Abstract
All living organisms are constantly exposed to stresses from internal biological processes and surrounding environments, which induce many adaptive changes in cellular physiology and gene expression programs. Unexpectedly, constitutive heterochromatin, which is generally associated with the stable maintenance of gene silencing, is also dynamically regulated in response to stimuli. In this review we will discuss the mechanism of constitutive heterochromatin assembly, its dynamic nature, and its responses to environmental changes.
Keywords: constitutive heterochromatin, dynamics, stress, epigenetic adaptation
Constitutive heterochromatin is traditionally viewed as static chromatin structures
In eukaryotes, genomic DNA wraps around histones to form chromatin. According to its compaction levels, chromatin is classified into two categories: gene-rich, less condensed euchromatin and gene-poor, highly condensed heterochromatin. Among heterochromatin regions, facultative heterochromatin often forms at developmentally regulated genes and its level of compaction changes in response to developmental cues and/or environmental signals [1]. In contrast, constitutive heterochromatin preferentially assembles at repetitive elements such as satellite DNA and transposons and maintains high compaction levels [2]. The stability of constitutive heterochromatin is exemplified by the classical position effect variegation (PEV) in Drosophila, in which the white gene is variably silenced when a chromosome translocation event places it adjacent to pericentric heterochromatin [3]. Remarkably, once a founder cell establishes heterochromatin at the white gene during early stages of development, silencing is maintained in all of the derivative cells through adulthood, resulting in patches of cells without pigmentation in the adult eye [4]. Such stability is essential for repressing recombination between repeat elements and limiting transcription of active transposons to maintain genome integrity. However, recent studies show that constitutive heterochromatin is also dynamically regulated and responsive to stimuli. While these changes could potentially help organisms adapt to new environments, in certain cases they also could cause human diseases. In the following sections, we will highlight a few examples of the dynamic regulation of constitutive heterochromatin domains.
Mechanism of heterochromatin assembly
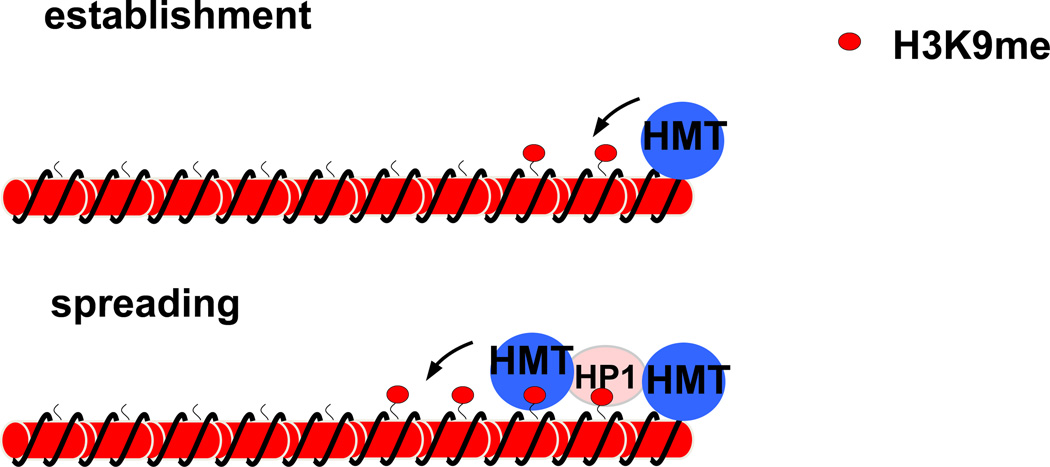
Constitutive heterochromatin harbors distinct chromatin modification profiles. For example, in constitutive heterochromatic regions histones are generally hypo-acetylated and hyper-methylated at H3 lysine 9 (H3K9me) [5–7]. The formation of these heterochromatin domains requires concerted actions of chromatin-modifying enzymes and is distinguished into three steps: initiation, spreading, and maintenance [8] (Fig. 1). Heterochromatin is initiated at nucleation centers by sequence-specific DNA binding proteins or non-coding RNAs, both of which recruit histone deacetylases (HDACs) and the SUV39 family histone H3K9 methyltransferases (HMTs), resulting in hypo-acetylation of histones and hyper-methylation of H3K9 at the nucleation sites [9–12]. These methyltransferases often contain a chromodomain that binds to existing H3K9me, forming a self-propagating mechanism to methylate adjacent nucleosomes [13, 14]. Methylated H3K9 also binds heterochromatin protein HP1, which in turn serves as a scaffold to further recruit chromatin modifiers, including H3K9 methyltransferases and histone deacetylases [15–19]. These combined actions lead to the spreading of heterochromatin along a large domain of chromatin in a DNA sequence-independent manner [8, 14, 20–22]. During DNA replication, parental histones, which contain existing modifications, are randomly incorporated into both daughter strands behind the replication fork [23]. H3K9me-mediated recruitment of H3K9 methyltransferases restores heterochromatin domains in both daughter strands, leading to the stable maintenance of this epigenetic state through generations [20, 24–26].
Figure 1.
