Abstract
Most Enterococcus faecalis and E. faecium are harmless to humans; however, strains harboring virulence genes, including esp, gelE, cylA, asa1, and hyl, have been associated with human infections. E. faecalis and E. faecium are present in beach waters worldwide, yet little is known about their virulence potential. Here, multiplex PCR was used to compare the distribution of virulence genes among E. faecalis and E. faecium isolated from beaches in Southern California and Puerto Rico to isolates from potential sources including humans, animals, birds, and plants. All five virulence genes were found in E. faecalis and E. faecium from beach water, mostly among E. faecalis. gelE was the most common among isolates from all source types. There was a lower incidence of asa1, esp, cylA, and hyl genes among isolates from beach water, sewage, septage, urban runoff, sea wrack, and eelgrass as compared to human isolates, indicating that virulent strains of E. faecalis and E. faecium may not be widely disseminated at beaches. A higher frequency of asa1 and esp among E. faecalis from dogs and of asa1 among birds (mostly seagull) suggests that further studies on the distribution and virulence potential of strains carrying these genes may be warranted.
1. Introduction
Enterococcus faecalis and Enterococcus faecium are commonly found in the intestinal tracts of humans and animals and also ubiquitous in the environment [1]. While generally considered to be harmless, certain strains of E. faecalis and E. faecium are among the leading causes of nosocomial infections including urinary tract infections, abdominal and wound infections, endocarditis, and bacteremia [2–5]. E. faecalis and E. faecium isolated from patients in hospital settings have been shown to harbor a higher frequency of gelE (gelatinase), asa1 (aggregation substance), esp (enterococcal surface protein), cylA (cytolysin activator), and hyl (glycoside-hydrolase) as compared to strains found in nonhospitalized individuals, animals, and food [6–12]. Commensal, that is, harmless, E. faecalis and E. faecium can become opportunistic pathogens by acquiring antibiotic resistant and putative virulent genes from other bacteria via horizontal gene transfer [2, 4, 13–17].
E. faecalis and E. faecium are among the most common species of enterococci found in the beach environment [18–21]. Enterococci found in the beach environment can include naturalized populations existing in soil and vegetation as well as strains from humans, sewage, animals, birds, reptiles, and insects [1]. Presumably, potentially pathogenic E. faecalis and E. faecium in human fecal waste would harbor higher numbers of virulence genes as compared to strains from animal and environmental sources.
In Puerto Rico, beaches receive storm flows containing contaminated septage and agricultural runoff potentially carrying enterococci derived from human and animal fecal waste. In Southern California, urban runoff, beach sand, and sea wrack (macroalgae on beach sand) have been identified as important sources of enterococci to beach water [22–25].
Previous studies showed that E. faecalis and E. faecium from the beach water and sand harbor antibiotic resistant genes suggesting a potential health risk for beach goers [26–30]; however, the frequency of other virulence factors was not determined. Here, we compared the frequency of putative enterococcal virulence genes (esp, gelE, cylA, asa1, and hyl) among E. faecalis and E. faecium from beaches in Southern California and Puerto Rico impacted by different enterococci source inputs to assess beaches as an environmental reservoir of potentially virulent enterococci.
2. Materials and Methods
2.1. Sources of E. faecalis and E. faecium Isolates
2.1.1. Southern California
A total of 170 Enterococcus (91 E. faecalis and 79 E. faecium) isolates were screened for putative enterococcal virulence genes (Table 1). The environmental isolates were randomly selected from a collection of strains obtained from previous studies [20, 31] including beach water, eelgrass (Zostera marina), wrack (mainly Macrocystis pyrifera), sand, creek, or storm drain runoff upstream of beaches and sewage influent (untreated waste) and effluent. Isolates from animals were obtained from bird (mostly seagull) stools on beach and dog stools. Human (nonclinical) strains of E. faecalis and E. faecium were isolated from urine and fecal samples from 18 healthy (nonhospitalized) individuals residing in Southern California. Human fecal and urine specimens obtained from healthy individuals were considered representative of strains that could be found in beach water due to human shedding or contamination from sewage and/or septage. Ten E. faecium isolates identified as vancomycin resistant enterococci (VRE), 5 clinical strains of E. faecium (non-VRE), and 10 E. faecalis isolated from rectal swabs (3), urine (1), blood (1), abscess (1), ascites (2), vagina (1), and joint (1) were provided by Orange County Public Health Laboratory (OCPHL). Clinical isolates were included for comparison to strains with enhanced virulence potential.
Table 1.
