Abstract
BACKGROUND & AIMS
The rs58542926 C>T variant of the transmembrane 6 superfamily member 2 gene (TM6SF2), encoding an E167K amino acid substitution, has been correlated with reduced total cholesterol (TC) and cardiovascular disease. However, little is known about the role of TM6SF2 in metabolism. We investigated the long-term effects of altered TM6SF2 levels in cholesterol metabolism.
METHODS
C57BL/6 mice (controls), mice that expressed TM6SF2 specifically in the liver, and mice with CRISPR/Cas9-mediated knockout of Tm6sf2 were fed chow or high-fat diets (HFD). Blood samples were collected from all mice and plasma levels of TC, low-density lipoprotein cholesterol (LDL-c), high-density lipoprotein cholesterol, and triglycerides were measured. Liver tissues were collected and analyzed by histology, real-time PCR, and immunoblot assays. Adenovirus vectors were used to express transgenes in cultured Hep3B hepatocytes.
RESULTS
Liver-specific expression of TM6SF2 increased plasma levels of TC and LDL-c, compared with controls, and altered liver expression of genes that regulate cholesterol metabolism. Tm6sf2-knockout mice had decreased plasma levels of TC and LDL-c, compared with controls, and consistent changes expression of genes that regulate cholesterol metabolism. Expression of TM6SF2 promoted cholesterol biosynthesis in hepatocytes.
CONCLUSIONS
TM6SF2 regulates cholesterol metabolism in mice and might be a therapeutic target for cardiovascular disease.
Keywords: translational research, gene regulation, lipid disorder, membrane proteins
Although tremendous advance in the research and clinical practice has been made over the past decades, cardiovascular diseases (CVDs) remain the leading cause of morbidity and mortality in the world. Increased plasma total cholesterol (TC), low-density-lipoprotein cholesterol (LDL-c), triglycerides (TG) and HDL cholesterol efflux capacity are among the most important risk factors for CVD.1 Indeed, lipid abnormality is associated with atherosclerosis, which causes the majority of cardiovascular events such as heart attack, stroke, and carotid artery stenosis.2
Discovery of molecular genetics basis for lipid disorders and understanding of the biological function of potential targets and molecular mechanisms underlying the regulation of lipid and cholesterol metabolism have revealed promising targets for treating and preventing cardiovascular diseases associated with lipid disorders.3 Using exome-wide association studies, we and other laboratories independently identified a coding variant (E167K) in the human TM6SF2 gene exhibiting significant correlation with total cholesterol (TC), myocardial infarction (MI), and non-alcoholic fatty liver disease (NAFLD).4–6 In human populations, the carriers of E167K presented lower TC and LDL-c in blood.4, 7–11 TM6SF2 is highly expressed in the liver, and localizes to the endoplasmic reticulum.12 Although the effect of transient modulation of TM6SF2 expression on cholesterol metabolism has been studied in mice,4, 5 little is known about the long-term role of TM6SF2 in these physiological and pathophysiological processes. In the current study, we aimed to understand the role of TM6SF2 in cholesterol metabolism utilizing novel genetically engineered transgenic mouse models.
Materials and Methods
Animal Procedures
Liver-specific transgenic mice on C57BL/6 background were generated expressing human TM6SF2 under the control of the mouse albumin enhancer/promoter as describe previously.13 The pBluescript II containing the albumin (Alb) promoter was kindly provided by Dr. Liangyou Rui (University of Michigan). We performed genotyping in the transgenic mice with primers: Forward, 5′-GAACCAATGAAATGCGAGGT-3; Reverse, 5′-AGAAGCCAGCAAGGATGAGA-3′, which recognize the mouse Alb enhancer/promoter and human TM6SF2 inserted sequence, respectively, yielding an 832 base pairs product. We generated Tm6sf2 KO mice (C57BL/6 background) with CRISPR/Cas9 technology, a well-established approach in our lab.14 The guide RNA targeted the mouse Tm6sf2 coding region sequence 5′-GACATCCCGCCGC-3′. The genotyping of the KO mice was performed by PCR with primers: Forward, 5′-CTGAAAACTGGGAAAGGACGCT-3′; Reverse, 5′-TGGAGAGGGATTTCTGCTTGCA-3′. The expression levels of the TM6SF2 transgene were determined by PCR with primers fully recognizing both human and mouse coding region of TM6SF2. Primer sequences were: Forward, 5′-TTCTACACCAAGGAGGGTGAGC-3′; Reverse, 5′-AACACCAGGATGCTCATGGCGA-3′. The PCR products were 182 base pairs in both human and mouse. The amplified DNA fragments containing the mutation (a C/G base pair inserted into the coding region right after start codon) were sequenced with primer: 5′-GTTCCTGGCAACTCAGAGCTCTGAC-3′.
Liver-specific TM6SF2 transgene (Alb-TM6SF2) mice, Tm6sf2 KO mice and their control mice were fed a high fat diet (HFD, 17.3% protein, 21.2% fat, 48.5% carbohydrate, 0.2% cholesterol by mass, 42% calories from fat. Harlan, TD.88137) or normal chow diet (22.5% protein, 11.8% fat and 52% carbohydrate by mass) for 10 or 12 weeks, as indicated. All mice were maintained on a 12-h light/dark cycle and had access to water and food ad libitum. Mice were fasted for 4 h before euthanasia. All animal work was performed in accordance with the University of Michigan Animal Care and Use Committee.
Lipid Profile
Plasma TC, LDL-c, HDL-c and TGs were measured with a Cobas Mira Plus chemistry analyzer (Roche) at the Michigan Diabetes Research and Training Center Chemistry Laboratory, University of Michigan. Lipids were measured by technicians blinded to the mouse experimental or control status.
Statistical Analysis
Statistical analyses between two groups were performed by two-tailed unpaired Student’s t test, and among three groups or more they were performed by one-way analysis of variance followed by a Newman-Keuls test using GraphPad Prism version 6.0. A P value of < .05 was considered statistically significant. Unless indicated otherwise, values are presented as mean ± S.D.
