Summary
Pili are proteinaceous polymers of linked pilins that protrude from the cell surface of many bacteria and often mediate adherence and virulence. We investigated a set of 20 Bacteroidia pilins from the human microbiome whose structures and mechanism of assembly were unknown. Crystal structures and biochemical data revealed a diverse protein superfamily with a common Greek-key β-sandwich fold with two transthyretin-like repeats that polymerize into a pilus through a strand-exchange mechanism. The assembly mechanism of the central, structural pilins involves proteinase-assisted removal of their N-terminal β-strand, creating an extended hydrophobic groove that binds the C-terminal donor strands of the incoming pilin. Accessory pilins at the tip and base have unique structural features specific to their location, allowing initiation or termination of the assembly. The bacteroidia pilus therefore has a biogenesis mechanism that is distinct from other known pili and likely represents a different type of bacterial pilus.
Introduction
Studies of the human microbiome are uncovering the importance and extent of the complex symbiotic relationships between the human host and the microbiota that inhabit various cavities (e.g. oral, gut, vagina) and exposed surfaces (skin). Many of these colonizing bacteria have long proteinaceous filaments up to several micrometers in length on their cell surface called pili (also known as fimbriae) that serve as probes or anchors for interaction with host cells. These extended appendages also often function as major virulence factors in pathogenic bacteria and are involved in biofilm formation. Structural studies of individual pilin subunits and their assembly have shed significant insights into the mechanism of pilus biogenesis in several model organisms (Allen et al., 2012; Choudhury et al., 1999; Kang et al., 2007; Li et al., 2009; Parge et al., 1995; Proft and Baker, 2009; Sauer et al., 1999). For example, in the well-characterized chaperone-usher pili (type I) of E. coli, precursors of the structural subunits are exported to the periplasm where they are processed into their mature forms by signal peptidases. The pilus filaments are then assembled from head-to-tail from the structural subunits (pilins) with the aid of a chaperone and an usher (Allen et al., 2012; Proft and Baker, 2009). In Gram-negative bacteria, pili are assembled via noncovalent interactions and directly inserted into the bacterial cell wall, whereas in Gram-positive bacteria, the pilus subunits are connected by intermolecular isopeptide bonds and the entire assembly tethered to a cell-wall peptidoglycan.
Porphyromonas gingivalis is a major oral pathogen associated with severe adult periodontitis (Holt and Ebersole, 2005). P. gingivalis pili are key virulence factors that are essential for host colonization and evasion of innate defenses (Amano, 2010; Hajishengallis et al., 2007; Hajishengallis et al., 2008). They are also involved in binding a wide array of oral or epithelial substrates and extracellular matrix proteins (Amano, 2003; Hajishengallis, 2007), in addition to co-aggregation with other pathogens, such as Streptococcus gordonii (Park et al., 2005). Two types of morphologically distinct pili have been identified in P. gingivalis: major or long (0.3 to 1.6 μm) and minor or short (80 to 120 nm) (Hamada et al., 1996; Yoshimura et al., 1984). These pili are encoded by similar operons (Figure 1A) that contain genes for the main structural pilins [FimA (major) or Mfa1 (minor)] that form the stalk of the pili (Park et al., 2005; Sojar et al., 1991), followed by genes for the anchors pilins [FimB (major) and Mfa2 (minor)] and other ancillary pilins or regulatory elements (Hasegawa et al., 2009; Nagano et al., 2010). The FimA pilins from various P. gingivalis strains are classified into at least five different subtypes (FimA1-FimA5) based on sequence and immunogenic properties. The best characterized FimA1 of strain 33277 (referred to as FimA hereafter) shares 50-85% sequence identity with other subtypes of FimA. Similar pili have been identified in the intestinal Bacteroides fragilis (van Doorn et al., 1987; van Doorn et al., 1992).
Figure 1. Operons, sequence motifs, crystal structures and clustering of the FimA pilin superfamily.
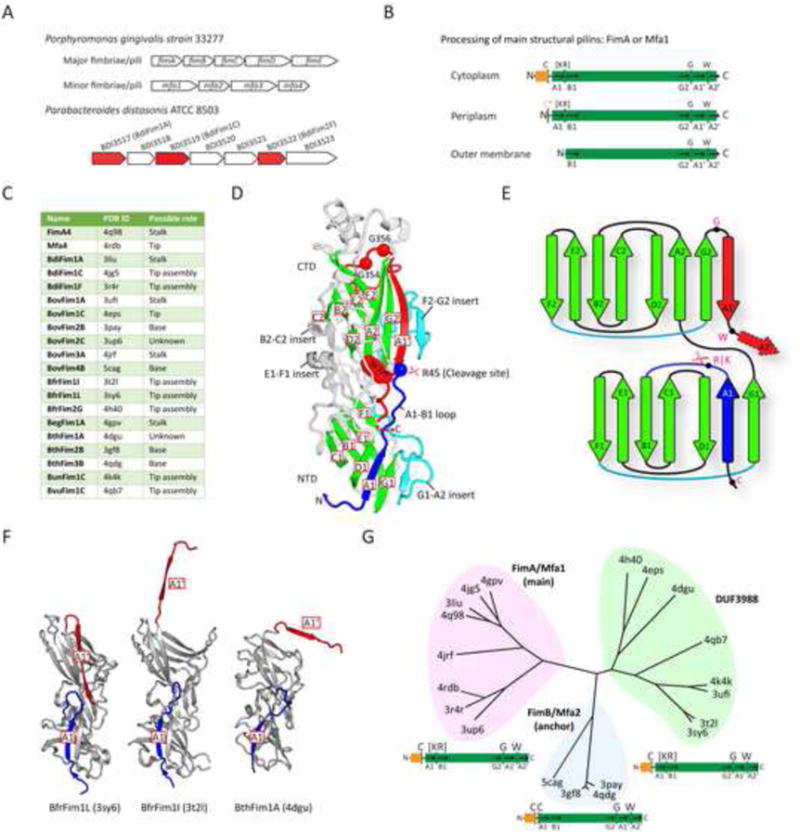
(A) Operons encoding the major and minor pili in P. gingivalis strain 33277, compared to an analogous typical pilin encoding operon from the gut microbiome, exemplified by P. distasonis. (P. distasonis. structures determined in this study are colored red).
(B) Sequential maturation of pilins via proteolytic and covalent modifications during export from the cytoplasm to the outer membrane. Conserved residues are marked on the top, while secondary structures are labeled at the bottom. Orange box: the lipoprotein signal peptide; asterisk: lipidated site.
(C) Pilin structures determined and their possible roles (see Table S1).
(D) Crystal structure of FimA4. The Arg/Lys specific cleavage site between A1 and B1 is marked by a scissor symbol. The conserved β-sheet core is colored in green, the A1 strand in blue, inserts in cyan, and the A1′ and A2′ appendage in red.
(E) A topological diagram of conserved secondary structures in FimA4 colored coded as in D.
(F) Crystal structures of DUF3988 members with the A1′ strand in “open” (BfrFim1I and BthFim1A) conformations, in comparison with BfrFim1L that is closely related to BfrFim1I, but containing a “closed” A1′ strand. Color coded as in D.
(G) Clustering of crystal structures into three families that form the FimA superfamily and sequence features below of each family.