Sources of E. faecalis and E. faecium isolated from Southern California.
| Source | Number of samples | Number of sites | Number of isolates | Total number of isolates | |
|---|---|---|---|---|---|
| E. faecalis | E. faecium | ||||
| Environmental | |||||
| Beach water | 5 | 5 | 8 | 10 | 18 |
| Urban runoff | 10 | 5 | 20 | 8 | 28 |
| Sand | 5 | 5 | 0 | 5 | 5 |
| Sea wrack | 5 | 1 | 5 | 5 | 10 |
| Eelgrass | 7 | 1 | 5 | 3 | 8 |
| Wastewater influent | 4 | 4 | 6 | 3 | 9 |
| Wastewater effluent | 5 | 2 | 6 | 11 | 17 |
| Human | |||||
| Human, healthy | 18 | NA | 15 | 8 | 23 |
| Human, clinical | 10 | Unk | 10 | 5 | 15 |
| Vancomycin resistant enterococci | Unk | Unk | 0 | 10 | 10 |
| Animal | |||||
| Dogs | 7 | 7 | 7 | 3 | 10 |
| Birds | 16 | 2 | 9 | 8 | 17 |
|
| |||||
| Total | 92 | 91 | 79 | 170 | |
NA = not applicable.
Unk = unknown.
2.1.2. Puerto Rico
A total of 247 Enterococcus (174 E. faecalis and 73 E. faecium) isolates from Puerto Rico were analyzed (Table 2). Enterococcal isolates from beach water were obtained from two beaches in Puerto Rico. Human (nonclinical) enterococcal strains were isolated from fresh fecal samples from nine healthy individuals from Mayaguez, Puerto Rico. Clinical enterococcal strains were isolated from urine specimens and identified to species level by a local hospital in Mayaguez. Six septage samples were obtained from individual houses or from septic tank trucks after emptying individual family tanks.
Table 2.
Sources of E. faecalis and E. faecium isolated from Puerto Rico.
| Source | Number of samples | Number of sites | Number of isolates | Total number of isolates | |
|---|---|---|---|---|---|
| E. faecalis | E. faecium | ||||
| Environmental | |||||
| Beach water | 9 | 2 | 108 | 32 | 140 |
| Septage | 6 | 6 | 19 | 26 | 45 |
| Human | |||||
| Human, healthy | 9 | 1 | 5 | 4 | 9 |
| Human, clinical | 53 | 1 | 42 | 11 | 53 |
|
| |||||
| Total | 77 | 174 | 73 | 247 | |
2.2. Isolation and Identification of Enterococci
2.2.1. Southern California
Enterococcal isolates from all samples (except for clinical specimens) were obtained using mEI agar and identified to species level using the Vitek II (bioMérieux) plus additional biochemical tests and pigment and motility as per Ferguson et al. [32]. Clinical strains were isolated by OCPHL using TSA with 5% sheep's blood; presumptive enterococcal colonies were gram-stained and identified using MicroScan (Siemens Healthcare) and/or API Strep 20 (bioMérieux). Up to 3 isolates per sample identified as E. faecalis and E. faecium were randomly selected for virulence gene analysis. Species identification of 8 different isolates obtained using biochemical methods was also confirmed by 16S rRNA sequencing conducted at GenoSeq, University of California, Los Angeles.
2.2.2. Puerto Rico
Enterococcal isolates from all samples (except for clinical specimens) were obtained using mE agar. All isolates were divided into four groups based on pigmentation and motility. The isolates were identified to the genus level based on growth in BHI with 6.5% NaCl, growth at 45°C, esculin hydrolysis, catalase, and PCR amplifying of the Tuf gene [33]. Species level identification was done by a double digestion of the PCR product of the ATP synthase α subunit gene in combination with a restriction fragment length polymorphism (RFLP) assay (paper in preparation). Clinical strains obtained from a local hospital were identified using MicroScan system (Siemens Health Care). Up to 12 isolates each of E. faecalis or E. faecium were randomly selected per sample for virulence gene analysis.
2.3. DNA Extraction
2.3.1. Southern California and Puerto Rico
E. faecalis and E. faecium strains were grown in BHI broth, incubated overnight at 37°C, and harvested by centrifugation (13,000 RPM for 5 min). The cells were washed three times in TE buffer and resuspended in 200 μL 1x TE (10 mM Tris-HCl; 1 mM EDTA, pH 8.0) and lysed by heating at 95°C for 10 min. The lysed cells were transferred to tubes with glass beads, subjected to bead beating for five minutes, and centrifuged as before.