Results
Hepatic TM6SF2 Transgene Increased Plasma TC and LDL-c in Mice
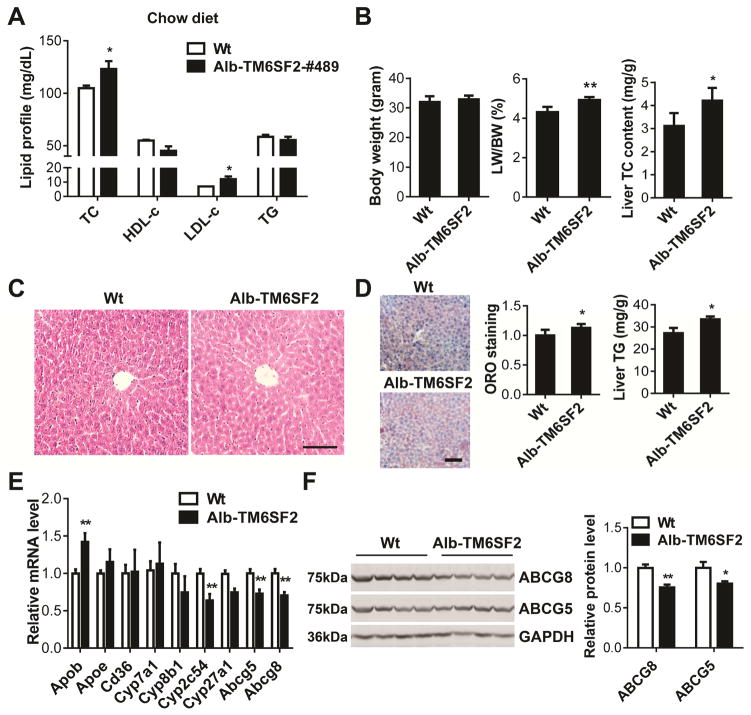
HFD leads to dysregulation of hepatic lipid metabolism in mice.15 We determined the effects of HFD on TM6SF2 expression in liver. TM6SF2 was downregulated at the mRNA and protein levels (Supplementary Figure 1A–B) in livers of mice fed a high fat diet (HFD), indicating that TM6SF2 is involved in the complicated liver metabolism network. Our previous work demonstrated that transient overexpression or knockdown of TM6SF2 modulates TC and LDL-c levels in C57BL/6 mice.4 To further gain insight into the long-term effects of TM6SF2 on hepatic cholesterol metabolism, we generated liver-specific human TM6SF2 (Alb-TM6SF2) transgenic mice (C57BL/6 background) with expression driven by the albumin (Alb) promoter (Supplementary Figure 2A). The transgene in two founder lines of transgenic mice (Alb-TM6SF2-#489 and #469) was genotyped by PCR (Supplementary Figure 2B). The effective expression of transgene was confirmed by both Real-time PCR (Supplementary Figures 2C) and Western blot (Supplementary Figure 2D). In agreement with our previous observation in mice transiently overexpressing human TM6SF2, the Alb-TM6SF2-#489 mice exhibited increased TC and LDL-c levels under chow diet conditions (Figure 1A). The body weights (BW) of mice showed no significant difference but the ratio of liver weight (LW) to BW was increased by 14.2% in the Alb-TM6SF2-#489 mice when compared with that of control mice. The cholesterol content in the liver was higher in the transgenic mice (Figure 1B). There were no obvious changes in the liver histology in wild type and transgenic mice (Figure 1C). Yet, increased local triglyceride (TG) accumulation was observed in livers of transgenic mice compared with those of wild type mice (Figure 1D).
Figure 1.
Increased TC and LDL-c in Alb-TM6SF2 mice fed a chow diet. (A), The plasma levels of cholesterol and TG in Alb-TM6SF2-#489 mice and littermate control mice (12-week-old male) were determined enzymatically (n =8 per group). (B), The BW, ratio of LW/BW and liver TC content were determined. (C), H&E staining of liver sections from Alb-TM6SF2 and control mice. (D), Liver sections were stained with Oil Red O (ORO) and quantitatively analyzed. Liver TG content was determined by a colorimetric assay. (E), The expression of genes related to cholesterol metabolism was determined by Real-time PCR. (F), The expression of ABCG5 and ABCG8 was determined by Western blot. The band intensity was quantitatively analyzed and normalized to GAPDH. Scale bar=100 μm. Values are mean ± SD. *, P <.05;**, P < .01.
The increases in TC and LDL-c in TM6SF2 transgenic mice may be attributed, amongst other factors, to altered bile acid metabolism, lipoprotein and cholesterol biosynthesis.16 Noteworthy, some bile acid and cholesterol related genes were dysregulated in livers of the Alb-TM6SF2 mice (Figure 1E). Cytochrome P450, family 2, subfamily c, polypeptide 54 (Cyp2c54) was downregulated in liver of the transgenic mice. Moreover, the TM6SF2 transgene decreased ATP-binding cassette, subfamily G, member 5 (ABCG5) and ABCG8 at both mRNA and protein levels (Figure 1E–F).