P. gingivalis pili are synthesized using a lipoprotein-proteinase pathway (Figure S1). Although the secretion apparatus remains to be fully characterized, the initial processing and transport of the pilins are assumed to piggyback on the lipoprotein sorting machinery (Shoji et al., 2004). FimA and Mfa1 prepilins possess significantly longer leader peptides than other bacterial pilins and are exported as lipoprotein precursors (Nakayama et al., 1996; Shoji et al., 2004; Shoji et al., 2010b). FimA is lipidated on the invariant cysteine at the C-terminus of the lipoprotein signal peptide (lipobox), which is subsequently removed in the periplasm by a type II signal proteinase (Shoji et al., 2004). A second required proteolytic step, performed by an outer membrane (OM) trypsin-like arginine-specific proteinase (R-gingipain or Rgp) (Nakayama et al., 1996), removes an additional N-terminal fragment, including the lipidated cysteine, to generate the mature pilin that assembles into the pilus filament (Figure 1B). This pilin maturation process is expected to be conserved in other related pili, such as the B. fragilis pili (Figure S1). The P. gingivalis pilus biogenesis pathway shares some similarity with that of the type I pilus, but with at least two important differences (Figure S1): involvement of lipoprotein precursors and requirement for an additional OM proteinase.
However, as P. gingivalis FimA-related pilins and other characterized pilins have no recognizable sequence homology and little was known about their evolutionary origin, distribution, three-dimensional structures, and assembly process, we investigated the structure and function of this family of Bacteroidia pilins. From crystal structures of a diverse set of FimA-like pilins from Bacteroidia and corroborating biochemical data, we define here a distinct type of pilus (type V) and propose a mechanism for its assembly. These pilins are ubiquitous in human gut microbiota and represent a large and diverse superfamily of proteins that may confer adaptive advantages to certain species of bacteria that colonize the human gut.
Results
Structure determination and overall structures
Bacteria from the Bacteroidia class form a significant component of the human microbiome and play a vital role in human health and disease (Cho and Blaser, 2012; Pflughoeft and Versalovic, 2012). To explore and better comprehend the complex interplay between the human gut microbiome and its host, we utilized the JCSG high-throughput structural biology pipeline (Elsliger et al., 2010) to investigate the Bacteroidia secretome. Structural targets were selected primarily from protein families, most of which had no known function, that were specific to, or enriched in, the human gut microbiome (Ellrott et al., 2010). We determined and analyzed crystal structures of 20 Bacteroidia putative pilus components [including BthFim2B from a previous study (Xu et al., 2010)], that included P. gingivalis FimA4 (type 4 structural pilin of the major pili) and Mfa4 (tip pilin of the minor pili), and derived their probable roles (Figure 1C, Tables S1-S2). Most pilins from the gut microbiome are encoded by operons similar to those in P. gingivalis, as exemplified in Parabacteroides distasonis (Figure 1A). For crystallographic studies, the predicted lipoprotein signal peptide (Hayashi and Wu, 1990) was omitted. Thus, the structures correspond to a periplasmic intermediate state and are mainly monomers in the crystals.
All structures (Figure S2) contain an N-terminal domain (NTD) and a slightly larger C-terminal domain (CTD). Each domain has a transthyretin-like fold that contains seven core β-strands arranged in two β-sheets (DAG and CBEF), as represented by FimA4 (Figure 1D-E). The NTD fold is prototypical (A1-G1), whereas the CTD (except for Mfa4) has an extra conserved “appendage” of two amphipathic β-strands (A1′ and A2′) attached to the C-terminus of the core (A2-G2). Two significantly different conformations are observed for this appendage. In BfrFim1I, BthFim1A, BovFim1C, and BovFim4B (Figures 1F and S2), it adopts an extended “open” conformation, while in other structures (e.g. FimA4 in Figure 1D), it folds back into the CTD, extending the DAG β-sheet (“closed” conformation). The “open” C-terminal strands of BfrFim1I and BthFimA1 are actually domain-swapped in the crystal, while the equivalent “open” strand of BovFim1C links to an additional lectin domain. In BovFim4B, the “open” C-terminal strand extends into the solvent and is not stabilized by crystal contacts and, thus, is mostly disordered. The last ∼10 C-terminal residues, containing β-strand A2′, are disordered in most structures (Figure S2). The NTD and the CTD are related mostly by translational symmetry to form an elongated molecule with dimensions of ∼90 Å × 45 Å × 40 Å. The domain interface is mainly stabilized by extensive interactions between two loop insertions: the NTD E1-F1 loop and CTD B2-C2 loop (Figure 1D). The loops that decorate the main fold vary greatly among the different structures, ranging from minimal length in BfrFim1I to more elaborate loops in FimA4 and BovFim3A (Figure S2).
Structural clustering of the FimA superfamily
The pilin structures are highly divergent despite a common core, with pairwise RMSDs ranging from 2.1 to 6.3 Å and sequence identities of 6% to 32% (Table S3). Clustering analysis based on overall structural similarity (Z-score) reveals clusters corresponding to three Pfam families: P_gingi_FimA (PF06321), Mfa2 (PF08842), and DUF3988 (PF13149) (Figure 1G). The P_gingi_FimA family contains structural (e.g. FimA and Mfa1) and tip (e.g. Mfa4) pilins, while the Mfa2 family includes the anchor pilins, such as Mfa2 and FimB. FimA4 and BovFim3A are representatives of the two size variations observed in the P_gingi_FimA family, which can differ by ∼100-150 residues. Most prominently, the larger BovFim3A contains a helical insertion between CTD strands E2 and F2, resulting in increased length of the long axis of the pilin subunit (Figure S2). DUF3988 is a large family whose members contain a domain of unknown function (DUF) and was identified in the gut microbiome (Ellrott et al., 2010). Structure analysis indicates that this family is related to the other two Pfam families (Table S3), although the homology is challenging to detect at the sequence level.
P_gingi_FimA and DUF3988 family members often have a conserved N-terminal proteinase cleavage site (between [Ser/Thr][Lys/Arg] and a residue with a small side-chain) that is not present in anchor pilins (Figures 1G and S2), which instead often have an additional conserved cysteine ∼10 residues downstream of the characteristic cysteine of the lipoprotein signal peptide. The CTD appendage features are also highly conserved in the superfamily, with one or more glycines between strands G2 and A1′, and a tryptophan between A1′ and A2′.
Mature FimA structural pilin lacks the A1 strand
Unexpectedly, the proteinase cleavage site (between Arg and Ala) in the second stage of FimA maturation is in a flexible loop between the first and second β-strands (A1 and B1) (Figures 1D-E and S3). This flexible A1-B1 loop carrying a conserved proteinase cleavage site is a common feature of both P_gingi_FimA and DUF3988 family members (Figure S2). The cleavage site is positioned at the junction of an “L”-shaped A1-B1 loop, which protrudes into solvent, providing access to the incoming proteinase (Figure 1D). Interestingly, the NTD groove occupied by the A1 strand extends perfectly into a CTD groove that is occupied by the amphipathic A1′ strand. Together, the grooves form an extended surface that spans the entire length of the pilin (Figure 2A-B).
Figure 2. Structure of FimA highlighting the A1 and A1′ regions and pilus-like assemblies in the crystal lattices.