2.4. Multiplex PCR for the Detection of Enterococcal Virulence Genes
Total DNA extracted from all isolates obtained from California and Puerto Rico was screened for enterococcal virulence genes (gelE, asa1, esp, cylA, and hyl) using PCR primers and multiplex method developed by Vankerckhoven et al. [34] with the following modifications: we used Promega Flexi Taq DNA polymerase instead of Hot-StarTaq DNA polymerase in the master mix; the initial activation step was done at 95°C for 2 min, followed by 35 cycles of denaturation (95°C for 30 sec), annealing (49.5°C for 30 sec), and extension (72°C for 2 min) and 1 cycle of elongation at 72°C for 10 min. Each set of primers has a characteristic product size to differentiate within the five virulence genes (asa1 at 375 bp, gelE at 213 bp, cylA at 688 bp, esp at 510 bp, and hyl at 276 bp). PCR products obtained by the Puerto Rico laboratory were confirmed by 1.8% agarose-gel electrophoresis (90 v, 2.5 hrs), stained with ethidium bromide, and visualized by UV transillumination (VersaDoc MP 4000). In Southern California, PCR products were visualized using the FlashGel® (Lonza) system. 2 μL of extracted DNA was diluted in 2 μL FlashGel loading dye and inserted into 12 + 1-cassette wells. A 50 bp–1.5 kb DNA ladder (Lonza) was used as a molecular size marker. FlashGels were run at 150 V for up to 13 minutes. Each PCR run included a no-template control; the positive control strain used for gelE, esp, asa1, and cylA was E. faecalis MMH594 kindly donated by N. Shankar, Department of Medicinal Chemistry and Pharmaceutics, University of Oklahoma Health Sciences Center, Oklahoma City [14].
3. Results
A total of 170 E. faecalis and E. faecium isolates from Southern California (SC) and 247 isolates from Puerto Rico (PR) from beach water and potential sources of enterococci to beaches were analyzed for enterococcal virulence genes gelE, asa1, esp, cylA, and hyl.
Eighty-seven (80.6%) E. faecalis isolates from PR beach water harbored one or more of the following genes: gelE (98.1%), asa1 (44.4%), esp (11.1%), and cylA (3.3%) (Table 3). Five (26.3%) E. faecalis isolates from septage contained gelE (21.0%), asa1 (5.3%), cylA (5.3%), and hyl (5.3%). Eighteen (91.7%) E. faecalis isolates from human specimens (clinical and nonclinical) contained gelE (17.4–100%), asa1 (50–100%), esp (33.3–40%), and cylA (19–60%).
Table 3.
Distribution of virulence factor genes among E. faecalis isolates from Southern California (SC) and Puerto Rico (PR).
| Source (number) of isolates | % (number) of isolates for the following virulence factor genes: | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gelE | asa1 | esp | cylA | hyl | None | |||||||
| PR beach water (108) | 98.1% | (106) | 44.4% | (48) | 11.1% | (12) | 3.3% | (3) | 0.0% | (0) | 19.4% | (21) |
| SC beach water (8) | 100.0% | (8) | 12.5% | (1) | 0.0% | (0) | 12.5% | (1) | 0.0% | (0) | 0.0% | (0) |
| SC wrack (5) | 100.0% | (5) | 20.0% | (1) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) |
| SC eelgrass (5) | 80.0% | (4) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 20.0% | (1) |
| SC urban runoff (20) | 70.0% | (14) | 10.0% | (2) | 5.0% | (1) | 0.0% | (0) | 0.0% | (0) | 30.0% | (6) |
| SC sewage influent (6) | 50.0% | (3) | 50.0% | (3) | 16.7% | (1) | 16.7% | (1) | 0.0% | (0) | 33.3% | (2) |
| SC sewage effluent (6) | 83.3% | (5) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 16.7% | (1) |
| PR septage (19) | 21.0% | (4) | 5.3% | (1) | 0.0% | (0) | 5.3% | (1) | 5.3% | (1) | 73.7% | (14) |
| SC dog (7) | 85.7% | (6) | 42.9% | (3) | 28.6% | (2) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) |
| SC bird (9) | 100.0% | (9) | 55.5% | (5) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) |
| PR human, nonclinical (5) | 100.0% | (5) | 100.0% | (5) | 40.0% | (2) | 60.0% | (3) | 0.0% | (0) | 44.4% | (2) |
| SC human, nonclinical (15) | 100.0% | (15) | 66.7% | (10) | 33.3% | (5) | 13.3% | (2) | 0.0% | (0) | 0.0% | (0) |
| PR human, clinical (42) | 71.4% | (30) | 50.0% | (21) | 33.3% | (14) | 19.0% | (8) | 0.0% | (0) | 7.1% | (3) |
| SC human, clinical (10) | 90.0% | (9) | 90.0% | (9) | 30.0% | (3) | 70.0% | (7) | 0.0% | (0) | 10.0% | (1) |
Eight (100%) E. faecalis isolates from SC beach water contained one or more of the following genes: gelE (100%), asa1 (12.5%), and cylA (12.5%) (Table 3). Fourteen (70%) E. faecalis isolates from urban runoff contained gelE (70%), asa1 (10%), and esp (1%). gelE was present among wrack and eelgrass; however, the other virulence genes were rare or absent in occurrence. E. faecalis from dogs contained gelE (85.7%), asa1 (42.9%), and esp (28.6%); isolates from birds contained gelE (100%) and asa1 (55.5%).