A step-wise model of heterochromatin assembly. Histone H3K9 methyltransferases (HMTs) are first recruited to heterochromatin nucleation centers, leading to H3K9 methylation (red dots associated with the chromosome), which is subsequently recognized and bound by heterochromatin protein 1 (HP1). Additional HMTases are recruited either directly through recognizing H3K9me or association with HP1, leading to the methylation of adjacent nucleosomes. The repetition of such binding-methylation cycles results in heterochromatin spreading in a sequence-independent manner.
The budding yeast Saccharomyces cerevisiae lacks the H3K9 methylation-HP1 system, but assembles functional heterochromatin using the silent information regulator (SIR) complex, composed of Sir2, Sir3, and Sir4 [27, 28]. Sir2 is a histone deacetylase with activity mainly towards H4K16 [29–31], the acetylation status of which directly regulates higher-order chromatin folding in vitro [32] and plays a major role in heterochromatin function in vivo [6, 33]. Sir3 and Sir4 preferentially interact with histone tails devoid of H4K16ac [34–36]. Heterochromatin formation starts with the recruitment of Sir2 by factors that recognize specific DNA sequences. Sir2 subsequently deacetylates histone H4K16, allowing Sir3 and Sir4 to bind. Sir3 oligomerizes and recruits more Sir2 to deacetylate H4K16 of the adjacent nucleosomes and thus facilitates the spread of the entire Sir complex [27, 28].
Even though the protein factors involved in heterochromatin assembly in many organisms are diverse, similar mechanisms of self-propagation underlie heterochromatin spreading. The step-wise spreading model is supported by the fact that heterochromatin assembly factors cover the entire heterochromatin domain, the distance of spreading is sensitive to the dosage of heterochromatin proteins, and silencing spreads continuously [5, 20, 37–41]. However, heterochromatin sometimes skips certain genomic regions, suggesting additional mechanisms, such as looping, also aid heterochromatin spreading [21, 42–44].
The dynamics of HP1
Originally identified in Drosophila, HP1 belongs to a highly conserved family of chromatin proteins, with homologues found from fission yeast (Swi6 and Chp2) to humans (HP1α, HP1β, and HP1γ) [45]. While the majority of these HP1 proteins are localized to heterochromatin, some isoforms have diverged from heterochromatin functions. For example, human HP1α and HP1β are distributed mainly at pericentric heterochromatin, and HP1γ is localized to discrete euchromatic regions. HP1 proteins are composed of a chromodomain, a chromo shadow domain, and a flexible hinge region [45, 46]. The chromodomain binds H3K9me [15, 16], while the chromo shadow domain mediates dimerization of HP1 [47]. These interactions result in the formation of an HP1 protein network that locks up chromatin in a highly compacted state [48]. Moreover, HP1 proteins recruit a diverse range of factors to further modify heterochromatin [2].
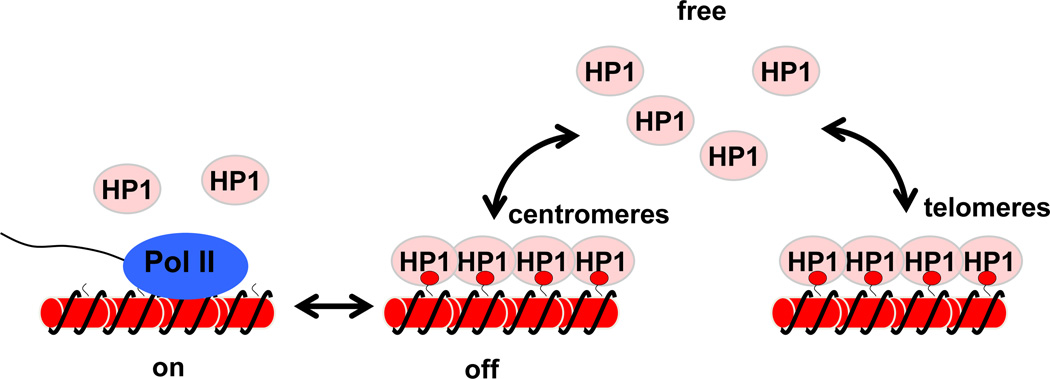
Given its essential structural roles in heterochromatin formation, HP1 is expected to stably associate with heterochromatin domains. However, Fluorescence Recovery After Photobleaching (FRAP) analyses of all three HP1 isoforms in human cells and Swi6 in fission yeast illustrate that binding of HP1 to chromatin is very dynamic, exchanging rapidly between chromatin bound and nucleoplasm forms and also among different heterochromatin domains [49–52] (Fig. 2). Interestingly, the dynamics of HP1 proteins also change during cell differentiation. For example, human HP1β is more mobile in embryonic stem cells and induced pluripotent stem cells compared to fibroblasts. Similarly, mouse embryonic stem cells are also characterized by higher mobility of HP1 proteins [53, 54]. Such differences suggest the existence of regulatory mechanisms for HP1 dynamics, although the molecular basis of such dynamic behavior remains unclear. It is also possible that the increased HP1 mobility in embryonic stem cells is the indirect result of less compact chromatin structure in these cells.
Figure 2.