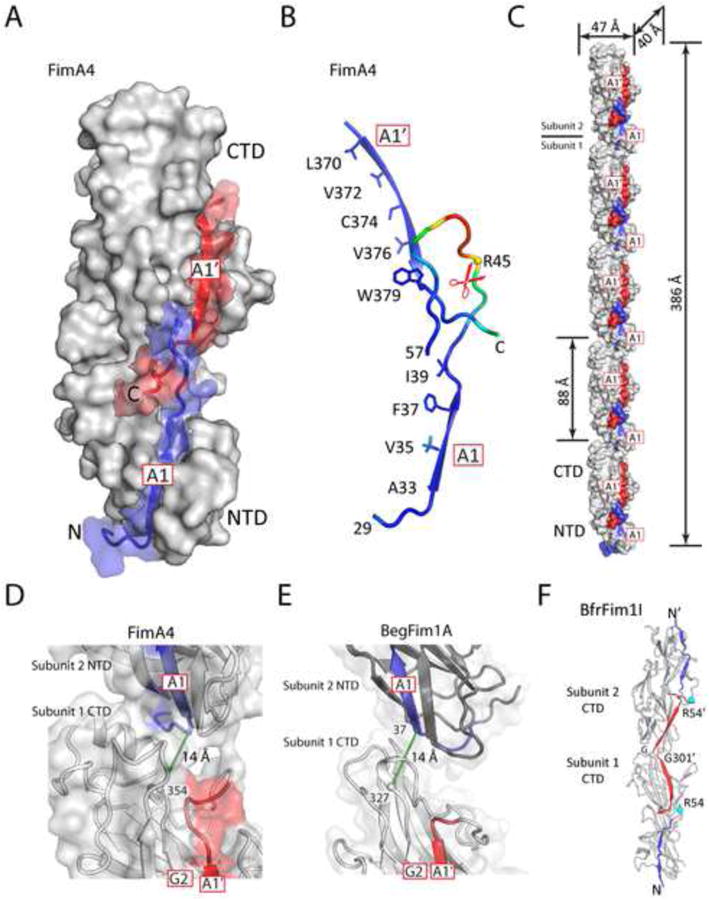
(A) Grooves for binding strands A1 (blue) and A1′ (red) align with one another and form a continuous groove, as shown here using FimA4 as an example.
(B) Arrangement of the two amphipathic strands A1, A1′, and the A1-B1 loop containing the cleavage site. Hydrophobic residues on the buried face of both strands are shown as sticks. The structure is colored by a B-value gradient from blue (low B-value) to red (high B-value).
(C) FimA4 packs in head-to-tail columns of subunits in the crystal. The A1 and A1′ regions are colored in blue and red.
(D-E) Close-up view of the interface between two subunits in one FimA4 (D) or BdiFim1A (E) column. Distances between a point near the C-terminus of G2 and a point near N-terminus of A1 of the adjacent pilin are shown as green dashed lines.
(F) An example of head-to-head crystallographic dimers formed by domain swapping of the C-terminal A1′ strand (red).
Pilus filaments are typically 40-50 Å in width (van Doorn et al., 1987; Yoshimura et al., 1984), which corresponds to the width of an individual pilin monomer (but ∼ ½ of its length). Thus, the pilin subunits must be assembled along their long axis, in a head-to-tail manner, to form the pilus filament. Our structural evidence suggests that the acceptor groove on the NTD is involved in pilus assembly, most likely providing a docking site for the C-terminal “donor” β-strands (A1′ and A2′) of the incoming pilin. To bind an adjacent “acceptor” subunit, the A1′ strand must switch from a configuration in which it is an integral part of the FimA CTD fold to one where it is extruded from the groove and no longer part of the β-sheet, as observed in BfrFim1I and BthFim1A (Figure 1F).
Pilus-like filaments in crystals
FimA4 crystallized in space group P21 with two molecules in the asymmetric unit. Strikingly, the molecules are stacked head-to-tail, forming pilus-like columns extending throughout the crystal (Figure 2C), where the G2 strand of one molecule aligns with the A1 strand of the next molecule (Figure 2D), consistent with our proposed mode of pilus assembly. A similar arrangement was observed in the BegFim1A crystal (Figure 2E) indicating some inherent affinity between pilin subunits at high concentrations, which would be further enhanced by the inter-subunit strand insertion.
In the BfrFim1l and BthFim1A crystals, we also observed tail-to-tail dimerization of two pilins through a domain-swapping mechanism (Figure 2F) that involves stacking of the transthyretin-like domains, but cannot accommodate a mechanism for pilus extension. These dimers are most likely crystallization artifacts, as the A1 strand groove of the prepilin is unavailable to the binding partner and size exclusion chromatography confirmed that BthFim1A is a monomer in solution.
The C-terminus of P. gingivalis FimA is critical for polymerization
The predicted C-terminal amphipathic strand A2′ contains a highly conserved hydrophobic face in P. gingivalis pilins (Figure 3A). To probe the roles of the C-terminal region in polymerization, we created a series of C-terminal truncation mutants of the mature FimA (Figures 3B and S3). Furthermore, we also generated a mutant that replaced the conserved tryptophan between A1′ and A2′ with an alanine (W328A). Plasmids containing the mutations were transformed into a P. gingivalis 33277 mutant strain in which fimA was disrupted, and pili production was analyzed by immunoblotting and electron microscopy. The WT and the knockout strain complemented the fimA produced pili that are characteristic of the native pili (Figure 3C-D). However, all C-terminal deletion mutants (as few as three amino acids) failed to produce pili. The W328A mutant could still produce pili that are similar to WT, but at a seemingly reduced level (Figure 3D). We also obtained similar results for additional truncation mutants that lack the C-terminal strands (A2′, or both A1′ and A2′) when tested for in vitro autopolymerization (Figure S3). Overall, these results support the critical role of the C-terminal region of FimA in polymerization.
Figure 3. The C-terminal region of FimA is critical for polymerization.
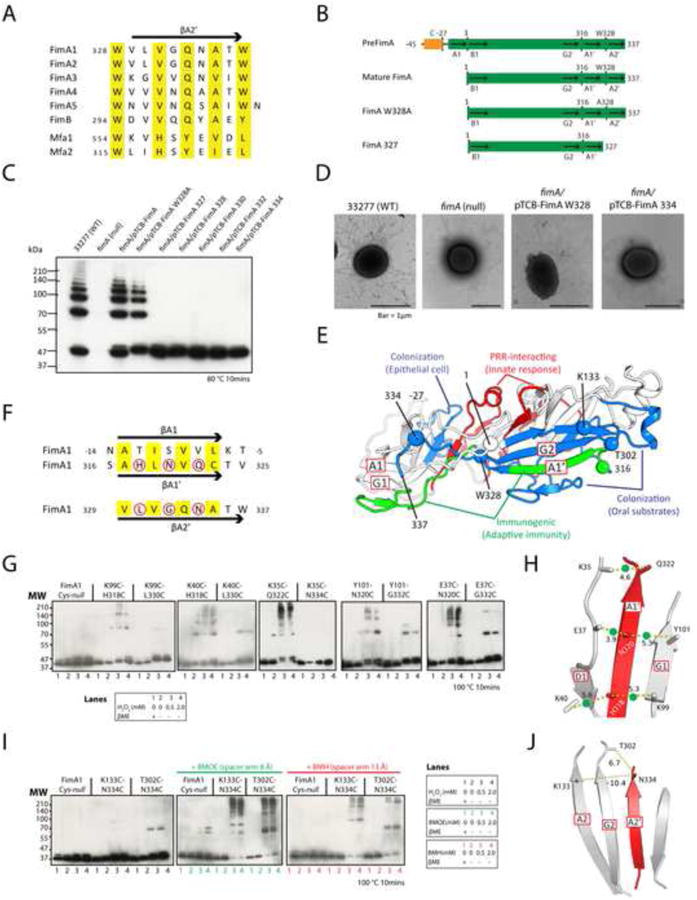
(A) Alignment of the C-terminal sequences for the A2′ strands of P. gingivalis FimA genotypes (FimA1-5), FimB (strain W83), Mfa1, and Mfa2. The sequence numbering of FimA is based on the mature pilin, whereas other proteins are numbered based on the prepilin forms.
(B) FimA mutants and their sequence features compared to pre- and mature FimA. PreFimA, prepilin with N-terminal signal peptide. Mature FimA, mature FimA pilin. FimA W328A, mutant of mature FimA. FimA327, mature FimA truncated after residue 327 (deletion of Trp328 and A2′).
(C) Immunoblot of FimA C-terminal mutants.