The frequency of enterococcal virulence genes differed between E. faecalis and E. faecium. Virulence genes were absent among the majority of E. faecium beach water isolates from PR and SC (62.5% and 100%), respectively (Table 4). Interestingly, enterococcal virulence genes were also rare among E. faecium from sewage from SC and septage from PR. esp was the most common virulence gene found among E. faecium from humans (12.5% to 83.3%). Ten clinical isolates of E. faecium from SC that were identified as vancomycin resistant strains by OCPHL were positive for the esp (80%) and hyl (10%) genes. In SC, none of the 5 virulence genes were detected among E. faecium isolates obtained from dog stools, wrack, and beach sand. gelE was the only virulence gene found among E. faecium from eelgrass.
Table 4.
Distribution of virulence factor genes among E. faecium isolates from Southern California (SC) and Puerto Rico (PR).
| Source (number) of isolates | % (number) of isolates for the following virulence factor genes: | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gelE | asa1 | esp | cylA | hyl | None | |||||||
| SC beach water (10) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (1) |
| PR beach water (32) | 37.5% | (12) | 25.0% | (8) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 62.5% | (20) |
| SC wrack (5) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (5) |
| SC eelgrass (3) | 33.0% | (1) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 67.0% | (2) |
| SC sand (5) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (5) |
| SC urban runoff (8) | 50.0% | (4) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 50.0% | (4) |
| SC sewage influent (3) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (3) |
| SC sewage effluent (11) | 9.1% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 90.9% | (10) |
| PR septage (26) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (26) |
| SC dog (3) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (3) |
| SC bird (8) | 0.0% | (0) | 0.0% | (0) | 37.5% | (3) | 0.0% | (0) | 0.0% | (0) | 62.5% | (5) |
| PR human, nonclinical (4) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) | 100% | (4) |
| SC human, nonclinical (8) | 0.0% | (0) | 0.0% | (0) | 12.5% | (1) | 0.0% | (0) | 0.0% | (0) | 87.5% | (7) |
| PR human, clinical (11) | 0.0% | (0) | 0.0% | (0) | 72.7% | (8) | 0.0% | (0) | 0.0% | (0) | 0.0% | (0) |
| SC human, clinical (5) | 0.0% | (0) | 0.0% | (0) | 83.3% | (4) | 0.0% | (0) | 0.0% | (0) | 16.7% | (1) |
| SC vancomycin resistant enterococci (10) | 0.0% | (0) | 0.0% | (0) | 80.0% | (8) | 0.0% | (0) | 10.0% | (1) | 10.0% | (1) |
The distribution of enterococcal virulence genes was also compared based on categorizing the source of E. faecalis and E. faecium isolates as environmental, animal, and human. E. faecalis isolates from human specimens (clinical and nonclinical) from both geographic locations had a higher frequency of the five virulence genes overall (Figure 1). gelE was the most abundant virulence gene found among E. faecalis isolates from human, animal, and environmental sources (59.6% to 95%), followed by asa1 (15.4% to 78.4%). cylA was found among 19.0% to 41.7% of E. faecalis human isolates and 4.9% to 19.0% environmental isolates and not detected in animal isolates.
Figure 1.
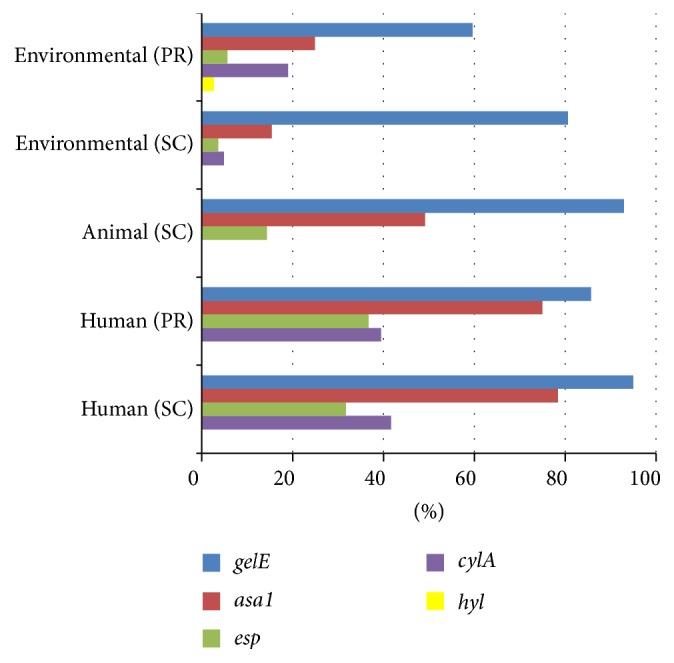
Distribution of virulence factor genes among E. faecalis isolates from environmental, animal, and human sources in Puerto Rico (PR) and Southern California (SC).