Hepatic TM6SF2 Increased Plasma TC and LDL-c under the Condition of HFD
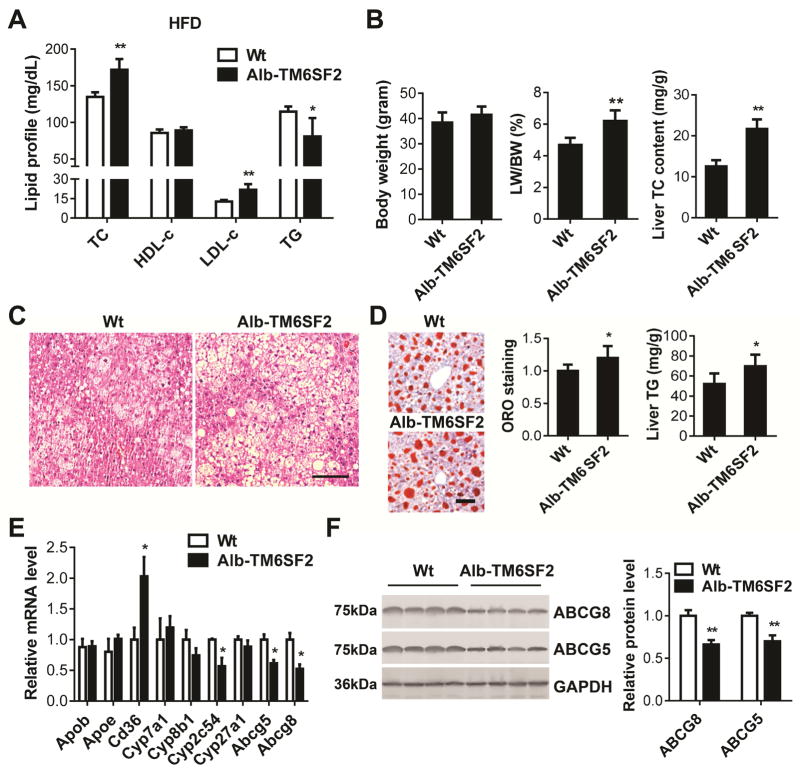
Intriguingly, the Alb-TM6SF2-#489 mice fed a HFD for 10 weeks showed TC, LDL-c significantly increased by 27.6% and 70.9%, respectively, compared with the littermate controls (Figure 2A). The body weights (BW) of mice had no significant difference but the ratio of liver weight (LW) to BW was increased by 47.8% in the Alb-TM6SF2-#489 mice when compared with the control mice. Moreover, the cholesterol content in the liver was higher in the transgenic mice (Figure 2B). The hepatic triglyceride (TG) accumulation was assessed by H&E and ORO staining of liver sections (Figure 2C–D) and the liver TG content was measured by a colorimetric assay (Figure 2D). The Alb-TM6SF2 mice fed a HFD exhibited higher local TG accumulation when compared with the control mice. To exclude a potential effect of random integration of the TM6SF2 transgene, we determined the lipid parameters in another transgenic line (Alb-TM6SF2-#469). Comparable increases in the TC and LDL-c levels, LW/BW, liver TC content and local TG accumulation were observed in the Alb-TM6SF2-#469 mice fed a HFD (Supplementary Figure 3). However, it was unexpected to find that plasma TG levels were not increased in the transgenic mice (Figure 2A and Supplementary Figure 3A), which will merit future investigation. Profiling of key genes indicated that the expression of Cyp2c54 was significantly decreased in livers of transgenic mice fed a HFD, while Cd36 was significantly upregulated (Figure 2E). Abcg5 and Abcg8 were downregulated at both mRNA and protein levels in livers of Alb-TM6SF2 mice (Figure 2E–F).
Figure 2.
Increased TC and LDL-c in Alb-TM6SF2 mice fed a HFD. Eight-week-old male Alb-TM6SF2-#489 and control mice were fed a HFD for 10 weeks. (A), The plasma levels of cholesterol and TG in the mice were determined enzymatically (n = 7–8 per group). (B), The BW and ratio of LW/BW were determined and TC in the liver was detected by a colorimetric assay. (C), H&E staining of liver sections from Alb-TM6SF2 and control mice fed a HFD. (D), Liver sections were stained with ORO and quantitatively analyzed. Liver TG content was determined by a colorimetric assay. (E), The expression of genes related to cholesterol metabolism was determined by Real-time PCR. (F), The expression of ABCG5 and ABCG8 was determined by Western blot. The band intensity was quantitatively analyzed and normalized to GAPDH. Scale bar=100 μm. Values are mean ± SD.*, P <.05;**, P < .01.
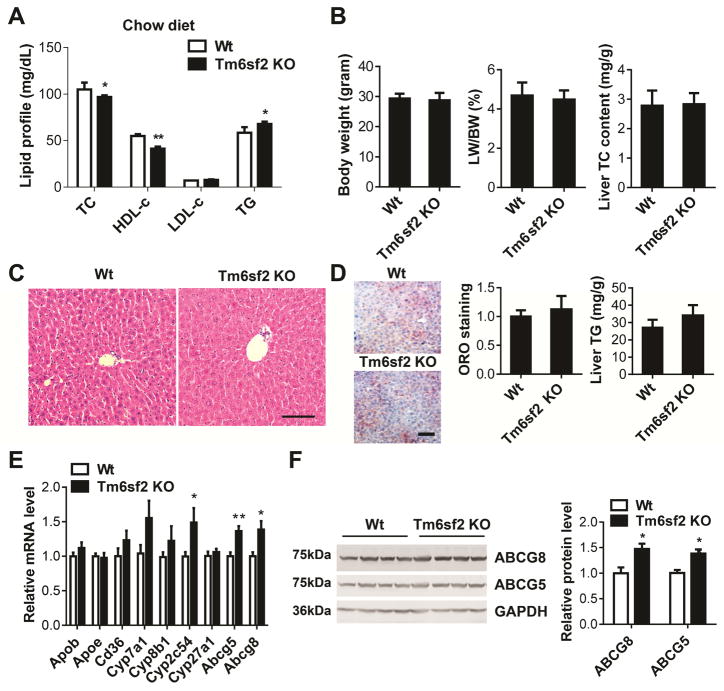
Tm6sf2 Knockout (KO) Mice Fed a Chow Diet Exhibited Decreased Plasma TC
In a complementary approach, we generated Tm6sf2 KO mice by CRISPR/Cas9 technology. A C/G base pair was inserted into the coding region to disrupt the open reading frame (Supplementary Figure 4A). The knockout efficacy was confirmed by Western blot (Supplementary Figure 4B). TM6SF2 is abundantly expressed in liver and small intestine in both human and mouse.4, 5, 12 Here, we determined the relative expression level of Tm6sf2 in different tissues by Real-time PCR. The mRNA level of Tm6sf2 is high in small intestine but moderate in colon (Supplementary Figure 4D). No obvious histological changes were observed in mouse brain, kidney, intestine and colon (Supplementary Figure 4E).