(D) EM micrographs of WT and fimA mutants.
(E) Functional mapping of adhesion and immune epitopes onto a homology model of FimA.
(F) Sequence comparison between the A1, A1′, and A2′ strands of the FimA pilin. Conserved residues that are predicted to be buried in the grooves are colored yellow. Surface residues mutated to cysteines are marked by red circles.
(G) Formation of disulfide bonds between cysteine pairs introduced on the A1′ strand and NTD, respectively. Lane 1: reducing reagent β ME, 2: no β ME or H2O2, 3, 4: oxidizing reagent H2O2 in two concentrations.
(H) A model of the A1′ strand replacing the A1 strand in the NTD. The Cβ-Cβ distances (Å) of Cys-Cys pairs are shown. The cross-linked Cys-Cys pairs observed are marked by green dots.
(I) Formation of cross-links between the CTD and A2′ strand.
(J) A model of the A2′ strand replacing the A1′ strand in the CTD.
As expected, FimA sequence variability between different P. gingivalis strains mostly map to the protein surface while the core is highly conserved, consistent with differences in immunogenic properties (Figure S4). Interactions between the major pili of P. gingivalis 33277 (including pilin FimA) and host molecules were studied extensively using peptide mapping (Hajishengallis, 2007). We mapped the adhesion and immune recognition sites onto a homology model of FimA, using FimA4 as a template (Figure 3E). Indeed, all known functional epitopes map to the FimA surface. The surfaces for binding substrates from the oral cavity (such as proline-rich protein 1 and statherin), extracellular matrix protein fibronectin, and other bacteria (such as early colonizer Streptococcus oralis) are clustered to a region on the CTD. The interacting regions for epithelial cells or pattern recognition receptors are both located near the NTD-CTD interface. The B cell epitopes are contributed by the G1-A2 insert and the A1′ strand, which are topographically separated on the prepilin (Figure 3E). However, the two regions would be co-located in our pilus assembly model where A1′ strand docks into the A1 groove on the NTD of the neighboring pilin. The buried faces of strands A1′ and A1 of FimA are highly conserved (Figure 3F), indicating that the A1 groove can easily accommodate the A1′ strand; in the alternative option, the sequence conservation of the A2′ strand is lower (Figure 3F) and less complementary to the A1 groove.
The FimA A1′ strand interacts with the NTD groove in the pilus assembly
We employed cysteine-based cross-linking experiments to experimentally determine if the donor C-terminal strand (A1′ or A2′) binds to the NTD groove of the acceptor pilin. First, we mutated all three cysteines (all buried) in the mature FimA to alanines (Cys-null) so that they would not interfere with cross-linking. Subsequently, we introduced a series of cysteine pairs: one cysteine was located at the edge of the NTD groove (on strand D1 or G1), and the other on the exposed surface of the putative donor strand (A1′ or A2′, Figure 3F). The fimA genes expressing various Cys-substituted FimAs were then introduced, one at a time, into a P. gingivalis mutant strain in which the genes expressing the WT FimA and Mfa1 were disrupted. Cys-null and cross-linking mutants generated pili that were observed by negative-stain electron microscopy (data not shown). Mutant pili display immunoblot ladder patterns similar to those observed for WT pili under similar conditions (sample incubated at 80°C for 10 mins). In the presence of reducing reagent β-mercaptoethanol (βME), WT or mutant dissociated into monomers when heated at 100°C for 10 minutes (Figure S3), indicating no disulfide bond formation under these conditions. However, in absence of βME or in presence of oxidizing reagent H2O2, higher molecular weight multimers, with a ladder migration pattern resembling that of WT pili, were clearly detected for mutants that contain a cysteine on the A1′ strand, but not for cysteine mutants of the A2′ strand (Figure 3G). Linked dimers (and a few oligomers) were detected for some A2′ mutants, but the nature of these oligomers is currently not clear. Overall, these results strongly support the notion that the A1′ strand binds the NTD groove in the pilus assembly (Figure 3H).
We then probed the interaction between the A2′ strand and CTD. Two mutants, T302CN334C and K133C-N334C, were selected such that the first residue is located on the CTD surface (Thr302 or Lys133) and the second residue (Asn334) is located on the A2′ strand (C α-C α distance in each pair >37 Å in the monomeric pilin) (Figure 3E). Both mutants did not directly form disulfide bridges in the pilus assembly, but could readily cross-link in presence of short-arm cross-linkers (Figure 3I), indicating A2′ is very likely associated with the CTD in the pilus assembly, presumably in the location occupied by the strand A1′ in the prepilins (Figure 3J).
Major and minor pili of P. gingivalis are homologous
The relationship between the various pilins in P. gingivalis was not previously detected due to very low pairwise sequence identities (<20%). Re-analysis using a highly sensitive profiled-based homology detection method (Soding et al., 2005) show now that all of the structural subunits of both types of P. gingivalis pili are homologous and adopt a conserved fold (Table S4). Additionally, we identified a new homolog (PGN1808) that is not part of major or minor pili loci and, although it is likely processed by Rgp, it does not form pili, but localizes to vesicles. (data not shown).
Due to conservation of the overall structure and maturation process (Shoji et al., 2004), we expect the assembly mechanism to be conserved in the minor pili of P. gingivalis. The mature pilin of Mfa1 also lacks the first strand of the NTD (Figure S5). Furthermore, the C-terminal regions of the minor pilins (Mfa1 and Mfa2) are also highly conserved, despite weak overall sequence similarity (Figure 3A). Indeed, we confirmed that deletion of three residues from the C-terminus of Mfa1 abolished its capability to polymerize (data not shown).
Structures of anchor pilins
Previous studies showed that anchor pilins (Mfa2 and FimB) of both minor and major pili from P. gingivalis are located at the pili base and regulate their length (Hasegawa et al., 2009; Nagano et al., 2010). The lack of a functional FimB in P. gingivalis 33277, due to a nonsense mutation, results in so-called “long” (i.e. major) pili that only weakly attach to the cell surface (Nagano et al., 2010). These results suggest that the pilus is assembled from tip-to-base with the anchor pilin incorporated last. We therefore determined structures of four anchor pilins, BovFim2B, BthFim2B, BthFim3B, and BovFim4B, all of which are more closely related in sequence to Mfa2 than to FimB.
The anchor pilin structures are more similar to each other compared to structural pilins (Figure 1G and Table S3) and contain unique structural features that differentiate them from the main structural pilins, such as Fim4A (Figure 4A-B). The A1-B1 loops of anchor pilins contain no potential Rgp or Kgp cleavage sites. Structurally, they are well-ordered and protected by a helical F2-G2 insert that protrudes on one side of the pilin, shielding it from any potential cleavage. The F2-G2 insert in FimA4 forms a parallel strand with A1′ so that A1′ is held in place and directed towards the A1-B1 loop. In contrast, the C-terminal “appendage” strands (A1′ and A2′) of anchor pilins either form a short hairpin extending the G2 sheet (Figure 4A) or adopt an “open” conformation (BovFim4B, Figure 4B). As a consequence, the conserved tryptophan between A1′ and A2′ is solvent exposed in anchor pilins, rather than being buried within the domain interface as in FimA4.
Figure 4. Structure and function of anchor pilins.