Dynamics of HP1 proteins. Contrary to the general conception that HP1 is a static component of heterochromatin, it dynamically exchanges between the heterochromatin-bound and free forms, which gives other factors access to the underlying DNA, such as RNA Pol II shown here. HP1 also exchanges rapidly between heterochromatin domains, such as centromeres and telomeres, to buffer changes in heterochromatin stability.
The dynamic nature of HP1 protein binding provides windows of opportunity for other factors to access the underlying DNA. For example, heterochromatin-mediated gene silencing in yeast frequently changes between “on” and “off’ states, which might be a result of the dynamic binding of heterochromatin proteins [55, 56]. HP1 dynamics might also allow cells to maintain the balance between different heterochromatin domains. For example, in fission yeast, disruption of telomeric heterochromatin releases limiting heterochromatin factors such as Swi6, which can restore defective pericentric heterochromatin due to the loss of RNAi components (see below)[57].
Transcription and heterochromatin assembly
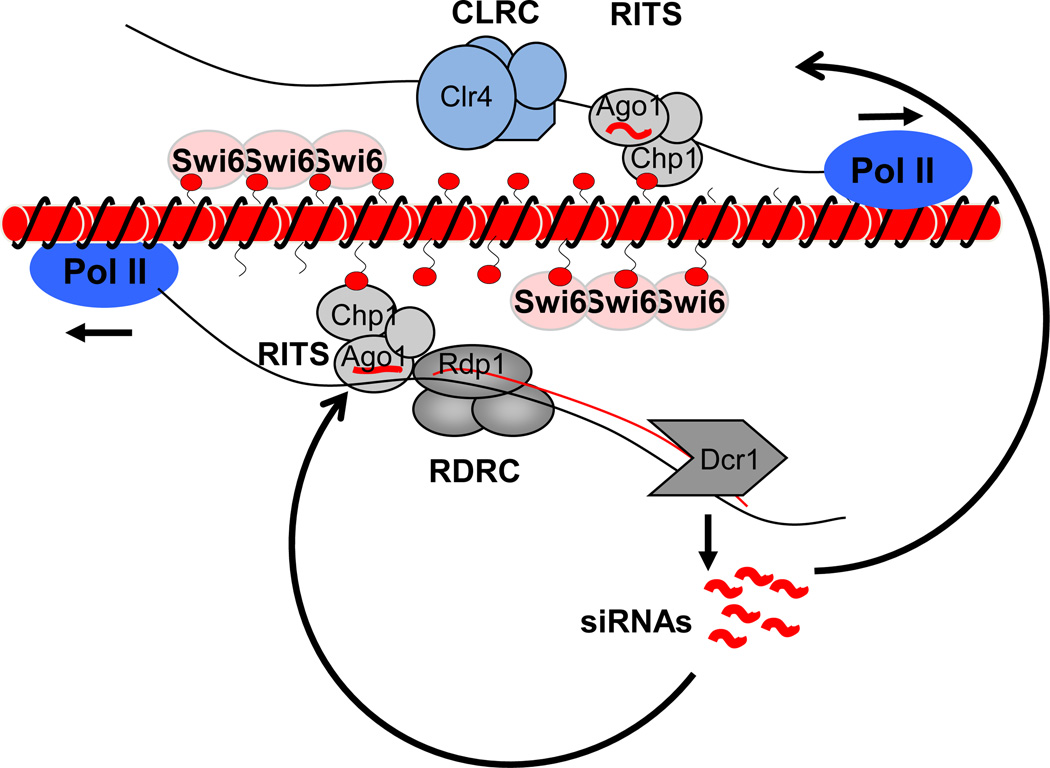
Another surprising aspect of the dynamic nature of heterochromatin is the involvement of transcription and non-coding RNAs during heterochromatin assembly [58, 59]. Heterochromatin usually forms at repetitive DNA elements and represses transcription of the underlying repeats. Therefore, it is counterintuitive that heterochromatin assembly actually requires transcription of these repeats. This process was first discovered and is best illustrated in the fission yeast (Fig. 3). In this organism, the repetitive sequences are transcribed during the S phase of the cell cycle [60, 61], possibly as a result of DNA replication, as the passage of DNA polymerases opens up the chromatin structure. These transcripts are converted to double-stranded RNAs with the help of the RNA-dependent RNA polymerase complex (RDRC), and are then processed by the ribonuclease Dicer into small interfering RNAs (siRNAs) [62, 63]. siRNAs are then loaded into the RITS (RNA-induced transcriptional silencing) complex, which contains the Argonaute protein Ago1 that uses siRNAs as guides to target the RITS complex to nascent transcripts originating from the repeats [64]. RITS then associates with the CLRC complex (containing H3K9 methyltransferase Clr4 and associated E3 ubiquitin ligase complex Cul4, Rik1, Raf1, and Raf2) to initiate H3K9 methylation [65–68]. The RITS subunit Chp1 contains a chromo domain that recognizes H3K9me, which stabilizes its association with heterochromatin, and forms a self-reinforcing loop of heterochromatin assembly and RNAi-mediated processing of repeat transcripts [62, 69].
Figure 3.
