):D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speakman J. R., Keijer J. Not so hot: optimal housing temperatures for mice to mimic the thermal environment of humans. Molecular Metabolism. 2013;2(1):5–9. doi: 10.1016/j.molmet.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satyanarayana A., Klarmann K. D., Gavrilova O., Keller J. R. Ablation of the transcriptional regulator Id1 enhances energy expenditure, increases insulin sensitivity, and protects against age and diet induced insulin resistance, and hepatosteatosis. The FASEB Journal. 2012;26(1):309–323. doi: 10.1096/fj.11-190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heusner A. A. Body size and energy metabolism. Annual Review of Nutrition. 1985;5:267–293. doi: 10.1146/annurev.nu.05.070185.001411. [DOI] [PubMed] [Google Scholar]
- 19.Duffield G. E. DNA microarray analyses of circadian timing: the genomic basis of biological time. Journal of Neuroendocrinology. 2003;15(10):991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- 20.Mandard S., Müller M., Kersten S. Peroxisome proliferator-activated receptor α target genes. Cellular and Molecular Life Sciences. 2004;61(4):393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valmaseda A., Carmona M. C., Barberá M. J., et al. Opposite regulation of PPAR-α and -γ gene expression by both their ligands and retinoic acid in brown adipocytes. Molecular and Cellular Endocrinology. 1999;154(1-2):101–109. doi: 10.1016/s0303-7207(99)00081-7. [DOI] [PubMed] [Google Scholar]
- 22.Villarroya F., Iglesias R., Giralt M. PPARs in the control of uncoupling proteins gene expression. PPAR Research. 2007;2007:12. doi: 10.1155/2007/74364.74364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barberá M. J., Schlüter A., Pedraza N., Iglesias R., Villarroya F., Giralt M. Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. Journal of Biological Chemistry. 2001;276(2):1486–1493. doi: 10.1074/jbc.m006246200. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 25.Hondares E., Rosell M., Díaz-Delfín J., et al. Peroxisome proliferator-activated receptor α (PPARα) induces PPARγ coactivator 1α (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: Involvement of PRDM16. Journal of Biological Chemistry. 2011;286(50):43112–43122. doi: 10.1074/jbc.m111.252775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harms M. J., Ishibashi J., Wang W., et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metabolism. 2014;19(4):593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seale P., Kajimura S., Yang W., et al. Transcriptional control of brown fat determination by PRDM16. Cell Metabolism. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Carmen González C., Corton J. C., Acero N., et al. Peroxisome proliferator-activated receptorα agonists differentially regulate inhibitor of DNA binding expression in rodents and human cells. PPAR Research. 2012;2012:9. doi: 10.1155/2012/483536.483536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajimura S., Seale P., Kubota K., et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-β transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3(5):333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Karaya K., Mori S., Kimoto H., et al. Regulation of Id2 expression by CCAAT/enhancer binding protein β . Nucleic Acids Research. 2005;33(6):1924–1934. doi: 10.1093/nar/gki339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell E., Kuhn P., Xu W. Nuclear receptor cofactors in PPARγ-mediated adipogenesis and adipocyte energy metabolism. PPAR Research. 2007;2007:11. doi: 10.1155/2007/53843.53843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen W.-J., Yu Z., Patel S., Jue D., Liu L.-F., Kraemer F. B. Hormone-sensitive lipase modulates adipose metabolism through PPARγ . Biochimica et Biophysica Acta. 2011;1811(1):9–16. doi: 10.1016/j.bbalip.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweiger M., Schreiber R., Haemmerle G., et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. Journal of Biological Chemistry. 2006;281(52):40236–40241. doi: 10.1074/jbc.m608048200. [DOI] [PubMed] [Google Scholar]