esp was the most commonly found virulence gene detected among E. faecium isolates (0% to 47.9%), followed by gelE (0% to 18.8%), asa1 (0% to 12.5%), and hyl (0% to 1.3%) (Figure 2). At both study sites, human derived E. faecium isolates had the highest frequency of esp (36.4% to 47.9%).
Figure 2.
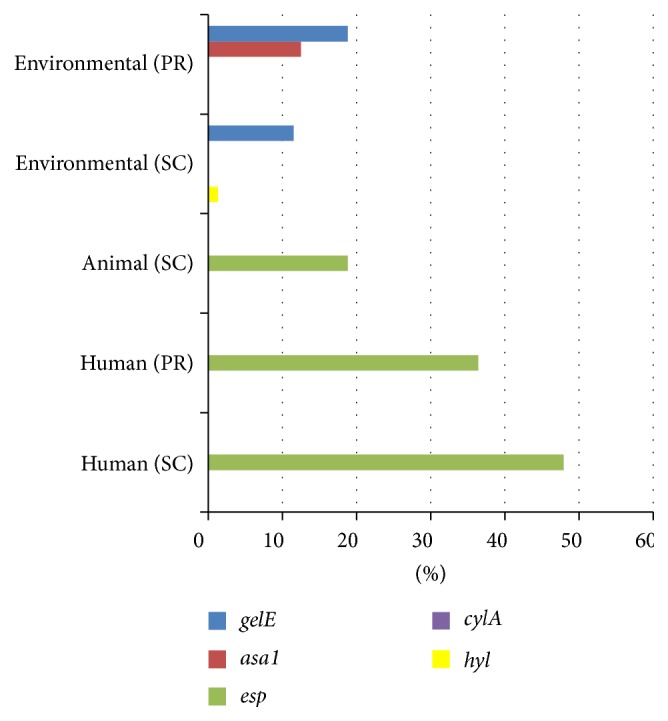
Distribution of virulence factor genes among E. faecium isolates from environmental, animal, and human sources in Puerto Rico (PR) and Southern California (SC).
4. Discussion
E. faecalis and E. faecium obtained from multiple sources, including the beach environment, humans (clinical and nonclinical), animals, and birds in Southern California and Puerto Rico, harbored putative enterococcal virulence genes that differed in frequency depending on source. At both study locations, there was a higher prevalence of virulence genes among E. faecalis as compared to E. faecium. Among both species groups, virulence genes were less abundant among beach strains overall compared to human isolates, which was also consistent with a similar study conducted in Australia [35].
Enterococcal virulence genes asa1 (aggregation substance) and cylA (cytolysin activator) were found among E. faecalis isolates from beach water, humans, dogs, and birds, indicative of strains with enhanced virulence potential. asa1 and cylA were first identified in the genome of multidrug resistant E. faecalis strain MMH594 and have also been associated with E. faecalis pathogenicity islands [16, 36]. Aggregation substance is encoded on a sex pheromone plasmid and mediates aggregation between bacteria, enabling the transfer of plasmids [37]. Cytolysins are toxins secreted by bacteria that damage cell membranes, facilitating the infection process. cylA can be carried on a plasmid or occur on the bacterial chromosome [38].
The distribution of asa1 and cylA among E. faecalis from human clinical specimens was 90% and 70%, respectively, of E. faecalis from SC as compared to 50% and 19%, respectively, of isolates from PR. These differences likely reflect variability in the types of clinical specimens analyzed from each study location; clinical isolates of E. faecalis from SC were obtained from rectal swabs, urine, blood, abscess, ascites, vagina, and joints; those from PR were obtained primarily from urine specimens.
asa1 and esp were also found among E. faecalis strains in dogs and birds (mostly seagull), suggesting that they may be important reservoirs of strains that could potentially be transferred to humans. esp is thought to aid enterococci in evading the immune system and also form biofilm [36, 39], which facilities colonization of E. faecalis in acute urinary tract infections [14]. Animals and birds have been suggested as potential sources of virulent strains to humans; gelE, asa1, esp, and cylA were detected in fecal E. faecalis isolated from dogs at veterinary hospitals [40, 41], poultry [42], and ducks and wild geese [43]. The presence of these genes among E. faecalis strains from dogs and birds warrants further studies to assess potential human health risks.