Both male and female KO mice fed a chow diet exhibited decreased plasma TC and HDL-c, but not LDL-c which was already at very low level in the C57BL/6 mice under basal conditions. Instead, the TG level was slightly increased in both genders of the transgenic mice (Figure 3A and Supplementary Figure 4C). The BW, ratio of LW/BW and liver cholesterol content between Tm6sf2 KO and control mice showed no significant difference (Figure 3B). We did not observe obvious histological changes in the liver sections from wild type and KO mice by H&E staining (Figure 3C). Unexpectedly, the KO mice had no significant changes in liver TG accumulation compared with control mice (Figure 3D). Additionally, we found that the expression pattern of Cyp2c54, Abcg5 and Abcg8 was opposite to that in the transgenic mice fed a chow diet (Figure 3E–F).
Figure 3.
Decreased TC levels in Tm6sf2 KO mice fed a chow diet. (A), The plasma levels of cholesterol and TG in KO and control mice (8–10-week-old male) were determined enzymatically (n = 7–8 per group). (B), The BW, ratio of LW/BW and liver TC content was determined. (C), H&E staining of liver sections from Tm6sf2 KO and control mice. (D), Liver sections were stained with ORO and quantitatively analyzed. Liver TG content was determined by a colorimetric assay. (E), The expression of genes related to cholesterol metabolism was determined by Real-time PCR. (F), The expression of ABCG5 and ABCG8 was determined by Western blot. The band intensity was quantitatively analyzed and normalized to GAPDH. Scale bar=100 μm. Values are mean ± SD.*, P <.05;**, P < .01.
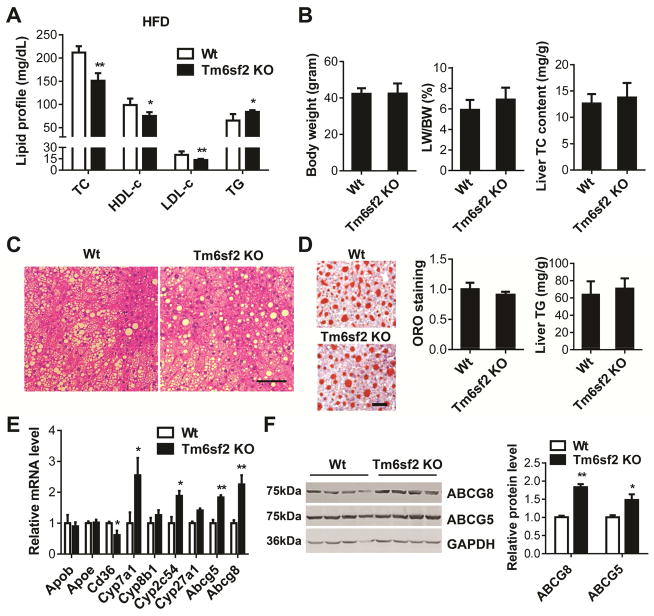
Loss of Tm6sf2 Decreased Plasma TC and LDL-c in the Mice Fed a HFD
After 12-week HFD feeding, the TC, LDL-c and HDL-c levels in Tm6sf2 KO mice were significantly decreased to 71.2%, 68.1% and 66.3%, respectively, when compared with control mice. However, the TG levels were significantly increased by 28.7% in the KO mice (Figure 4A). There was no significant difference in the BW, ratio of LW/BW and liver cholesterol content between Tm6sf2 KO and control mice (Figure 4B) and no significant histological changes were observed in the liver of KO mice compared with control mice (Figure 4C). The KO mice had no significant changes in liver TG accumulation (Figure 4D). However, we found that the expression pattern of Abcg5, Abcg8 and Cd36 was opposite to that in the transgenic mice fed a HFD (Figure 4E). ABCG5 and ABCG8 were upregulated in livers of KO mice compared with control mice (Figure 4F).
Figure 4.
Decreased TC and LDL-c levels in Tm6sf2 KO mice fed a HFD. Eight-week-old male Tm6sf2 KO and control mice were fed a HFD for 12 weeks (n = 7–8 per group). (A), The plasma levels of cholesterol and TG in KO and control mice were determined enzymatically. (B), The BW and ratio of LW/BW were determined and TC in the liver was detected by a colorimetric assay. (C), H&E staining of liver sections from Tm6sf2 KO and control mice fed a HFD. (D), Liver sections were stained with ORO and quantitatively analyzed. Liver TG content was determined by a colorimetric assay. (E), The expression of genes related to cholesterol metabolism was determined by Real-time PCR. (F), The expression of ABCG5 and ABCG8 was determined by Western blot. The band intensity was quantitatively analyzed and normalized to GAPDH. Scale bar=100 μm. Values are mean ± SD.*, P <.05;**, P < .01.
TM6SF2 Did Not Alter the Alanine Aminotransferase (ALT) Levels in the Transgenic and KO Mice Compared with Control Mice
Recent work reported that TM6SF2 E167K is positively correlated with steatosis, non-alcolholic steatohepatitis (NASH) and liver fibrosis.9, 17, 18 We detected the serum ALT and aspartate aminotransferase (AST) in the TM6SF2 genetically engineered mice. Our data suggested that the serum levels of ALT and AST showed no significant difference in both Alb-TM6SF2 and Tm6sf2 KO mice fed a chow diet or HFD compared with their respective control mice (Supplementary Figure 5A–D). We further determined fibrosis in the mice fed a HFD. Feeding with HFD for 10–12 weeks did not induce development of significant liver fibrosis measured by picrosirius red staining in either Alb-TM6SF2 mice expressing the wild type human allele (Supplementary Figures 5E) or Tm6sf2 KO mice (Supplementary Figure and 5F) compared with their respective controls. NASH is characterized by steatosis, hepatocellular hypertrophy, and mild diffuse lobular mixed acute and chronic inflammation.19, 20 In our study, H&E and ORO staining of liver sections showed steatosis in the mice fed a 10–12-week HFD (Figures 2C–D and 4C–D). Nevertheless, no significant inflammation was found in livers of Alb-TM6SF2, Tm6sf2 KO mice and their respective control mice by overall assessment of inflammatory foci in the H&E-stained sections (Supplementary Figure 6A and 6C). Tumor necrosis factors (TNF)-α and monocyte chemoattractant protein-1(MCP-1, CCL2) are critical factors in the development NASH in patients and murine models.21–23 In our study, the expression of Tnf-α and Mcp-1 in liver was not significantly increased after 10–12-week HFD feeding compared with chow diet and Tm6sf2 KO did not significantly altered the expression of Tnf-α and Mcp-1 in liver (Supplementary Figure 6D). However, TM6SF2 transgene significantly upregulated Mcp-1 in the mice fed a HFD compared with control mice, implicating a progression toward inflammation in TM6SF2 transgenic mice (Supplementary Figure 6B).