Transcription-dependent heterochromatin assembly at repetitive DNA elements in fission yeast. During S phase of the cell cycle, DNA repeats are transcribed by RNA polymerase II (Pol II). These transcripts are converted into double-stranded RNAs by the RNA-dependent RNA polymerase complex (RDRC) and then processed into small interfering RNAs (siRNAs) by the RNAi machinery. The siRNAs guide the RNA-induced transcriptional silencing (RITS) complex back to nascent transcripts. RITS associates with Clr4 methyltransferase complex (CLRC), which initiates H3K9 methylation and heterochromatin assembly.
Small RNA-mediated heterochromatin assembly has also been discovered in C. elegans and Drosophila. In addition, small RNAs also mediate DNA methylation in plants and a class of small RNAs termed piRNAs silences transposons in the germ line of animals. Since these topics have been extensively reviewed recently [58, 59], we will not discuss them in detail here. Although the chromatin modifying activities involved differ, a general theme of these different systems is that the nascent transcripts not only are the source of small RNAs, but also provide a scaffold for the recruitment of heterochromatin assembly factors, therefore explaining the need for transcription in heterochromatin assembly.
Heterochromatin changes during aging
The dynamics of constitutive heterochromatin is also reflected in the aging process. It was first found that somatic cells from patients with Hutchinson-Gilford Progeria Syndrome, a premature aging disease caused by a mutation in the nuclear membrane-associated lamin A protein, show a global loss of heterochromatin, as indicated by a reduction of H3K9me3 and HP1 protein staining [70, 71]. Similarly, mutations in the Werner Helicase (WRN) cause premature aging, and mesenchymal cells without this helicase also show a reduction in these heterochromatin markers [72]. Most importantly, cells from elderly individuals display a global loss of these heterochromatin marks, suggesting that loss of heterochromatin is part of the normal aging process [72, 73]. WRN directly associates with heterochromatin proteins such as the H3K9 methyltransferase SUV39H1 and HP1α as well as lamina-associated polypeptide LAP2β [72], suggesting a possible direct requirement of the nuclear membrane proteins and WRN in regulating heterochromatin assembly during aging.
These findings were corroborated by genetic studies of aging in Drosphila and C. elegans, where a global loss of heterochromatin also occurs during normal aging [74, 75]. Moreover, in Drosophila, overexpression of HP1 extends life span, whereas mutation in HP1 reduces it [76]. Similarly, in mammalian mesenchymal stem cells, loss of SUV39H1 activity results in premature aging phenotypes, while overexpression of HP1 proteins alleviates premature aging phenotypes caused by the loss of the WRN protein [72]. Even budding yeast, which assembles heterochromatin by way of Sir2-mediated histone deacetylation, experiences similar effects of modulating heterochromatin on life span [77]. Yet the role of heterochromatin changes during aging is unclear. It is possible that loss of heterochromatin results in the misregulation of gene expression, which contributes to the aging-associated phenotypes [78]. Alternatively, heterochromatin loss might affect other essential functions of heterochromatic domains, such as telomeres [77]. Despite the global loss of heterochromatin during aging, senescent cells often rearrange their chromosomes, leading to the formation of senescence-associated heterochromatin foci (SAHF), which colocalize with heterochromatin hallmarks such as H3K9me and HP1 [79, 80]. Therefore, the dynamic changes in heterochromatin might have other unappreciated roles in regulating the aging process.