Among the virulence genes analyzed, gelE was the most frequently detected and widely distributed among E. faecalis strains from multiple sources, including the environment which is consistent with previous studies [9, 44, 45]. gelE is thought to enhance survivability of enterococci in extraintestinal environments [46].
In the beach environment, E. faecium was rare among enterococci identified from eelgrass, sewage influent, and dog samples, thus limiting the number of isolates that could be analyzed for virulence genes. E. faecium and E. faecalis were also rare or not detected in composite fecal samples from horses, goats, and pigs from PR, which is consistent with studies showing the low prevalence of these species in livestock [43, 47, 48]. Birds were rarely observed at the study beaches in PR, which precluded efforts to obtain enterococci isolates.
It is important to note that the presence of virulent strains among E. faecalis and E. faecium alone is not predictive of infection as there may be other mediators of pathogenicity that have yet to be elucidated [49]. It has been suggested that pathogenicity is also related to the ability of virulent strains to grow in high densities in the intestinal tract and spread to other sites in the body [50]. Host factors, such as predisposing medical conditions, immune status, and exposure to antibiotics, are also thought to play a role in the ability of enterococci to establish infection [51].
5. Conclusion
The low incidence of asa1, esp, and cylA among E. faecalis and E. faecium from the PR and SC beaches indicates that these virulence genes were not widely disseminated among strains found here, suggesting low potential health risks to humans. Still, the presence of E. faecalis and E. faecium harboring asa1, esp, and cylA suggests humans, birds, and dogs as potential sources of enterococci to beach water. Future surveys of enterococcal virulence genes at beaches should include those with different source inputs and populations of enterococci.
Acknowledgments
The authors thank the laboratory staff at the Department of Biology, University of Puerto Rico Mayaguez. Special thanks are due to Dr. Nathan Shankar for providing E. faecalis strain MMH594 and Orange County Public Health Laboratory for providing clinical strains.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Byappanahalli M. N., Nevers M. B., Korajkic A., Staley Z. R., Harwood V. J. Enterococci in the environment. Microbiology and Molecular Biology Reviews. 2012;76(4):685–706. doi: 10.1128/mmbr.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray B. E. The life and times of the enterococcus. Clinical Microbiology Reviews. 1990;3(1):46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jett B. D., Huycke M. M., Gilmore M. S. Virulence of enterococci. Clinical Microbiology Reviews. 1994;7(4):462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias C. A., Panesso D., Singh K. V., Rice L. B., Murray B. E. Cotransfer of antibiotic resistance genes and a hyl Efm-containing virulence plasmid in Enterococcus faecium . Antimicrobial Agents and Chemotherapy. 2009;53(10):4240–4246. doi: 10.1128/AAC.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willems R. J. L., van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiology. 2009;4(9):1125–1135. doi: 10.2217/fmb.09.82. [DOI] [PubMed] [Google Scholar]
- 6.Eaton T. J., Gasson M. J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Applied and Environmental Microbiology. 2001;67(4):1628–1635. doi: 10.1128/aem.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coque T. M., Willems R., Cantón R., Del Campo R., Baquero F. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. Journal of Antimicrobial Chemotherapy. 2002;50(6):1035–1038. doi: 10.1093/jac/dkf229. [DOI] [PubMed] [Google Scholar]
- 8.Rice L. B., Carias L., Rudin S., et al. A potential virulence gene, hyl Efm, predominates in Enterococcus faecium of clinical origin. The Journal of Infectious Diseases. 2003;187(3):508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- 9.Creti R., Imperi M., Bertuccini L., et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. Journal of Medical Microbiology. 2004;53(1):13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- 10.Sheldon W. L., Macauley M. S., Taylor E. J., et al. Functional analysis of a group A streptococcal glycoside hydrolase Spy1600 from family 84 reveals it is a β-N-acetylglucosaminidase and not a hyaluronidase. Biochemical Journal. 2006;399(2):241–247. doi: 10.1042/bj20060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher K., Phillips C. The ecology, epidemiology and virulence of Enterococcus . Microbiology. 2009;155(6):1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 12.Hammerum A. M. Enterococci of animal origin and their significance for public health. Clinical Microbiology and Infection. 2012;18(7):619–625. doi: 10.1111/j.1469-0691.2012.03829.x. [DOI] [PubMed] [Google Scholar]