TM6SF2 Promoted Cholesterol Biosynthesis
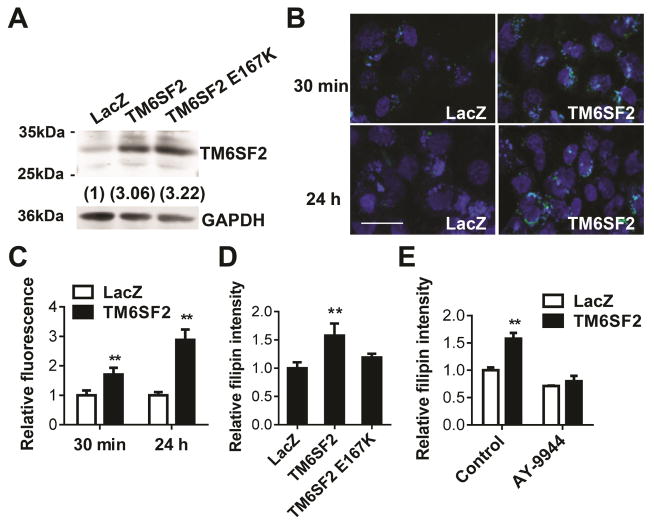
TM6SF2 was predicted to be an EBP-containing protein, which may enzymatically convert zymosterol to 5-α-cholesta-7,24-dien-3β-ol, a critical step in cholesterol biosynthesis.24 To determine the effects on cholesterol biosynthesis of TM6SF2 and the mutant allele identified in human genetic studies, we transfected Hep3B cells, a human hepatoma cell line, and confirmed expression of the wild type and mutant allele by Western blot (Figure 5A). TM6SF2 overexpression induced cholesterol accumulation under both normal and serum-free conditions (Figure 5B–C and Supplementary Figure 7A). Either in normal culture medium or serum-free medium in the presence of lanosterol and mevalonate, TM6SF2 overexpression increased cholesterol biosynthesis, while the E167K mutation attenuated the effect of TM6SF2 (Figure 5D and Supplementary Figure 7B). The effect of TM6SF2 could be abolished by AY-9944, an inhibitor of Δ7-dehydrocholesterol reductase (Figure 5E and Supplementary Figure 7C). Taken together, we documented for the first time that TM6SF2 is involved in the cholesterol biosynthesis.
Figure 5.
TM6SF2 promoted cholesterol biosynthesis. (A), Hep3B cells were infected with Ad-TM6SF2, Ad-TM6SF2-E167K, or Ad-LacZ (20 MOI) and 48h later, the expression level of TM6SF2 was detected by Western blot. (B), Hep3B cells were incubated with 3-dodecanoyl-NBD Cholesterol (15 μg/mL) for 30 min, or 3-dodecanoyl-NBD Cholesterol (1.5 μg/mL) for 24 h in 10% FBS medium. The intracellular NBD-cholesterol was detected by immunofluorescence. Scale bar = 50 μm. (C), The fluorescence intensity was quantitatively analyzed by NIH ImageJ. (D), Hep3B cells were infected with Ad-TM6SF2, Ad-TM6SF2-E167K, or Ad-LacZ (20 MOI), and 48 h later, the intracellular cholesterol was detected based on the intensity of Filipin III fluorescence. (E), Hep3B cells were infected with Ad-TM6SF2 or Ad-LacZ (20 MOI), and 48 h later, cells were treated with AY-9944 (10 μmol/L), an inhibitor of Δ7-dehydrocholesterol reductase for 24 h. The intracellular cholesterol was detected based on the intensity of Filipin III fluorescence. Values are mean ± SD.**, P < .01.
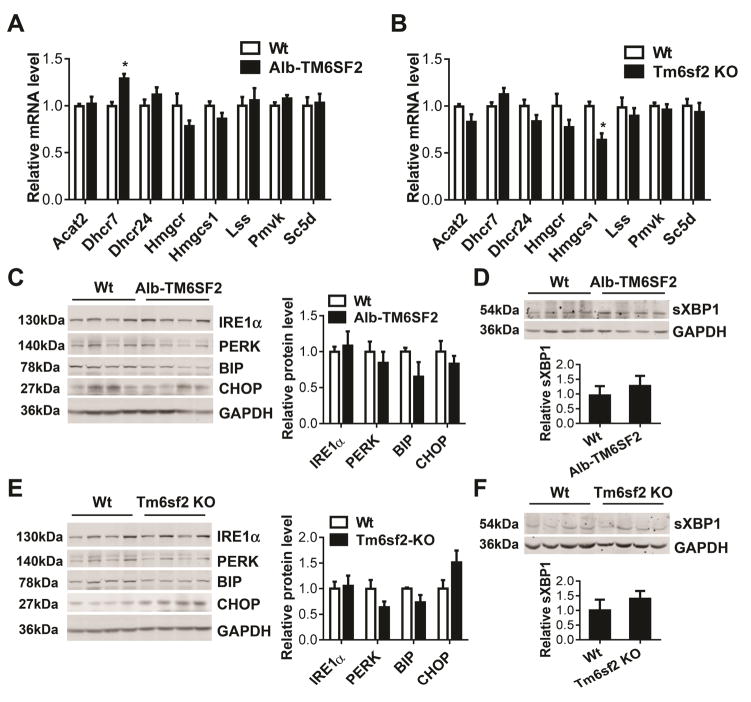
To investigate whether TM6SF2 impacts other genes related to cholesterol biosynthesis, we determined the expression of key genes encoding enzymes involved in the cholesterol biosynthesis. Our data suggest that TM6SF2 has a minor effect on expression of those genes. Although 7-dehydrocholesterol reductase (Dhcr7), an enzyme converting 7-dehydrocholesterol to cholesterol at the final step of cholesterol biosynthesis, was increased at the mRNA level in livers of Alb-TM6SF2 mice compared with control mice (Figure 6A), no significant changes were detected in the Tm6sf2 KO mice (Figure 6B). Collectively, our data suggest that TM6SF2 is critical to regulate intracellular cholesterol homeostasis but has limited effect on other genes directly involved in the cholesterol biosynthesis pathway.
