Abstract
Lipids comprise the bulk of the Meibomian gland secretion (meibum) which is produced by meibocytes. Complex arrays of lipogenic reactions in Meibomian glands, which we collectively call meibogenesis, have not been explored on molecular level yet. Our goals were to elucidate the possible biosynthetic pathways that underlie generation of meibum, reveal similarities in, and differences between, lipid metabolism in Meibomian glands and other organs and tissues, and integrate Meibomian gland studies into the field of general metabolomics. Specifically, we have conducted detailed analyses of human and mouse specimens using genomic, immunohistochemical, and lipidomic approaches. Among equally highly expressed genes found in Meibomian glands of both species were those related to fatty acid elongation, branching, desaturation, esterification, reduction of fatty acids to alcohols, and cholesterol biosynthesis. Importantly, corresponding lipid products were detected in meibum of both species using lipidomic approaches. For the first time, a cohesive, unifying biosynthetic scheme that connects genomic, lipidomic, and immunohistochemical observations is outlined and discussed.
Keywords: Lipidomics, meibocytes, meibogenesis, gene expression, extremely long chain lipids, branched lipids
1. INTRODUCTION
Meibomian glands (MG) are lipid-producing holocrine secretory glands that are present in humans [1] and most of studied mammals [2, 3]. MG are embedded in the tarsal plates (TP) of the lower and upper eyelids perpendicularly to the eyelid margins, and deliver their secretion (called meibum [4]) directly onto the ocular surface through a system of ducts and orifices. The cells that synthesize meibum in MG are called meibocytes. TP themselves are formed of a dense connective tissue with a rather complex anatomy, but relatively few, sparsely distributed cells, such as fibroblasts [5].
Meibum is considered an important part of the tear film/tear film lipid layer (TF/TFLL) defense system that protects eyes from microorganisms and desiccation [6–10]. TF/TFLL combination also serves as a lubricant [11] that smoothes movements of the eyeball and the eyelids, and improves visual acuity [12, 13]. In humans, diseases such as various forms of Dry Eye (DE) [14] and chalazion – an ocular lesion that typically originates from MG [15, 16] – have been linked to dysfunctionalities in MG, which result either in abnormal lipid secretions, or insufficient production or expression of meibum [17]. DE disease affects a large portion of the human population (between 8 and 30%, depending on the region and diagnostic criteria) [18, 19]. Alterations and deficiencies in meibomian lipids (ML) can have a profound negative impact on the TF/TFLL, and are currently being addressed with therapeutic (Restasis®, the only FDA-approved drug for DE in the USA [20]) or physiotherapeutic (LipiFlow® [21]) treatments, with rather limited information on their efficacy available. Thus, a clear understanding of the biochemistry and physiology of the ocular surface in general, and TF/TFLL specifically, is needed. A comprehensive elucidation of the biosynthetic pathways and regulatory and signaling mechanisms that underlie formation of meibum in MG (which we will collectively call hereafter meibogenesis), should help researchers in accomplishing this task.
Biochemically, human meibum (produced by mature, differentiated meibocytes in the acini of MG) is a complex and rather unusual secretion formed mostly of wax esters (WE), cholesteryl esters (CE), (O)-acylated ω-hydroxy fatty acids (OAHFA) and their cholesteryl esters (CHL-OAHFA), diacylated fatty α,ω-diols (DiAD), small amounts of triacylglycerols (TAG), free cholesterol (CHL) and possibly other sterols (STE), and traces of phospholipids (PL), sphingomyelins (SM), squalene (SQL) and ceramides (CER), with the latter two, possibly, originating from the epidermis of the eyelids (see [22–25] and references therein).
Available literature data suggest that the closest related lipid secretion – sebum, produced by sebaceous glands in the epidermis [26, 27] – differs markedly from meibum. For example, human sebum, compared to meibum, is highly enriched with triglycerides, SQL, and free fatty acids (FFA), has lower percentage of WE, and is depleted of CE (Table 1 and [22, 26]). These differences imply either tissue-specific alterations in the expression patterns and the substrate/product specificity of known enzymes (or their isoforms), or the expression and metabolic activity of still unknown enzymatic systems. Importantly, the enzymological processes that underlie formation of meibum have not yet been identified, explored or characterized in any species.
Table 1.