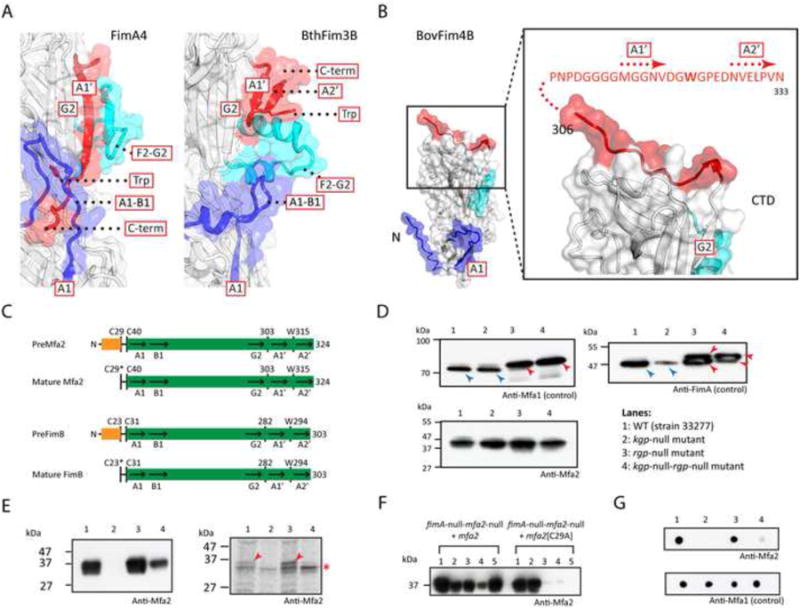
(A) Structures of anchor pilins (BthFim3B as example) differ significantly from structural pilins (FimA4 as example) in regions important for polymerization.
(B) The C-terminus of BovFim4B adopts an “open” conformation. The sequence of the appendage with the A1′ and A2′ strands and additional C-terminal disordered region is shown in red.
(C) Sequence features of anchor pilins of the minor and major pili, Mfa2 and FimB (see Figure 1B for caption).
(D) Immunoblot of cell lysates of P. gingivalis strains with anti-FimA, anti-Mfa1 and anti-Mfa2 antibodies. Precursor and mature forms of pilins are marked by red and blue arrows, respectively. In lanes 3 and 4, the bands correspond to previously identified precursor forms (Kadowaki et al., 1998; Shoji et al., 2004).
(E) Immunoblot with anti-Mfa2 antibody (left) and autoradiography of the SDS–PAGE gel (right) of the protein sample of P. gingivalis grown in the presence of C14-palmitic acid and immunoprecipitated with anti-Mfa2 antibody. Labeled Mfa2 are marked by red arrows (nonspecific bands marked by an asterisk). Lane 1: fimA-null, 2: fimA-null-mfa2-null, 3: fimA-null-mfa2-null complemented with mfa2+, 4: fimA-null-mfa2-null complemented with the mfa2[C29A] mutant.
(F) Fractionation analysis. Lane 1: whole cell lysate, 2: cytoplasm and periplasm, 3: total membrane fraction, 4: inner membrane (soluble fraction by Triton X-100), 5: OM (insoluble fraction by Triton X-100).
(G) Dot blot analysis with anti-Mfa2 antibody. Lanes as in (E).
Anchor pilins are cell-surface lipoproteins
The structures of anchor pilins therefore suggest they are not substrates for proteinases, such as the P. gingivalis arginine (Rgp) or lysine (Kgp) specific proteinase and, as a result, would remain as lipoproteins with their N-terminal cysteine lipidated. To test whether anchor pilin Mfa2 (Figure 4C) is processed by either Rgp or Kgp, we compared its state in P. gingivalis mutant strains in which Kgp or Rgp, or both, were knocked out. Mfa1 and FimA exist in prepilin forms with molecular weights higher than that of mature pilins in the absence of Rgp (Kadowaki et al., 1998; Shoji et al., 2004), whereas Mfa2 is found as a single state that is not dependent on proteinases (Figure 4D). To determine whether Mfa2 is modified by fatty acid, we grew P. gingivalis cells in the presence of C14-palmitic acid, and analyzed the results using immunoprecipitation with an anti-Mfa2 antibody and autoradiography. We constructed fimA-null (positive control) and fimA-null-mfa2-null (negative control) strains, and reintroduced mfa2+ or mfa2[C29A] on a mobilizable pTCB plasmid. Mfa2 was radiolabelled by C14-palmitic acid in fimA-null and fimA-null-mfa2-null/mfa2+ strains, but not in fimA-null-mfa2-null and fimA-null-mfa2-null/mfa2[C29A] strains, indicating that Cys29 of Mfa2 was lipidated (Figure 4E). Mfa2 was detected in the OM fraction and a dot blot analysis indicated that it was located on the cell surface, while the C29A mutant was located in predominantly in cytoplasmic or periplasmic fractions (Figure 4F-G). Similar results were obtained when we complemented P. gingivalis 33277 (fimB-null) with fimB+ from strain W83 (Figure S6). Overall, these results support the conclusion that Mfa2 and FimB are both exported to cell surface as lipoproteins, but do not undergo the second proteolytic step for maturation of the structural pilins (Figure 4C).
The C-terminus of the anchor pilin is necessary for its incorporation into pili
The C-termini of the P. gingivalis anchor pilins are highly conserved (Figures 3A and S5). To assess the role of the C-terminus of Mfa2 in Mfa1assembly (i.e. minor pili), we disrupted the mfa2 gene of P. gingivalis 33277 and then restored mfa2+ or C-terminal truncation mutants with a plasmid (Figure 5A). The expression of fimA was also disrupted in all mutants so that observation of Mfa1 pili is not obscured by FimA1 pili. Three C-terminal truncation mutants were constructed: C-terminal deletion of three residues (1-321), C-terminal deletion of A2′ (1-314), and C-terminal deletion of both A1′ and A2′ (1-303). Expression was confirmed by the Mfa2 antibody against cell lysate, except for the mfa2-null mutant (Figure 5B). Mfa2 proteins were all exported to the cell surface, except for the C29A mutant (Figure 5C). However, surface presence of the two-strand deletion mutant (1-303) appears to be reduced compared to the others, suggesting that deletion of the A1′ region may impact transmembrane export and/or structure integrity. Consistent with previous results (Hasegawa et al., 2009), 3-4 times longer Mfa1 pili are produced by mutants without Mfa2, compared to WT (Figure 5D-E). Furthermore, the average pili length of all truncation mutants are comparable to the mfa2-null mutants, indicating the Mfa2 mutants are not incorporated into Mfa1 pili, consistent with immunoprecipitation analysis that showed that Mfa1 and Mfa2 do not interact in absence of the C-terminus of Mfa2 (Figure 5F). Similar results were obtained when we restored fimB+ (from strain W83) or its C-terminal truncation mutants into P. gingivalis 33277 without mfa1 (data not shown). These data support the conclusion that the C-termini of anchor pilins are critical for their incorporation into pili.
Figure 5. The C-terminus of Mfa2 is essential for its incorporation into the Mfa1 pili.
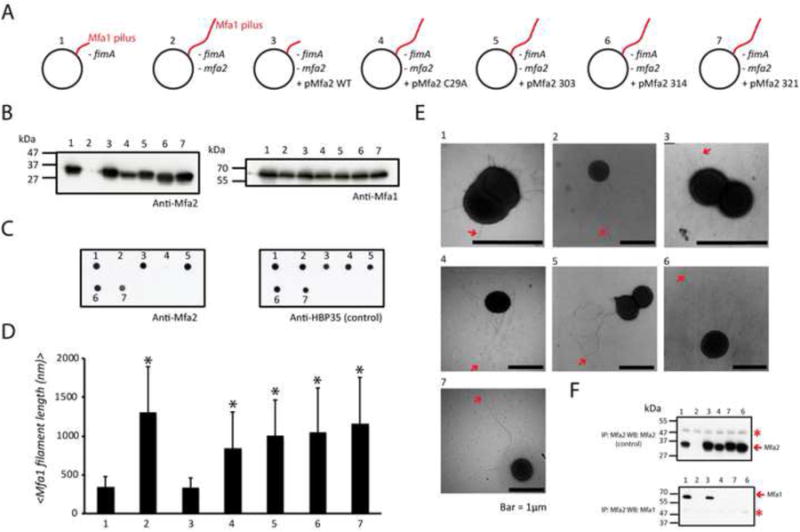
(A) fimA-null mutants constructed to study the impact of truncating the C-terminus of Mfa2 on the length of the Mfa1 pili. 1: fimA-null of P. gingivalis 33277, 2: mfa2-null of the mutant 1, 3: Mutant 2 complemented with mfa2+ on a plasmid, 4: Mutant 2 complemented with mfa2[C29A] on a plasmid, and 5-7: Mutant 2 complemented C-terminal truncated of mfa2 (residues 1-303, 1-314 or 1-321).