Heterochromatin dynamics in response to stress
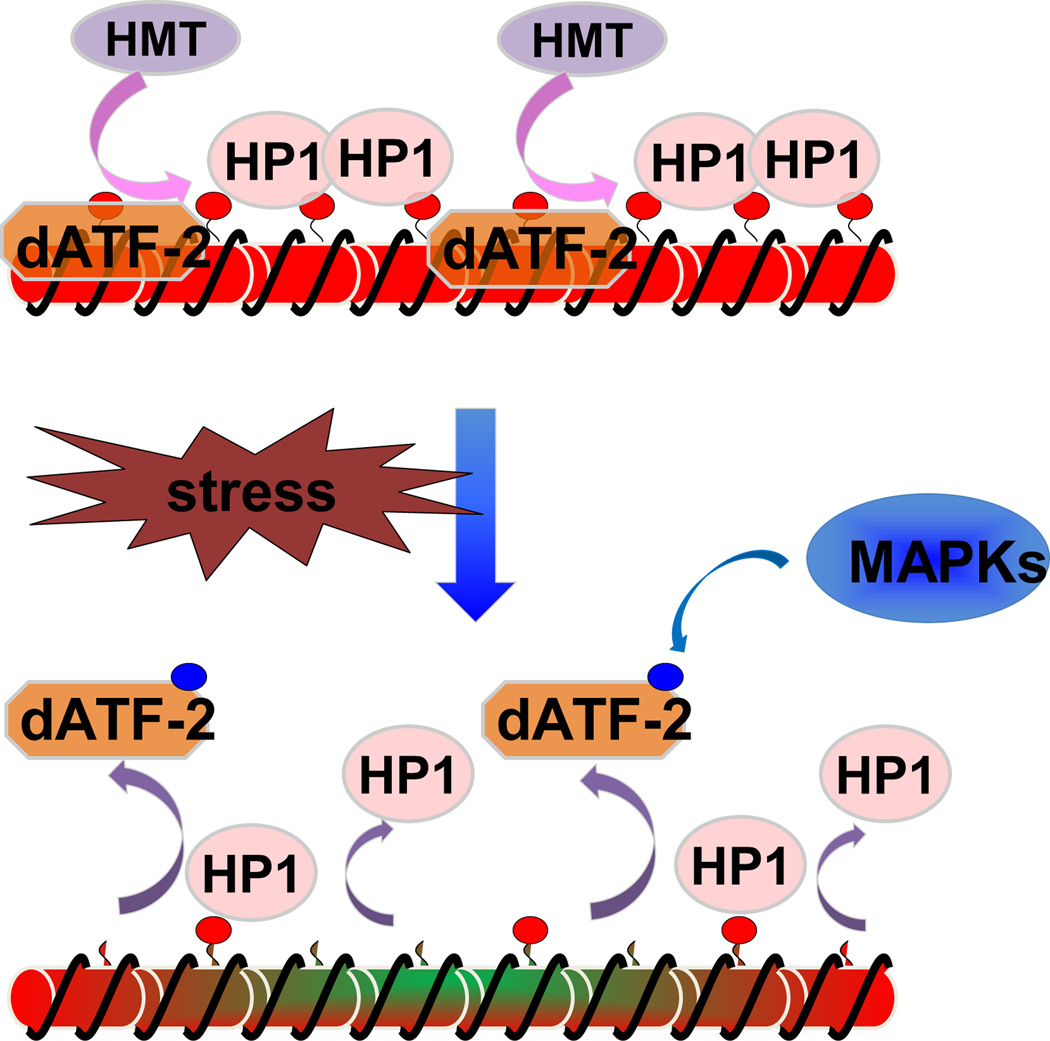
Constitutive heterochromatin domains also change their stability in response to environmental stimuli. For example, gene silencing at constitutive heterochromatin in fission yeast is less stable at elevated temperatures [81]. Interestingly, stress responsive transcription factors are directly involved in heterochromatin assembly, providing a possible mechanism for temperature-mediated effects on heterochromatin. The activating transcription factor/cyclic AMP response element binding protein (ATF/CREB) family transcription factors (Atf1/Pcr1), which regulate the expression of stress response genes, bind to a specific DNA element within the silent mating-type locus and recruit histone deacetylases and the histone methyltransferase Clr4 to initiate heterochromatin assembly [10, 11, 82]. Under stress, Atf1 is activated by MAP kinase-mediated phosphorylation to directly activate transcription of stress response genes [83]. The fact that the disruption of MAP kinase stabilizes heterochromatin indicates that phosphorylation of Atf1 by MAP kinase may destabilize heterochromatin as part of the stress response [11], although there is no direct evidence to support that phosphorylated Atf1 affects heterochromatin formation in fission yeast. However, a recent study in fruit flies provides a direct link of stress response to heterochromatin assembly. Similar to that in fission yeast, heterochromatin in flies is also sensitive to temperature fluctuations [84]. dATF-2, the homologue of fission yeast Atf1, colocalizes with HP1 and recruits HP1 to pericentric heterochromatin regions that contain dATF-2 binding sites under normal conditions (Fig. 4) [85]. Under stress conditions, dATF-2 is phosphorylated by MAP kinase and released from pericentric heterochromatin, which in turn abolishes HP1 enrichment, resulting in the disruption of heterochromatin [85]. Similar phenomena of stress-mediated modulation of heterochromatin have also been observed in human cells. Upon heat shock, heat shock transcription factor 1 (HSF1) redistributes to a few nuclear foci of pericentric heterochromatin. HSF1 binds directly to satellite repeats and facilitates transcription of ncRNA from these regions [86–88]. In addition, HSF1 regulates global histone acetylation levels by recruiting histone deacetylases HDAC1 and HDAC2 [89]. Therefore, direct involvement of stress-response transcription factors in heterochromatin assembly seems to be a highly conserved process that allows cells to modify constitutive heterochromatin domains in response to environmental stress.
Figure 4.
Pericentric heterochromatin disassembly in response to stresses in Drosophila. Under normal conditions, the transcription factor dATF-2 is in a hypophosphorylated form, which recruits HP1 to establish constitutive heterochromatin at pericentric regions. In response to stress, the MAPK pathway phosphorylates dATF-2, thus reducing its binding to pericentric heterochromatin and releasing HP1.