Figure 6.
The effect of TM6SF2 on ER stress pathway. (A–B), Eight-week-old Alb-TM6SF2-#489 and control mice (A) and Tm6sf2 KO and control mice (B) were fed a chow diet (n = 5–6 per group). The cholesterol biosynthesis genes were determined by Real-time PCR. (C-D), Eight-week-old Alb-TM6SF2-#489 and control mice were fed a chow diet. The expression of ER stress proteins (C) and spliced XBP1 (sXBP1) (D) was determined by Western blot and quantitatively analyzed. (E–F), Eight-week-old Tm6sf2 KO mice and control mice were fed a chow diet. The expression of ER stress genes (E) and sXBP1 (F) was determined by Western blot and quantitatively analyzed (n = 4–5 per group). Values are mean ± SD.*, P < .05.
TM6SF2 Did Not Alter Endoplasmic Reticulum (ER) Stress Response in the Liver of Genetically Engineered Mice
TM6SF2 mainly localizes to the ER membrane.12 ER is an active subcellular organelle for cholesterol metabolism and modulating physiological and pathological insults. Thus, we determined whether TM6SF2 has other effects beside cholesterol synthesis in the ER. The ER stress proteins IRE1α, PERK, BIP and CHOP were detected by Western blot. However, we did not find significant difference in expression of these proteins in either transgenic mice (Figure 6C and Supplementary Figure 8A) or KO mice (Figure 6E and Supplementary Figure 8C) when compared with their respective control mice under both chow diet and HFD conditions. The spliced XBP1 (sXBP1) mRNA encodes an active and stable transcription factor (54 kDa, as a result of a frameshift), which regulates unfolded protein response.25 In our study, TM6SF2 did not significantly change the spliced form of XBP1 in the liver of transgenic mice (Figure 6D and Supplementary Figure 8B) and KO mice (Figure 6F and Supplementary Figure 8D) when compared with their respective control mice under both chow diet and HFD conditions.
Discussion
TM6SF2 protein is predicted as an EBP containing enzyme homolog involved in cholesterol biosynthesis.24 The human and mouse TM6SF2 protein have high similarity with a 78% sequence identity. More significantly, there is a conserved E167 in TM6SF2 of both species and the TM6SF2 E167K mutation is negatively associated with total cholesterol, LDL-c7–11 and myocardial infarction.4 In this study, we have revealed a critical role of TM6SF2 in cholesterol metabolism that is consistent with the genetic discovery that TM6SF2 E167K reduces total cholesterol and LDL-c. In this study, the liver-specific human TM6SF2 transgene in mice elevated plasma TC and LDL-c and enhanced cholesterol biosynthesis in hepatocytes. Conversely, loss of TM6SF2 in the Tm6sf2 KO mice decreased plasma TC and LDL-c. Additionally, these data suggest that human and mouse TM6SF2 have a similar function, at least in cholesterol metabolism, in models of chronic alteration in expression levels of this gene, which likely constitute a more physiological approach to those involving transient expression as reported before.4
The role of TM6SF2 in liver cholesterol metabolism is also supported by the regulation of cholesterol metabolism related genes. Cyp2c54 was consistently regulated in livers from both TM6SF2 transgenic (downregulation) and KO mice (upregulation). Cyp2c54 is involved in liver detoxification functions and metabolizes linoleic acid to epoxyoctadecenoic acids.26, 27 Mutations in ABCG5 and ABCG8 cause sitosterolemia28 and sterol accumulation in the liver, 29 and are associated with TC and LDL-c in humans.30 CD36, a scavenger receptor that binds modified forms of LDL,31 was upregulated in TM6SF2 transgenic mice and downregulated in Tm6sf2 KO mice fed a HFD. Hepatic CD36 is associated with increased steatosis in NASH and chronic hepatitis C.32 Loss of Cd36 in mice inhibits hepatic steatosis.33
TM6SF2 was predicted to be involved in cholesterol biosynthesis.24 Our data suggest that TM6SF2 increases cholesterol biosynthesis and indicates that the E167K mutation may result in partial loss-of-function. Understanding the functions of TM6SF2 and its variants will help in the development of new therapeutic approaches useful in lipid disorders. Future studies are warranted to explore whether TM6SF2 directly enzymatically converts the substrate zymosterol into the downstream product 5-α-cholesta-7, 24-dien-3β-ol. Although TM6SF2 has limited effect on the expression of genes encoding enzymes in cholesterol biosynthesis, the regulation of cholesterol metabolism related genes and promotion of cholesterol biosynthesis might contribute cooperatively to the TM6SF2 modulation of plasma TC and LDL-c in mice.