(B) Expression of Mfa2 and Mfa1 in mutants 1-7 detected by antibodies.
(C) Cell surface presence of Mfa2 analyzed by dot blots. Cell surface protein HBP35 was used as a control.
(D) Average length (nm) of the Mfa1 pili in mutants 1-7. Error bars represent the standard deviation. * P<0.001.
(E) Representative EM images of mutants 1-7 (pili are marked by arrows).
(F) Mfa1 and Mfa2 interactions in mutants analyzed by western blots (Mfa2-C29A does not interact with Mfa1 since it cannot reach the OM). Asterisk, non-specific bands.
Structures of tip pilins
Mfa4 and BovFim1C are two representative tip pilins. Mfa4 contains no additional C-terminal domain beyond the assembly component (Figure 6A), while BovFim1C (Figure 6B) is a concatenation of a conserved pilus assembly component and a C-terminal C-type lectin domain (CTLD), which is a predominant metazoan protein module also found in bacterial and virus invasins (Zelensky and Gready, 2005). Correspondingly, Mfa4 does not contain C-terminal A1′ and A2′ strands (Figure 6A), which is consistent with Mfa4 being located at the tip of Mfa pili (Hasegawa et al., 2013; Nagano et al., 2015) where these strands are no longer necessary for assembly. In contrast, in BovFim1C, the A1′ and A2′ strands of the assembly domain adopt an extended conformation, with the A1′ strand augmenting a β-hairpin insert (strands A3 and B3) between CTD strands A2 and B2 and the A2′ strand extending an edge-strand of the CTLD (Figure 6B). Thus, BovFim1C t uses its donor strands to interact with the CTLD. The lipoprotein signal peptide and a conserved proteinase cleavage site in the disordered A1-B1 loop region is present in both Mfa4 and BovFim1C, suggesting they undergo the same proteolytic processing as the main structural pilins. However, the C-terminus of BovFim1C is buried inside the CTLD, which thereby caps the structure and produces a surface cavity akin to a ligand binding site (Figure 6C).
Figure 6. Representative structures of tip pilins.
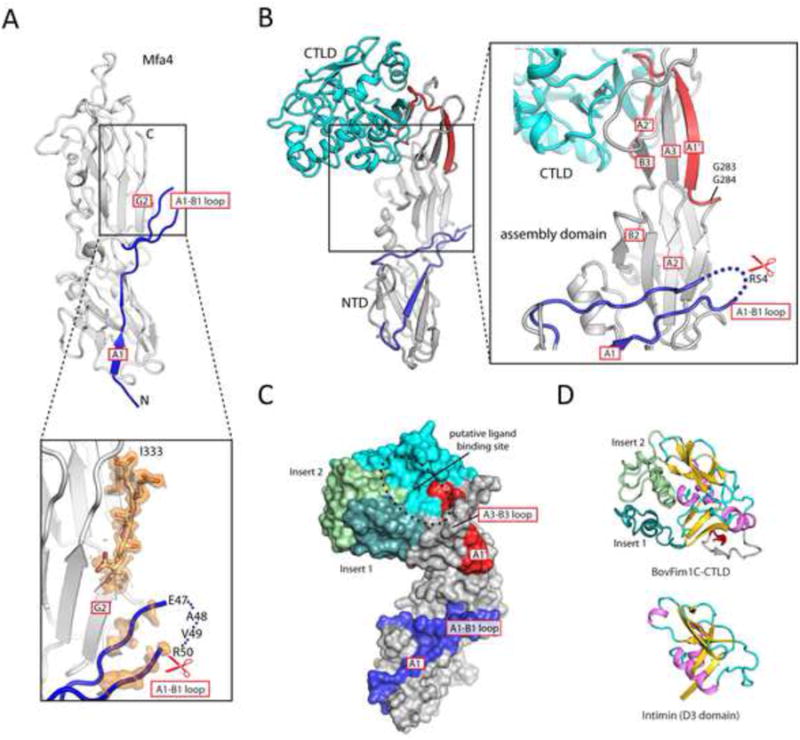
(A) Crystal structure of the tip pilin Mfa4. The unbiased experimental electron density map (solvent modified) of the A1-B1 loop and the C-terminus are shown (contoured at 1 σ, orange).
(B) The structure of BovFim1C consists of a prototypical pilus assembly domain (gray/blue/red) connected to a C-terminal CTLD (cyan).
(C) Surface representation of BovFim1C color coded as in B with inserts in green.
(D) Structural comparison of the CTLD domain of BovFim1C with another bacterial CTLD. The common core regions are colored in violet/gold/cyan.
The relationship of the BovFim1C CTLD to other CTLDs can only be identified by structure-based similarity searches due to paucity of sequence similarity. The tip pilin CTLD possesses two long insertions that are absent in prototypical CTLDs, such as the D3 domain of intimin (Figure 6D). These insertions, together with the A3-B3 and A1′-A2′ loops, form a putative ligand binding cavity. When these two insertions are removed, the CTLD of BovFim1C is clearly related to DUF1566 and Fib_succ_major protein families, either of which are often found fused to the C-terminus of a DUF3988 domain. These newly identified bacterial CTLDs are widely distributed and may have diverse roles.
Discussion
Assembly mechanism of Type V pili
Based on the above results, we propose a proteinase-mediated donor strand exchange mechanism for type V pilus assembly (Figure 7A-C). In the prepilin conformation, two intramolecular peptide segments (strands A1 and A1′) occupy hydrophobic grooves that extend from the NTD to the CTD (Figures 1E and 2A). Cleavage of the pilus subunit during the maturation process cleaves off the A1 strand and enables release of the A1′ strand and the rest of the C-terminal appendage so that it can adopt an extended conformation. During pilus assembly, the extended surface groove created by the cleavage and maturation event can now be filled by the extended C-terminal appendage of the incoming subunit, where the A1′ strand of the CTD of the assembling subunit would dock into the NTD groove of the terminal subunit to re-form its seven-stranded β-sheet core, and the A2′ strand would dock into the CTD groove (parallel to G2) that was previously occupied by its own A1′ and A2′ strands (Figure 7A-B). A highly conserved tryptophan residue between strands A1′ and A2′ would occupy the same position in the domain interface in the mature pilin, and may facilitate correct alignment of the two donor strands into the two grooves during assembly. In addition, some pilins contain additional structural features that likely enhance the interaction between the donor strands and the grooves. For example, two hairpin inserts in FimA4, contributed by loops G1-A2 and F2-G2 respectively (Figure 1D), could help to further enclose the groove and provide additional contacts to the donor strands, thereby clamping them in place. Molecular modeling suggests that a twist of ∼90° between two consecutive subunits is necessary to maximize surface complementarity between FimA pilin subunits, in order to generate a filament with a right-handed twist (∼4.5 monomers per turn), consistent with previous observations (Shoji et al., 2010b) (Figure 7B).
Figure 7. A proposed assembly mechanism for type V pili.