In addition to the involvement of stress-response pathways in heterochromatin assembly, other signal transduction pathways also directly modify heterochromatin proteins to control heterochromatin stability. The diverse histone-modifying activities involved in heterochromatin formation provide ample opportunities to incorporate environmental signals to regulate the stability of heterochromatin domains. One of the key proteins to receive such signals is HP1. For instance, casein kinase 2 (CK2)-mediated phosphorylation of fission yeast Swi6, Drosophila HP1a, and mammalian HP1α all promote binding to H3K9me3 nucleosomes [90], and such phosphorylation is required for heterochromatin function in fission yeast and Drosophila [91, 92]. Interestingly, a different region of mammalian HP1β is also phosphorylated by CK2 upon DNA damage, resulting in the rapid release of HP1β from the damaged site, facilitating access of DNA repair machinery [93]. These results suggest that the functions of HP1 phosphorylation are diverse in response to distinct stimuli, allowing cells to better adapt to environmental alterations. In addition, the histones themselves are targets of signal transduction pathways that impact heterochromatin stability. For example, in both fission yeast and mammals, the phosphorylation of H3S10 by Aurora kinase releases HP1 from chromatin during the M phase of the cell cycle, allowing proteins involved in chromosome condensation to access the underlying DNA [60, 82, 94, 95].
Heterochromatin-mediated epigenetic adaptation
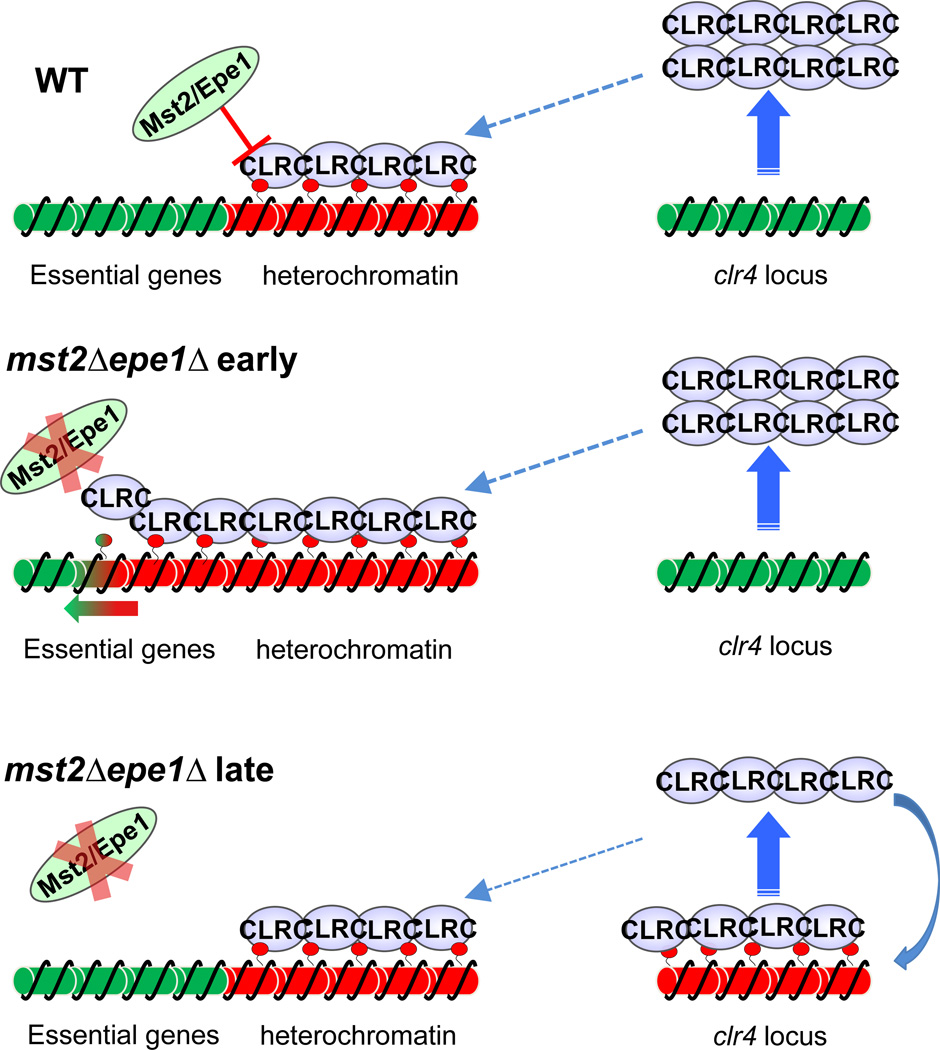
Although heterochromatin formation is highly organized and heterochromatin domains are well defined, recent findings suggest that heterochromatin formation is much more promiscuous than expected. In fission yeast, two factors, Epe1 and Mst2, negatively regulate heterochromatin assembly. Epe1 contains a JmjC domain, which is typically associated with histone demethylase activity [96]. Although the enzymatic activity of Epe1 has not been demonstrated in vitro, genetic evidence is consistent with the idea that Epe1 acts as a functional H3K9 demethylase, and loss of Epe1 results in the expansion and stabilization of heterochromatin domains [25, 26, 97–99]. Mst2 is a component of an acetyltransferase complex that is highly specific to histone H3 lysine 14 [100]. Loss of Mst2 results in a reduction of H3K14 acetylation, which in turn slows histone turnover to preserve parental histone modifications required for heterochromatin maintenance [24, 101]. In mst2Δ epe1Δ cells, there is massive up-regulation of heterochromatin, resulting in severe growth defects at the early stage of development due to the inactivation of essential genes (Fig. 5, Key Figure) [101]. Surprisingly, these cells quickly adapt to such a heterochromatic stress by accumulating heterochromatin at the clr4+ locus, which in turn down-regulates the levels of the Clr4 histone H3K9 methyltransferase to constrain heterochromatin domains in the genome. Such epigenetic changes can be inherited through mitosis and meiosis, conferring future generations greater resistance to heterochromatic stress. Interestingly, when heterochromatin is unable to form at the clr4+ locus via genetic manipulations, cells accumulate heterochromatin at the rik1+ locus, which encodes another subunit of CLRC complex required for its activity [101]. These observations indicate that epigenetic alteration is a rapid and efficient way to adapt to a new environment in response to stress.