- 13.Gilmore M. S., Coburn P. S., Nallapareddy S. R., Murray B. E. Enterococcus virulence. In: Gilmore M. S., Clewell D. B., Courvalin P., Dunny G. M., Murray B. E., Rice L. B., editors. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. 2002. p. p. 325. [Google Scholar]
- 14.Shankar V., Baghdayan A. S., Huycke M. M., Lindahl G., Gilmore M. S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infection and Immunity. 1999;67(1):193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coburn P. S., Baghdayan A. S., Dolan G. T., Shankar N. Horizontal transfer of virulence genes encoded on the Enterococcus faecalis pathogenicity island. Molecular Microbiology. 2007;63(2):530–544. doi: 10.1111/j.1365-2958.2006.05520.x. [DOI] [PubMed] [Google Scholar]
- 16.Manson J. M., Hancock L. E., Gilmore M. S. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(27):12269–12274. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Cesare A., Pasquaroli S., Vignaroli C., et al. The marine environment as a reservoir of enterococci carrying resistance and virulence genes strongly associated with clinical strains. Environmental Microbiology Reports. 2014;6(2):184–190. doi: 10.1111/1758-2229.12125. [DOI] [PubMed] [Google Scholar]
- 18.Bonilla T. D., Nowosielski K., Esiobu N., McCorquodale D. S., Rogerson A. Species assemblages of Enterococcus indicate potential sources of fecal bacteria at a south Florida recreational beach. Marine Pollution Bulletin. 2006;52(7):807–810. doi: 10.1016/j.marpolbul.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Badgley B. D., Thomas F. I. M., Harwood V. J. The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. Environmental Microbiology. 2010;12(5):1271–1281. doi: 10.1111/j.1462-2920.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson D. M., Griffith J. F., McGee C. D., Weisberg S. B., Hagedorn C. Comparison of enterococcus species diversity in marine water and wastewater using enterolert and EPA method 1600. Journal of Environmental and Public Health. 2013;2013:6. doi: 10.1155/2013/848049.848049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran Q., Badgley B. D., Dillon N., Dunny G. M., Sadowsky M. J. Occurrence, genetic diversity, and persistence of enterococci in a Lake Superior watershed. Applied and Environmental Microbiology. 2013;79(9):3067–3075. doi: 10.1128/aem.03908-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noble R. T., Weisberg S. B., Leecaster M. K., et al. Storm effects on regional beach water quality along the southern California shoreline. Journal of Water and Health. 2003;1(1):23–31. [PubMed] [Google Scholar]
- 23.Reeves R. L., Grant S. B., Mrse R. D., Oancea C. M. C., Sanders B. F., Boehm A. B. Scaling and management of fecal indicator bacteria in runoff from a coastal urban watershed in Southern California. Environmental Science and Technology. 2004;38(9):2637–2648. doi: 10.1021/es034797g. [DOI] [PubMed] [Google Scholar]
- 24.Yamahara K. M., Walters S. P., Boehm A. B. Growth of enterococci in unaltered, unseeded beach sands subjected to tidal wetting. Applied and Environmental Microbiology. 2009;75(6):1517–1524. doi: 10.1128/AEM.02278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura G. J., Thompson R. S., Boehm A. B., Jay J. A. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiology Ecology. 2011;77(1):40–49. doi: 10.1111/j.1574-6941.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 26.Choi S., Chu W., Brown J., Becker S. J., Harwood V. J., Jiang S. C. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Marine Pollution Bulletin. 2003;46(6):748–755. doi: 10.1016/S0025-326X(03)00046-8. [DOI] [PubMed] [Google Scholar]
- 27.Novais C., Coque T. M., Ferreira H., Sousa J. C., Peixe L. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Applied and Environmental Microbiology. 2005;71:3364–3368. doi: 10.1128/AEM.71.6.3364-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts M. C., Soge O. O., Giardino M. A., Mazengia E., Ma G., Meschke J. S. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. Journal of Applied Microbiology. 2009;107(1):300–307. doi: 10.1111/j.1365-2672.2009.04207.x. [DOI] [PubMed] [Google Scholar]
- 29.Moore D. F., Guzman J. A., McGee C. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. Journal of Applied Microbiology. 2008;105(4):1017–1025. doi: 10.1111/j.1365-2672.2008.03828.x. [DOI] [PubMed] [Google Scholar]
- 30.Rathnayake I., Hargreaves M., Huygens F. SNP diversity of Enterococcus faecalis and Enterococcus faecium in a South East Queensland waterway, Australia, and associated antibiotic resistance gene profiles. BMC Microbiology. 2011;11, article 201 doi: 10.1186/1471-2180-11-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson D. M., Weisberg S. B., Hagedorn C., et al. Enterococcus growth on eelgrass (Zostera marina); Implications for water quality. FEMS Microbiology Ecology. 2016;92(4) doi: 10.1093/femsec/fiw047. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson D. M., Moore D. F., Getrich M. A., Zhowandai M. H. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. Journal of Applied Microbiology. 2005;99(3):598–608. doi: 10.1111/j.1365-2672.2005.02660.x. [DOI] [PubMed] [Google Scholar]