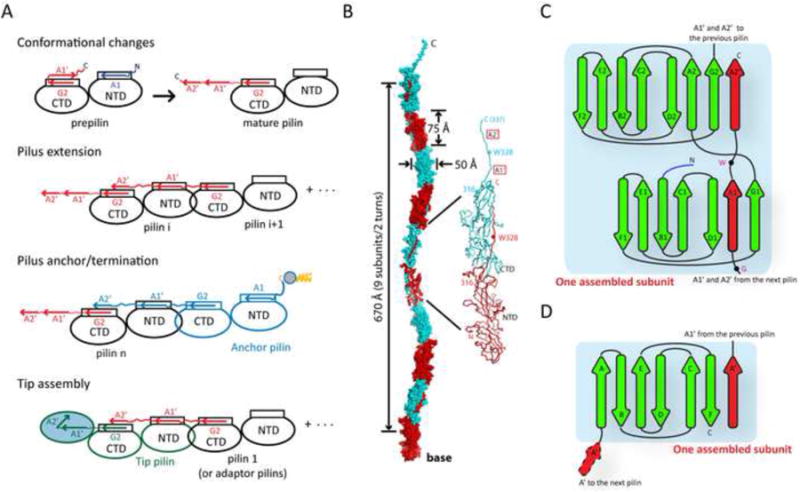
(A) A schematic model for pilus assembly.
(B) Proposed atomic model of a FimA pilus filament. FimA subunits are colored alternately in red and cyan.
(C-D) Topological diagram of one assembled subunit in a type V pilus assembly (C) compared to that of type I pilus (D). Donor strands are colored red.
The above mechanism accounts for the pilus elongation pilus but specialized pili are required to cap the pilus and to insert it into the bacterial membrane. The tip assembly is a manifestation of the same principle involved in pilin assembly. However, the C-terminal donor strands are now unavailable (absent or secluded by the CTLD domain), while its own assembly domain provides an accepting groove (Figure 7A). In some cases, it appears that additional minor pilins may connect the tip and the stalk (Hasegawa et al., 2013). Anchor pilins, on the other hand, contain no potential Rgp or Kgp cleavage site within the A1-B1 loop and, as a result, are not substrates for pilin maturation proteinases; therefore, the A1 strand remains anchored in the mature anchor pilin. Consequently, anchor pilins lack a groove for binding the donor strand, which would prevent further pilus extension. The donor strand region in the CTD C-terminus is still present, enabling donation of the A1′ and A2′ strands to the upstream pilin. Membrane attachment of the terminal anchor pilin is likely mediated via lipidation of the N-terminal cysteine, and potential association with other proteins. Thus, our analysis of a number and variety of the component pilin structures combined with biochemical results have enabled elucidation of a complete model for assembly of the type V pilus (Figures 7A and S1).
The exact timing of the conformational changes in the A1′ strand and the release (or displacement) of A1 strand from NTD remain to be elucidated. Interestingly, the C-termini of the P_gingi_FimA family prepilins are often sequestered beneath the A1-B1 loop and, in some cases, are actually threaded through the loop, with the conserved tryptophan between A1′ and A2′ in close proximity to the cleavage site (Figures 2B and S7). Thus, the cleavage of the A1 segment by Rgp or Kgp and conformational changes of A1′ are likely coupled and could possibly be influenced by additional factors. However, conformational changes in A1′ can occur spontaneously in solution since purified mature pilin can polymerize, but only into short filaments (Shoji et al., 2010b). In contrast, the C-termini of DUF3988 members and anchor pilins are not secured by the A1-B1 loop (Figure S2), which could explain the extended A1′ conformation observed in some of our crystal structures. For anchor pilins, only a conformational change of donor strands is needed, consistent with a different conformation of the A2′ region and conserved tryptophan (Figure 4A).
Relationship to other types of pili
Structural relationship of this pilin fold compared to other pilins was analyzed previously (Xu et al., 2010); the type V and type I pilins are not evolutionarily related. Pilins with similar domain architecture to FimA, but unrelated in sequence, have only been observed previously in Gram-positive bacteria (Kang et al., 2007; Krishnan et al., 2007). Nonetheless, our proposed mechanism of pilus assembly is analogous to the donor-strand exchange mechanism in the type I pilus system (Allen et al., 2012), where the prepilin has an incomplete IgG fold lacking the C-terminal G strand, but possesses an extended N-terminus with a propensity of forming a β-strand (A′ strand; Figure 7D). The donor strand of type I pilin is contributed by this N-terminal extension, which completes the Ig fold of the incoming pilin. Thus, the elements involved in the strand exchange are swapped in these pilins (Figure 7C-D). Interestingly, this “swap” is also observed at the fold level, as there is a largely inverse relationship between the transthyretin-like fold and the IgG fold on a topological level (Deivanayagam et al., 2000). Thus, our results suggest that the type V pilus represents a “hybrid” that resembles the Gram-positive pilus at the fold and inter-subunit arrangement level, and the Gram-negative type I pilus at the assembly mechanism level.
Prevalence of the FimA superfamily in the gut microbiome
The FimA superfamily currently consist of >1800 unique members, which are found predominately in the Bacteroidetes phylum and especially in the Bacteroidia class. Proteins in this superfamily are extremely divergent, and sequence homology is often only detectable using profile-profile based methods (Soding et al., 2005). To better characterize this diverse family, we systematically identified FimA superfamily members in representative bacteria with completed genome sequences, by searching profiles of proteins from each bacterium against the profiles of proteins with structures determined in this study (Table S5). Our results indicate that FimA superfamily of proteins is highly abundant in bacterial species in the gut microbiome, often making up ∼1-2.5% of a bacterial proteome (Figure S7). The genes encoding FimA superfamily proteins are often clustered in Bacteroides genomes as putative pilus biogenesis operons (Table S5), which are located in the neighborhood of transposases and integrases, a sign of horizontal gene transfer.
Our structural studies indicate the FimA superfamily arises from a common core that has diverged mainly through changes in variable loops between core β-strands. DUF3988 members generally have more prototypical structures with less sophisticated loop decorations (Figure S2), and are more widely distributed (e.g. 49 DUF3988 members in Spirochaeta coccoides, but none from Mfa2 or P_gingi_FimA families). Genes encoding DUF3988 ORFs often are not clustered in the genome in the same way as those encoding pilus components, e.g. in S. coccoides and Prevotella dentalis (Table S5). Instead, many of the genes are likely a part of polysaccharide utilization loci (e.g. 19 DUF3988 members from B. ovatus, Table S5). Furthermore, some members of DUF3988 exhibit more complex domain architectures, where additional C-terminal domains may contribute other functions as well as pathogenesis (Figure S7). Overall, these results suggest that DUF3988 family may have other functions beyond pilus, such as carbohydrate utilization, while Mfa2 and P_gingi_FimA families perform more specialized functions.
Our structural analysis indicates that this common pilin fold can give rise to extreme divergence, allowing it to adapt to different binding partners or other functional roles, reminiscent of the immunoglobulin fold of antibodies as a structural platform for recognition of diverse antigens. Thus, the FimA superfamily likely represents an important class of molecules that are involved in both symbiotic relationships and pathogenesis in the human microbiome. The pilus assembly mechanism uncovered here for Bacteroidia is distint from other known pili and may represent a more general mechanism in bacteria for assembling molecular probes that extend out to interact with their environment. This information can now be used to understand host-microbe interactions and for structure-based drug design and development of vaccines against pathogenic bacteria that colonize humans.