Normal sebum and meibum composition of humans
| Component | Human sebum [26] | Human meibum [22] |
|---|---|---|
| Percentage of total lipids, % | ||
| Acyl glycerols (TAG and DAG) | 30–63 | 1 (mostly, TAG) |
| Free fatty acids | 15 | ~0.1–1 |
| Squalene | 12–13 | traces |
| Cholesterol | 1.5 | 0.5 |
| Cholesteryl esters | 2 | 31 |
| Wax esters | 26 | 41 |
| Diacylated diols | n/r | Reported, not quantitated |
| OAHFA | n/r | 4 |
| Cholesteryl esters of OAHFA | n/r | 3 |
| Phospholipids | n/r | <0.1 |
| Ceramides | n/r | traces |
n/r – not reported
Direct in vivo experimentation with human TP, MG and TF/TFLL is, however, limited to non-invasive and/or minimally invasive techniques. On the other hand, mice are standard laboratory animals whose genome is well documented [28]. Moreover, in our initial studies of mouse MG [29, 30] we demonstrated that mouse meibum is close to the human one in terms of their overall lipid makeup – much more so than meibum of canines and rabbits [30].
The goals of this paper were to describe possible biosynthetic pathways that underlie generation of meibum in meibocytes of humans and animals (specifically, mice), reveal similarities in, and differences between, lipid metabolism in MG and other lipid-enriched organs and tissues, and integrate MG studies into the field of general metabolomics. For the first time, a cohesive, unifying biosynthetic scheme that connects genomic, lipidomic, and immunohistochemical observations is outlined and discussed.
2. MATERIALS AND METHODS
2.1. Reagents
Lipid standards used in this study were purchased from either Sigma Chemical Co. (St. Louis, MO) or Nu-Chek Prep. (Elysian, MN). Deuterochloroform (CDCl3) used for NMR studies was >99.96% (D) (Sigma-Aldrich, St. Louis, MO). Organic solvents were of chromatographic or spectroscopy grade. Anti-human ELOVL4 antibodies for immunohistochemical staining (IHC) were a gift from Dr. Robert Molday (University of British Columbia, Vancouver, Canada). Anti-mouse ELOVL4 antibodies were from Dr. Robert “Gene” Anderson (University of Oklahoma, OK). Anti-ELOVL3 antibodies (Cat#TA315865) were from OriGene (Rockville, MD). Antibodies against branched-chain alpha-keto acid dehydrogenase E1 (BCKDHA) and dihydrolipoamide branched chain transacylase E2 (DBT) were a gift from Dr. David Chuang (UT Southwestern Medical Center, Dallas, TX).
2.2. Software
HPLC-MS and GC-MS data were analyzed using Xcalibur (v. 2.2) software from Thermo Electron/Thermo Fisher Corp. (Waltham, MA). The NMR data were analyzed using Mnova (v.10.0) from MestreLab Research S.L. (Santiago de Compostela, Spain). Gene expression patterns were analyzed using Expression Console (v. 1.4) from Affymetrix (Santa Clara, CA). Statistical analyses were performed in SigmaStat (v. 3.5) from Systat Software (San Jose, CA) and Statistica (v.6.1) from StatSoft (Tulsa, OK).
2.3. Human and animal samples
All human study sample collection procedures were approved by the Institutional Review Board of the University of Texas Southwestern Medical Center (UTSW) and performed in accordance with the principles of the Declaration of Helsinki. Written informed consents were obtained from all study volunteers. The animal procedures were approved by UTSW Institutional Animal Care and Use Committee and were conducted in accordance with the Association for Research and Vision and Ophthalmology Statement for the use of animals in research.
2.4. Human meibum and sebum sample collection and processing
Human meibum had been extensively characterized in our previous publications in detail [16, 22–25, 30–39, and references therein] and was used in this study only to compare it with meibum of mice. Notably, inter-donor variability in the molar ratios of major tested lipids (WE and CE) in meibum of normal, non-DE humans was found to be rather low [36, 38, 39] and not exceeding ±15%. Two adult males and six adult females (mean age 41±12 yrs) with no clinically diagnosed dry eye, eye lid and conjunctiva inflammation, or any other ocular surface abnormalities were recruited. None of the volunteers was a contact lens wearer. Collection of human meibum samples was conducted as previously described [33, 36, 37]. Before collecting samples, the donor’s eyelid margins were cleaned with a cotton swab to remove any accumulated skin-derived debris, and then meibum was expressed by soft-squeezing the eyelids with two cotton swabs on each side of the eyelid, collected with a platinum microspatula, and dissolved in 1 mL of chloroform:methanol=2:1 (v/v) mixture (CM) which had been placed in a 2 mL HPLC-style glass vial with a Teflon-lined lid. The weight of the collected sample was measured on a microbalance as a difference between the weight of the empty, dry vial and the vial with the sample after the solvent had been evaporated under a gentle stream of nitrogen at about 35°C. The individual dry sample weight ranged from 0.2 to 2.5 mg.