Figure 5.
Heterochromatin-mediated epigenetic adaptation in fission yeast. The negative regulators of heterochromatin, Mst2 and Epe1, prevent promiscuous heterochromatin spreading in wild type cells. However, in mst2Δ epe1Δ cells, uncontrolled heterochromatin spreading inactivates essential genes, resulting in severe growth defects during early stage of development. Gradually, cells form heterochromatin at the clr4+ locus to reduce Clr4 expression levels, resulting in a new equilibrium that maintains heterochromatin at critical locations but minimizes heterochromatin spreading, leading to the normal growth during late stages of development.
In plants, a process termed vernalization silences the flowering repressor gene FLOWERING LOCUS C (FLC) in response to prolonged low temperature in winter, allowing flowering to occur in the following spring. Such silencing requires Polycomb Repressive Complex 2 (PRC2)-mediated H3K27 methylation, which forms facultative heterochromatin [102], demonstrating an important role of heterochromatin in integrating environmental signals. Similar epigenetic changes in heterochromatin might also enable tumor cells to survive certain therapies. For example, populations of tumor cells show both genetic and epigenetic heterogeneity, which contribute to the variations of almost every phenotype in these tumors [103]. A recent study demonstrates that in a non-small cell lung cancer cell line model, there are consistently subpopulations of drug resistant cells, which contribute to the development of drug resistance [104]. The establishment of drug resistance requires histone H3K4 demethylase KDM5A and histone deacetylase activities, indicating that a more repressive heterochromatic environment favors the adaptation of drug-resistant tumor cells. More importantly, these cells restore drug sensitivity after 20–30 passages, suggesting that this epigenetic adaptation is reversible [104]. Therefore, heterochromatin-mediated epigenetic adaptation offers greater flexibility for cells to tolerate environmental insults and seems to be an evolutionarily conserved phenomenon.
Concluding remarks
Constitutive heterochromatin has traditionally been viewed as a highly stable structure that represses transcription and recombination of repetitive DNA elements. However, recent studies have demonstrated that constitutive heterochromatin domains are also highly dynamic. The function of such dynamics is only beginning to be appreciated (see Outstanding Questions), and it might be part of the cellular response to outside stimuli by modifying chromatin structure, which cushions against adverse effects [105]. The silencing of gene expression by heterochromatin in a sequence-independent manner makes heterochromatin formation one of the most versatile forms of epigenetic changes. Changes of heterochromatin in response to numerous stresses are widely adapted from yeasts to humans. As a critical step of tumor development is the inactivation of tumor suppressor genes, the discoveries of epigenetic inactivation phenomena in different systems provide invaluable clues for studying the adaptation of tumor cells and designing new strategies to counteract such effects.
What is the molecular mechanism that regulates HP1 dynamics?
What is the mechanism of small RNAs-mediated chromatin-modifications in higher eukaryotes?
What are the signals that trigger heterochromatin changes during normal aging? What are the roles of these changes during aging?
What are the roles of heterochromatin changes in cellular adaptation to stress?
What is the molecular mechanism of ectopic heterochromatin formation in response to uncontrollable heterochromatin spreading? Is it programed or the selection of low frequency events?
Are there chromatin-based mechanisms for transgenerational inheritance in higher eukaryotes?
Constitutive heterochromatin has long been considered as a highly stable state. However, recent studies show that it is highly dynamic in nature.
HP1 proteins are in a dynamic equilibrium between chromatin bound and free forms.
Transcription is essential for RNAi-mediated heterochromatin assembly by providing the source of small RNAs and the scaffold for the recruitment of chromatin modifying enzymes.
Heterochromatin changes during the normal aging process and responds to environmental stresses.
Promiscuous heterochromatin formation is a novel way to adapt to external stresses and allows for transgenerational inheritance.