Experimental Procedures
Crystallography
The cloning, expression, crystallization, and structure determination of each protein are described in detail in the Supplemental Information. Briefly, the selenomethionine derivative of each protein was expressed in E. coli with an N-terminal, TEV-cleavable, His-tag and purified by metal affinity chromatography. Crystals were obtained using the JCSG HTP crystallization pipeline and were screened for diffraction to identify the best crystals for structure determination. MAD or SAD data were collected at SSRL or ALS, and structures were solved, refined and deposited into PDB. See Table S1 for summary of the crystal structures presented in the studies, and Table S2 for summary of the crystallization conditions, data collection, processing, and refinement statistics.
Bacterial strains and plasmids
The bacterial strains, plasmids and primers used in this study are listed in Table S6. Construction of recombinant strains is described in the Supplemental Information.
Gel electrophoresis and immunoblot analysis
SDS-PAGE and immunoblot analysis were performed as described previously (Shoji et al., 2004; Shoji et al., 2010a).
Cys-Cys cross-linking
To detect the Cys-Cys disulfide bridges of mutated FimA protein, cell pellets were collected then gently suspended water including 0, 0.5, and 2.0 mM of H2O2 and incubated at room temperature for 1 h. Cell pellets were then collected, and resuspended in PBS, and then mixed with SDS sample buffer with or without βME. All samples were incubated at 100 °C for 10 min. Further details are provided in Supplemental Information.
Labelling of P. gingivalis cells with [14C]-palmitic acid
P. gingivalis cells were incubated for 24 h in enriched 5 ml BHI medium containing [1-14C]-palmitic acid. Cells were harvested, washed with PBS buffer and dissolved in BugBuster protein extraction reagent. The samples were then immunoprecipitated with protein G agarose beads with anti-Mfa2 or anti-FimB antiserum. The resulting precipitates were dissolved in Laemmli sample buffer and analyzed by SDS-PAGE. Radiolabelled lipoproteins were detected with a fluoro-image analyzer. Further details are provided in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank the other members of the JCSG for their contribution to this work. This work was supported by the NIH NIGMS Protein Structure Initiative (U54 GM094586 to IAW) and Ministry of Education, Culture, Sports, Science and Technology of Japan (24117006 and 25293375 to KN). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Accession Numbers: The coordinates and structure factors for all pilin structures have been deposited in the PDB with accession numbers listed in Figure 1C.
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, 8 figures and 6 tables, and can be found with this article online at http://dx.doi.org/….
Author Contributions: Conceived and designed the experiments: QX, MS, AMD, AG, MAE, SAL, KN, and IAW. Performed the experiments and analyzed the data: QX and MS. Contributed reagents, materials, or analysis tools: SS, MN, KS, JG, HLA, HJC, CLF, LJ, MWK, and MAC. Analyzed results and wrote or edited the paper: QX, MS, MAE, AMD, KN, and IAW.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen WJ, Phan G, Waksman G. Pilus biogenesis at the outer membrane of Gram-negative bacterial pathogens. Curr Opin Struct Biol. 2012;22:500–506. doi: 10.1016/j.sbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Amano A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003;74:90–96. doi: 10.1902/jop.2003.74.1.90. [DOI] [PubMed] [Google Scholar]
- Amano A. Bacterial adhesins to host components in periodontitis. Periodontol. 2010;200052:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Hook M, Narayana SV. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- Ellrott K, Jaroszewski L, Li W, Wooley JC, Godzik A. Expansion of the protein repertoire in newly explored environments: human gut microbiome specific protein families. PLoS Comput Biol. 2010;6:e1000798. doi: 10.1371/journal.pcbi.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wooley J, Wuthrich K, Wilson IA. The JCSG high-throughput structural biology pipeline. Acta Crystallogr F Struct Biol Cryst Commun. 2010;66:1137–1142. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Peptide Mapping of a Functionally Versatile Fimbrial Adhesin from Porphyromonas gingivalis. Int J Pept Res Ther. 2007;13:533–546. [Google Scholar]
- Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359–2367. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci USA. 2008;105:13532–13537. doi: 10.1073/pnas.0803852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Sojar HT, Cho MI, Genco RJ. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect Immun. 1996;64:4788–4794. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Iwami J, Sato K, Park Y, Nishikawa K, Atsumi T, Moriguchi K, Murakami Y, Lamont RJ, Nakamura H, et al. Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology. 2009;155:3333–3347. doi: 10.1099/mic.0.028928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Nagano K, Ikai R, Izumigawa M, Yoshida Y, Kitai N, Lamont RJ, Murakami Y, Yoshimura F. Localization and function of the accessory protein Mfa3 in Porphyromonas gingivalis Mfa1 fimbriae. Mol Oral Microbiol. 2013;28:467–480. doi: 10.1111/omi.12040. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2005;200038:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Nakayama K, Yoshimura F, Okamoto K, Abe N, Yamamoto K. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J Biol Chem. 1998;273:29072–29076. doi: 10.1074/jbc.273.44.29072. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Poole S, Nishio K, Jang K, Rasulova F, McVeigh A, Savarino SJ, Xia D, Bullitt E. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 2009;106:10793–10798. doi: 10.1073/pnas.0812843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Hasegawa Y, Murakami Y, Nishiyama S, Yoshimura F. FimB regulates FimA fimbriation in Porphyromonas gingivalis. J Dent Res. 2010;89:903–908. doi: 10.1177/0022034510370089. [DOI] [PubMed] [Google Scholar]
- Nagano K, Hasegawa Y, Yoshida Y, Yoshimura F. A Major Fimbrilin Variant of Mfa1 Fimbriae in Porphyromonas gingivalis. J Dent Res. 2015;94:1143–1148. doi: 10.1177/0022034515588275. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- Shoji M, Naito M, Yukitake H, Sato K, Sakai E, Ohara N, Nakayama K. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol Microbiol. 2004;52:1513–1525. doi: 10.1111/j.1365-2958.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- Shoji M, Shibata Y, Shiroza T, Yukitake H, Peng B, Chen YY, Sato K, Naito M, Abiko Y, Reynolds EC, et al. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010a;10:152. doi: 10.1186/1471-2180-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Yoshimura A, Yoshioka H, Takade A, Takuma Y, Yukitake H, Naito M, Hara Y, Yoshida S, Nakayama K. Recombinant Porphyromonas gingivalis FimA preproprotein expressed in Escherichia coli is lipidated and the mature or processed recombinant FimA protein forms a short filament in vitro. Can J Microbiol. 2010b;56:959–967. doi: 10.1139/w10-084. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojar HT, Lee JY, Bedi GS, Cho MI, Genco RJ. Purification, characterization and immunolocalization of fimbrial protein from Porphyromonas (bacteroides) gingivalis. Biochem Biophys Res Commun. 1991;175:713–719. doi: 10.1016/0006-291x(91)91624-l. [DOI] [PubMed] [Google Scholar]
- van Doorn J, Mooi FR, Verweij-van Vught AM, MacLaren DM. Characterization of fimbriae from Bacteroides fragilis. Microb Pathog. 1987;3:87–95. doi: 10.1016/0882-4010(87)90067-2. [DOI] [PubMed] [Google Scholar]
- van Doorn J, Oudega B, MacLaren DM. Characterization and detection of the 40 kDa fimbrial subunit of Bacteroides fragilis BE1. Microb Pathog. 1992;13:75–79. doi: 10.1016/0882-4010(92)90033-k. [DOI] [PubMed] [Google Scholar]
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, et al. A conserved fold for fimbrial components revealed by the crystal structure of a putative fimbrial assembly protein (BT1062) from Bacteroides thetaiotaomicron at 2.2 Å resolution. Acta Crystallogr F Struct Biol Cryst Commun. 2010;66:1281–1286. doi: 10.1107/S1744309110006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F, Takahashi K, Nodasaka Y, Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J Bacteriol. 1984;160:949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