- 35.Moldes M., Boizard M., Liepvre X. L., Fève B., Dugail I., Pairault J. Functional antagonism between inhibitor of DNA binding (Id) and adipocyte determination and differentiation factor 1/sterol regulatory element-binding protein-1c (ADD1/SREBP-1c) trans-factors for the regulation of fatty acid synthase promoter in adipocytes. Biochemical Journal. 1999;344(3):873–880. doi: 10.1042/0264-6021:3440873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grønning L. M., Tingsabadh R., Hardy K., et al. Glucose induces increases in levels of the transcriptional repressor Id2 via the hexosamine pathway. American Journal of Physiology—Endocrinology and Metabolism. 2006;290(4):E599–E606. doi: 10.1152/ajpendo.00242.2005. [DOI] [PubMed] [Google Scholar]
- 37.Miki H., Yamauchi T., Suzuki R., et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Molecular and Cellular Biology. 2001;21(7):2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng Y.-H., Kriauciunas K. M., Kokkotou E., Kahn C. R. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Molecular and Cellular Biology. 2004;24(5):1918–1929. doi: 10.1128/mcb.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navarro M., Valentinis B., Belletti B., Romano G., Reiss K., Baserga R. Regulation of Id2 gene expression by the type 1 IGF receptor and the insulin receptor substrate-1. Endocrinology. 2001;142(12):5149–5157. doi: 10.1210/en.142.12.5149. [DOI] [PubMed] [Google Scholar]
- 40.Hammarstedt A., Hedjazifar S., Jenndahl L., et al. WISP2 regulates preadipocyte commitment and PPARγ activation by BMP4. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(7):2563–2568. doi: 10.1073/pnas.1211255110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elsen M., Raschke S., Tennagels N., et al. BMP4 and BMP7 induce the white-to-brown transition of primary human adipose stem cells. American Journal of Physiology-Cell Physiology. 2014;306(5):C431–C440. doi: 10.1152/ajpcell.00290.2013. [DOI] [PubMed] [Google Scholar]
- 42.Bowers R. R., Lane M. D. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle. 2007;6(4):385–389. doi: 10.4161/cc.6.4.3804. [DOI] [PubMed] [Google Scholar]
- 43.Nakahiro T., Kurooka H., Mori K., Sano K., Yokota Y. Identification of BMP-responsive elements in the mouse Id2 gene. Biochemical and Biophysical Research Communications. 2010;399(3):416–421. doi: 10.1016/j.bbrc.2010.07.090. [DOI] [PubMed] [Google Scholar]
- 44.Pan D., Fujimoto M., Lopes A., Wang Y.-X. Twist-1 is a PPARδ-inducible, negative-feedback regulator of PGC-1α in brown fat metabolism. Cell. 2009;137(1):73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isenmann S., Arthur A., Zannettino A. C. W., et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27(10):2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 46.Trayhurn P., Alomar S. Y. Oxygen deprivation and the cellular response to hypoxia in adipocytes—perspectives on white and brown adipose tissues in obesity. Frontiers in Endocrinology. 2015;6, article 19 doi: 10.3389/fendo.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S. B., Frattini V., Bansal M., et al. An ID2-dependent mechanism for VHL inactivation in cancer. Nature. 2016;529(7585):172–177. doi: 10.1038/nature16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Löfstedt T., Jögi A., Sigvardsson M., et al. Induction of ID2 expression by hypoxia-inducible factor-1. A role in dedifferentiation of hypoxic neuroblastoma cells. Journal of Biological Chemistry. 2004;279(38):39223–39231. doi: 10.1074/jbc.M402904200. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Nishina P. M., Naggert J. K. Degradation of IRS1 leads to impaired glucose uptake in adipose tissue of the type 2 diabetes mouse model TALLYHO/Jng. Journal of Endocrinology. 2009;203(1):65–74. doi: 10.1677/JOE-09-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamauchi T., Kamon J., Waki H., et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. Journal of Biological Chemistry. 2001;276(44):41245–41254. doi: 10.1074/jbc.m103241200. [DOI] [PubMed] [Google Scholar]
- 51.Miles P. D. G., Barak Y., He W., Evans R. M., Olefsky J. M. Improved insulin-sensitivity in mice heterozygous for PPAR-γ deficiency. Journal of Clinical Investigation. 2000;105(3):287–292. doi: 10.1172/jci8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 summarizes the regression analysis of time-of-day representative core body temperatures (day and night).
Supplementary Table 2 shows the mean ± SEM body mass of wild type and Id2−/− mice.