Acknowledgments
We thank members of the Jia lab for comments on the manuscript. Work in the Jia lab is supported by NIH grant R01-GM085145.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 3.Muller HJ, Altenburg E. The Frequency of Translocations Produced by X-Rays in Drosophila. Genetics. 1930;15:283–311. doi: 10.1093/genetics/15.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. In: Allis CD, et al., editors. Epigenetics. Cold Spring Harbor Press; 2007. pp. 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noma K, et al. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 6.Suka N, et al. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 7.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. The EMBO journal. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci. 2014;71:4841–4852. doi: 10.1007/s00018-014-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 10.Jia S, et al. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 11.Kim HS, et al. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. The Journal of biological chemistry. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- 12.Bulut-Karslioglu A, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nature structural & molecular biology. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Histone methylation in transcriptional control. Current opinion in genetics & development. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K, et al. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nature structural & molecular biology. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 15.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 16.Lachner M, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama J, et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 18.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. The EMBO journal. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart MD, et al. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Molecular and cellular biology. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 21.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 22.Al-Sady B, et al. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Probst AV, et al. Epigenetic inheritance during the cell cycle. Nature reviews. Molecular cell biology. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 24.Reddy BD, et al. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 2011;25:214–219. doi: 10.1101/gad.1993611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragunathan K, et al. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2014 doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audergon PN, et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348:132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusche LN, et al. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 28.Kueng S, et al. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 29.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 30.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 33.Kimura A, et al. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 34.Hecht A, et al. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 35.Liou GG, et al. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 36.Onishi M, et al. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Renauld H, et al. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 38.Hecht A, et al. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 39.Strahl-Bolsinger S, et al. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 40.Locke J, et al. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demerec M, Slizynska H. Mottled White 258-18 of Drosophila Melanogaster. Genetics. 1937;22:641–649. doi: 10.1093/genetics/22.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fourel G, et al. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. The EMBO journal. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. The EMBO journal. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbert PB, Henikoff S. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics. 2000;154:259–272. doi: 10.1093/genetics/154.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eissenberg JC, Elgin SC. HP1a: a structural chromosomal protein regulating transcription. Trends in genetics : TIG. 2014;30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canzio D, et al. Mechanisms of functional promiscuity by HP1 proteins. Trends in cell biology. 2014;24:377–386. doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowieson NP, et al. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Current biology : CB. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 48.Canzio D, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheutin T, et al. In vivo dynamics of Swi6 in yeast: evidence for a stochastic model of heterochromatin. Molecular and cellular biology. 2004;24:3157–3167. doi: 10.1128/MCB.24.8.3157-3167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 51.Festenstein R, et al. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 52.Stunnenberg R, et al. H3K9 methylation extends across natural boundaries of heterochromatin in the absence of an HP1 protein. The EMBO journal. 2015;34:2789–2803. doi: 10.15252/embj.201591320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manukyan M, Singh PB. Epigenome rejuvenation: HP1beta mobility as a measure of pluripotent and senescent chromatin ground states. Scientific reports. 2014;4:4789. doi: 10.1038/srep04789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 56.Dodson AE, Rine J. Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae. eLife. 2015;4:e05007. doi: 10.7554/eLife.05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tadeo X, et al. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 2013;27:2489–2499. doi: 10.1101/gad.226118.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 61.Kloc A, et al. RNA interference guides histone modification during the S phase of chromosomal replication. Current biology : CB. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motamedi MR, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 63.Sugiyama T, et al. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bayne EH, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong EJ, et al. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 67.Horn PJ, et al. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia S, et al. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 69.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 70.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang W, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haithcock E, et al. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brandt A, et al. The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging cell. 2008;7:541–551. doi: 10.1111/j.1474-9726.2008.00406.x. [DOI] [PubMed] [Google Scholar]
- 76.Larson K, et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS genetics. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsurumi A, Li WX. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 2012;7:680–688. doi: 10.4161/epi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 80.Corpet A, Stucki M. Chromatin maintenance and dynamics in senescence: a spotlight on SAHF formation and the epigenome of senescent cells. Chromosoma. 2014;123:423–436. doi: 10.1007/s00412-014-0469-6. [DOI] [PubMed] [Google Scholar]
- 81.Ayoub N, et al. Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics. 1999;152:495–508. doi: 10.1093/genetics/152.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamada T, et al. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 83.Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Molecular and cellular biology. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gowen JW, Gay EH. Effect of Temperature on Eversporting Eye Color in Drosophila Melanogaster. Science. 1933;77:312. doi: 10.1126/science.77.1995.312. [DOI] [PubMed] [Google Scholar]
- 85.Seong KH, et al. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 86.Jolly C, et al. Stress-induced transcription of satellite III repeats. The Journal of cell biology. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rizzi N, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Molecular biology of the cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biamonti G, Vourc’h C. Nuclear stress bodies. Cold Spring Harbor perspectives in biology. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fritah S, et al. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Molecular biology of the cell. 2009;20:4976–4984. doi: 10.1091/mbc.E09-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nishibuchi G, et al. N-terminal phosphorylation of HP1alpha increases its nucleosome-binding specificity. Nucleic acids research. 2014;42:12498–12511. doi: 10.1093/nar/gku995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimada A, et al. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009;23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao T, Eissenberg JC. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. The Journal of biological chemistry. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 93.Ayoub N, et al. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 94.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 95.Hirota T, et al. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 96.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 97.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 98.Trewick SC, et al. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. The EMBO journal. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y, et al. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. The Journal of biological chemistry. 2012;287:4386–4393. doi: 10.1074/jbc.M111.329417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang J, et al. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife. 2015:4. doi: 10.7554/eLife.06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 103.Marusyk A, et al. Intra-tumour heterogeneity: a looking glass for cancer? Nature reviews. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 104.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]