Accumulated population genetic studies have suggested that the E167K mutation in TM6SF2 is positively correlated with different stages of NAFLD: steatosis, NASH and fibrosis.5, 9, 10, 17, 34–37 More recently, the same variant in the TM6SF2 gene has been identified as a novel risk locus for alcohol-related cirrhosis in the European descent and validated in two independent European cohorts.38 The global Tm6sf2 KO mice and liver-specific transgenic mice allowed us to determine the long-term effect of TM6SF2 on lipid metabolism in the liver. In this study, we used HFD mouse model which exhibited elevated plasma TC and LDL-c levels after HFD feeding in mice and that recapitulated the discoveries from the human studies regarding circulating TC and LDL-c. However, TM6SF2 gain-of-function unexpectedly induced steatosis in mouse liver, while the loss-of-function had a minor effect on TG accumulation. Although we did find that TG accumulated in livers of the mice fed a HFD for 10–12 weeks, we did not observed significant NASH-like pathological changes in livers. Indeed, the same HFD protocol we used in this study induced hepatic steatosis at 12 weeks in C57BL/6 mice but a longer feeding period of HFD was required to develop the conditions associated with NASH and fibrosis in mice.39 Thus, to determine the effect of TM6SF2 on the pathological process of NASH and liver fibrosis, long-term of HFD-feeding, which will be highly facilitated with these new TM6SF2 animal models, should be applied to these mice in follow up studies. Other mouse feeding models such as methionine and choline diet,40, 41 and cholesterol and cholate diet42 may be better choices to further investigate the direct role of TM6SF2 in liver steatosis, NASH and fibrosis.43
Additionally, we recognize that there are other limitations in interpreting the divergence in LW/BW and TG accumulation in liver-specific TM6SF2 transgenic mice and Tm6sf2 KO mice because loss of Tm6sf2 in extrahepatic tissues, such as small intestine, colon, skeletal muscle and adipose tissue, may also have contributed to the steatosis phenotype. Future studies are necessary to determine whether and how modulation of TM6SF2 in other organs or tissues regulates hepatic steatosis. The difference between the findings in the mouse studies and the discovery from the human genetic studies may be attributable to the difference between coding variants, alteration in gene expression and possible species difference. In addition, aging promotes NASH and leads to increased hepatocellular injury and inflammation.44 Therefore, the role of TM6SF2 in hepatic metabolism should be determined in aging mice in future studies. As a matter of fact, recent studies have linked the E167K mutation to increased steatosis in children, thus indicating a complex interaction of TM6SF2 with aging and obesity.8, 45 Indeed, liver-specific TM6SF2 KO mice would be a better tool to dissect the exact role of TM6SF2 in liver and TM6SF2 E167K mutation knock-in mice are under development to dissect the exact role of this mutation in cholesterol and TG metabolism as well as to shed further insight on the biochemical function of TM6SF2.
This study adds to a rapidly accumulating body of work that defines TM6SF2 as both a resilience factor for CVD and a risk factor for other liver disorders. Thus, it is likely that the allelic status of TM6SF2 in combination with that of other synergistic factors contributing to liver metabolism will likely contribute to better patient care within the emerging paradigm of personalized medicine. Understanding the physiological and pathophysiological roles of genes like TM6SF2 is highly relevant to translational research towards individualized outcomes.
Conclusions
The current study is the first reporting long-term biological effects of TM6SF2 in the liver cholesterol metabolism, which are consistent with prior human studies and further establishes TM6SF2 as a critical regulator of cholesterol homeostasis and a likely cardiovascular resilience factor. Consequently, these findings define TM6SF2 as a potential molecular target for treatment of lipid disorders associated with cardiovascular disease.
Supplementary Material
Acknowledgments
Grant support: This work was supported, in whole or in part, by National Institutes of Health Grants HL068878, HL105114, and HL088391 (to Y.E.C.); The National Natural Science Foundation of China (81428004); American Heart Association grants 14SDG19880014 (Y.F.) and 15SDG24470155 (to Y.G.).
Abbreviations
- TM6SF2
transmembrane 6 superfamily member 2
- KO
knockout
- TC
total cholesterol
- TG
triglyceride
- HFD
high fat diet
- MI
myocardial infarction
- Alb
albumin
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Author contributions: All authors have been involved in critical review of the manuscript; Fan Y, Lu H, Guo Y obtained, contributed and analyzed the data. Zhu T provided technical and material support. The manuscript was drafted by Fan Y, and then critically reviewed, including comments and feedback from Chen YE, Zhang J, Garcia-Barrio MT, Jiang Z, and Willer CJ. The study was conceived and designed by Chen YE, Fan Y, and Zhang J.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rohatgi A, Khera A, Berry JD, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371:2383–93. doi: 10.1056/NEJMoa1409065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotto AM., Jr Evolving concepts of dyslipidemia, atherosclerosis, and cardiovascular disease: the Louis F. Bishop Lecture J Am Coll Cardiol. 2005;46:1219–24. doi: 10.1016/j.jacc.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 3.Rached FH, Chapman MJ, Kontush A. An overview of the new frontiers in the treatment of atherogenic dyslipidemias. Clin Pharmacol Ther. 2014;96:57–63. doi: 10.1038/clpt.2014.85. [DOI] [PubMed] [Google Scholar]
- 4.Holmen OL, Zhang H, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345–51. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–6. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahali B, Liu YL, Daly AK, et al. TM6SF2: catch-22 in the fight against nonalcoholic fatty liver disease and cardiovascular disease? Gastroenterology. 2015;148:679–84. doi: 10.1053/j.gastro.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Wong VW, Wong GL, Tse CH, et al. Prevalence of the TM6SF2 variant and non-alcoholic fatty liver disease in Chinese. J Hepatol. 2014;61:708–9. doi: 10.1016/j.jhep.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 8.Grandone A, Cozzolino D, Marzuillo P, et al. TM6SF2 Glu167Lys polymorphism is associated with low levels of LDL-cholesterol and increased liver injury in obese children. Pediatr Obes. 2015 doi: 10.1111/ijpo.12032. [DOI] [PubMed] [Google Scholar]