- 33.Ke D., Picard F. J., Martineau F., et al. Development of a PCR assay for rapid detection of enterococci . Journal of Clinical Microbiology. 1999;37(11):3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vankerckhoven V., Van Autgaerden T., Vael C., et al. Development of a multiplex PCR for the detection of asaI, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among european hospital isolates of Enterococcus faecium. Journal of Clinical Microbiology. 2004;42(10):4473–4479. doi: 10.1128/jcm.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathnayake I. U., Hargreaves M., Huygens F. Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. Systematic and Applied Microbiology. 2012;35(5):326–333. doi: 10.1016/j.syapm.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Upadhyaya G. P., Lingadevaru U. B., Lingegowda R. K. Comparative study among clinical and commensal isolates of Enterococcus faecalisfor presence of esp gene and biofilm production. The Journal of Infection in Developing Countries. 2011;5(5):365–369. doi: 10.3855/jidc.1385. [DOI] [PubMed] [Google Scholar]
- 37.Süßmuth S. D., Muscholl-Silberhorn A., Wirth R., Susa M., Marre R., Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infection and Immunity. 2000;68(9):4900–4906. doi: 10.1128/iai.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biendo M., Adjide C., Castelain S., et al. Molecular characterization of glycopeptide-resistant enterococci from hospitals of the picardy region (France) International Journal of Microbiology. 2010;2010:8. doi: 10.1155/2010/150464.150464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo-Arana A., Valle J., Solano C., et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Applied and Environmental Microbiology. 2001;67(10):4538–4545. doi: 10.1128/aem.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh A., Dowd S. E., Zurek L. Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0022451.e22451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon K. H., Hwang S. Y., Moon B. Y., et al. Occurrence of antimicrobial resistance and virulence genes, and distribution of enterococcal clonal complex 17 from animals and human beings in Korea. Journal of Veterinary Diagnostic Investigation. 2012;24(5):924–931. doi: 10.1177/1040638712455634. [DOI] [PubMed] [Google Scholar]
- 42.Poeta P., Costa D., Rodrigues J., Torres C. Detection of genes encoding virulence factors and bacteriocins in fecal enterococci of poultry in Portugal. Avian Diseases. 2006;50(1):64–68. doi: 10.1637/7394-061505R.1. [DOI] [PubMed] [Google Scholar]
- 43.Han D., Unno T., Jang J., et al. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. International Journal of Food Microbiology. 2011;144(3):387–392. doi: 10.1016/j.ijfoodmicro.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Abriouel H., Omar N. B., Molinos A. C., et al. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. International Journal of Food Microbiology. 2008;123(1-2):38–49. doi: 10.1016/j.ijfoodmicro.2007.11.067. [DOI] [PubMed] [Google Scholar]
- 45.Ahmed W., Sidhu J. P. S., Toze S. Speciation and frequency of virulence genes of Enterococcus spp. isolated from rainwater tank samples in southeast Queensland, Australia. Environmental Science and Technology. 2012;46(12):6843–6850. doi: 10.1021/es300595g. [DOI] [PubMed] [Google Scholar]
- 46.McBride S. M., Fischetti V. A., LeBlanc D. J., Moellering R. C., Jr., Gilmore M. S. Genetic diversity among Enterococcus faecalis . PLoS ONE. 2007;2, article e582 doi: 10.1371/journal.pone.0000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franz C. M. A. P., Holzapfel W. H., Stiles M. E. Enterococci at the crossroads of food safety? International Journal of Food Microbiology. 1999;47(1-2):1–24. doi: 10.1016/s0168-1605(99)00007-0. [DOI] [PubMed] [Google Scholar]
- 48.Kühn I., Iversen A., Burman L. G., et al. Comparison of enterococcal populations in animals, humans, and the environment —a European study. International Journal of Food Microbiology. 2003;88(2-3):133–145. doi: 10.1016/s0168-1605(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 49.Kim E. B., Marco M. L. Non-clinical and clinical Enterococcus faecium but not Enterococcus faecalis have distinct structural and functional genomic features. Applied and Environmental Microbiology. 2013;80:154–165. doi: 10.1128/AEM.03108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mundy L. M., Sahm D. F., Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clinical Microbiology Reviews. 2000;13(4):513–522. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arias C. A., Murray B. E. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]