- 9.Dongiovanni P, Petta S, Maglio C, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–14. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 10.Goffredo M, Caprio S, Feldstein AE, et al. Role of the TM6SF2 rs58542926 in the pathogenesis of non-alcoholic pediatric fatty liver disease (NAFLD): A multiethnic study. Hepatology. 2015 doi: 10.1002/hep.28283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology. 2015 doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 12.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinkert CA, Ornitz DM, Brinster RL, et al. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987;1:268–76. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- 14.Yang D, Xu J, Zhu T, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6:97–9. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buettner R, Scholmerich J, Bollheimer LC. High-fat diets: modeling the metabolic disorders of human obesity in rodents. Obesity (Silver Spring) 2007;15:798–808. doi: 10.1038/oby.2007.608. [DOI] [PubMed] [Google Scholar]
- 16.Thomas C, Pellicciari R, Pruzanski M, et al. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–93. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 17.Liu YL, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roh YS, Loomba R, Seki E. The TM6SF2 variants, novel genetic predictors for nonalcoholic steatohepatitis. Gastroenterology. 2015;148:252–4. doi: 10.1053/j.gastro.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunt EM. Nonalcoholic steatohepatitis. Semin Liver Dis. 2004;24:3–20. doi: 10.1055/s-2004-823098. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner DE, Bedossa P. Liver Histology and Clinical Trials for Nonalcoholic Steatohepatitis-Perspectives From 2 Pathologists. Gastroenterology. 2015;149:1305–8. doi: 10.1053/j.gastro.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Baeck C, Wehr A, Karlmark KR, et al. Pharmacological inhibition of the chemokine CCL2 (MCP-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut. 2012;61:416–26. doi: 10.1136/gutjnl-2011-300304. [DOI] [PubMed] [Google Scholar]
- 22.Haukeland JW, Damas JK, Konopski Z, et al. Systemic inflammation in nonalcoholic fatty liver disease is characterized by elevated levels of CCL2. J Hepatol. 2006;44:1167–74. doi: 10.1016/j.jhep.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Braunersreuther V, Viviani GL, Mach F, et al. Role of cytokines and chemokines in non-alcoholic fatty liver disease. World J Gastroenterol. 2012;18:727–35. doi: 10.3748/wjg.v18.i8.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Pulido L, Ponting CP. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front Genet. 2014;5:439. doi: 10.3389/fgene.2014.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida H, Matsui T, Yamamoto A, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Zhao Y, Bradbury JA, et al. Cloning, expression, and characterization of three new mouse cytochrome p450 enzymes and partial characterization of their fatty acid oxidation activities. Mol Pharmacol. 2004;65:1148–58. doi: 10.1124/mol.65.5.1148. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Yuan B, Lo KA, et al. Adiponectin regulates expression of hepatic genes critical for glucose and lipid metabolism. Proc Natl Acad Sci U S A. 2012;109:14568–73. doi: 10.1073/pnas.1211611109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubacek JA, Berge KE, Cohen JC, et al. Mutations in ATP-cassette binding proteins G5 (ABCG5) and G8 (ABCG8) causing sitosterolemia. Hum Mutat. 2001;18:359–60. doi: 10.1002/humu.1206. [DOI] [PubMed] [Google Scholar]
- 29.Yu L, von Bergmann K, Lutjohann D, et al. Selective sterol accumulation in ABCG5/ABCG8-deficient mice. J Lipid Res. 2004;45:301–7. doi: 10.1194/jlr.M300377-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jay AG, Chen AN, Paz MA, et al. CD36 binds oxidized low density lipoprotein (LDL) in a mechanism dependent upon fatty acid binding. J Biol Chem. 2015;290:4590–603. doi: 10.1074/jbc.M114.627026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miquilena-Colina ME, Lima-Cabello E, Sanchez-Campos S, et al. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 2011;60:1394–402. doi: 10.1136/gut.2010.222844. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Febbraio M, Wada T, et al. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 2008;134:556–67. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 34.Dongiovanni P, Romeo S, Valenti L. Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis. Biomed Res Int. 2015;2015:460190. doi: 10.1155/2015/460190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milano M, Aghemo A, Mancina RM, et al. Transmembrane 6 superfamily member 2 gene E167K variant impacts on steatosis and liver damage in chronic hepatitis C patients. Hepatology. 2015;62:111–7. doi: 10.1002/hep.27811. [DOI] [PubMed] [Google Scholar]
- 36.Daly AK, Day CP, Liu YL, et al. TM6SF2 as a genetic risk factor for fibrosis. Hepatology. 2014 doi: 10.1002/hep.27656. [DOI] [PubMed] [Google Scholar]
- 37.Coppola N, Rosa Z, Cirillo G, et al. TM6SF2 E167K variant is associated with severe steatosis in chronic hepatitis C, regardless of PNPLA3 polymorphism. Liver Int. 2015;35:1959–63. doi: 10.1111/liv.12781. [DOI] [PubMed] [Google Scholar]
- 38.Buch S, Stickel F, Trepo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–8. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 39.VanSaun MN, Lee IK, Washington MK, et al. High fat diet induced hepatic steatosis establishes a permissive microenvironment for colorectal metastases and promotes primary dysplasia in a murine model. Am J Pathol. 2009;175:355–64. doi: 10.2353/ajpath.2009.080703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dela Pena A, Leclercq I, Field J, et al. NF-kappaB activation, rather than TNF, mediates hepatic inflammation in a murine dietary model of steatohepatitis. Gastroenterology. 2005;129:1663–74. doi: 10.1053/j.gastro.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Ip E, Farrell G, Hall P, et al. Administration of the potent PPARalpha agonist, Wy-14,643, reverses nutritional fibrosis and steatohepatitis in mice. Hepatology. 2004;39:1286–96. doi: 10.1002/hep.20170. [DOI] [PubMed] [Google Scholar]
- 42.Matsuzawa N, Takamura T, Kurita S, et al. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 2007;46:1392–403. doi: 10.1002/hep.21874. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2300–8. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontana L, Zhao E, Amir M, et al. Aging promotes the development of diet-induced murine steatohepatitis but not steatosis. Hepatology. 2013;57:995–1004. doi: 10.1002/hep.26099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mancina RM, Sentinelli F, Incani M, et al. Transmembrane-6 superfamily member 2 (TM6SF2) E167K variant increases susceptibility to hepatic steatosis in obese children. Dig Liver Dis. 2015 doi: 10.1016/j.dld.2015.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.