Human sebum was collected from the upper eyelids of a 53 year old volunteer using SebuTape® (from CuDerm, Dallas, TX) [40, 41]. The samples were collected daily on five consecutive days. The SebuTape Skin Oil Tester patches were pressed against the skin for 5 min, and then the absorbed lipids were eluted with 3×2 mL of n-hexane:iso-propanol solvent mixture (1:1, v/v, HIP). The eluates from all patches were combined in a glass vial, and then the solvent was evaporated under a stream of nitrogen. The samples were pooled and analyzed using HPLC-MS and GC-MS similarly to meibum samples.
Initially, the quality of each individual sample was verified using HPLC-MS as described earlier [32, 36, 38], to determine if the samples were contaminated with typical impurities such as components of cosmetic products and plasticware (fatty acid amides, Irgafos and others [36]). These experiments were also used in attempt to detect outliers among the individual samples, which were not found – the lipid profiles of all samples were within ±16% range of the average for major lipid species observed in the samples [32, 36, 38].
For 13C- and 1H-NMR analyses, the individual samples were pooled to compensate for a relatively low sensitivity of NMR instruments in 13C-NMR experiments. Four randomly selected individual samples were combined to form the first study sample with a total mass of ~3 mg, while the other four samples were pooled to make the second pooled study sample with a total mass of 5 mg. The original CM solvent mixture was completely evaporated from the pooled samples under a stream of nitrogen, and the samples were re-dissolved in CDCl3. The samples then were kept at −80°C until use.
2.5. Human tarsal plate tissue collection
Two samples were obtained from females (25 and 51 year old) with no signs of DE conditions. The tissue samples were surgical discards obtained during scheduled eyelid surgeries at the Department of Ophthalmology of the UT Southwestern Medical Center. No abnormalities or pathological changes were observed in the tissue samples. After the fragments of TP with MG were dissected free from surrounding eyelid tissue under a dissecting microscope, and then either flash frozen in liquid nitrogen for mRNA extraction and lipid staining, fixed in paraformaldehyde for (immuno)histochemical staining, or paraffin-embedded for histological and IHC evaluation. The samples were stored at −80°C until used. Also, a fragment of MG-free TP tissue was collected and stored for a subsequent mRNA analysis.
2.6. Mouse meibum sample collection
Mouse meibum and skin lipids had been extensively characterized in our previous publications [29, 42, 43]. To recapitulate our original findings in this project, mouse meibum was collected from six adult mice (2 to 4 month old), which were an equal mix of both genders. The mice used were predominantly of 129SvEv type from a breeding colony whose derivation and evaluation were described in our earlier publications [29, 42, 43]. Meibum samples were collected from both upper and lower eyelids. Meibum was collected from the eyelid margins under a dissecting microscope, as described previously [30] and dissolved in the CM mixture. The samples were stored as described above for human samples.
2.7. Mouse tarsal plate tissue collection
The mouse tarsal tissue used for gene expression profiling experiments was a pooled sample collected from five male mice (median age – 3 months) on the same background as in Section 2.6. Animals were euthanized and all four eyelids surgically removed and placed on ice. Using a dissecting microscope, the tarsal plates were dissected free from epidermis, subcutaneous, muscle and conjunctival tissue. In addition, particular attention was taken to remove eyelid margin tissue. Immediately post dissection tarsal plates were flash frozen with liquid nitrogen and stored at −80 °C.
2.8. (Immuno)histochemical studies and Western blot experiments
Both mouse and human eyelid tissue was fixed in 4% paraformaldehyde and processed for ELOVL4 immunohistochemistry. Primary antibodies used were a rabbit polyclonal anti-mouse ELOVL4 (dilution 1:500) [44] and a monoclonal anti-human ELOVL4 antibody Elo 6F1 (hybridoma supernatant; dilution 1:5) [45]. Secondary antibodies were Alexa Fluor®488 conjugated goat anti-rabbit A11070 (dilution from 1:800; from Invitrogen) and donkey anti-mouse A21202 (dilution 1:1000; from Molecular Probes). ELOVL3 was detected in a paraffin-embedded human tarsal tissue and treated for antigen retrieval by heating 2 minutes in a pressure cooker in 50mM EDTA and 0.05% Tween20 prior to incubation with rabbit polyclonal anti-human ELOVL3 (TA315865, from OriGene; dilution 1:50) followed by 1) incubation with an horse reddish peroxidase-conjugated goat anti-rabbit secondary antibody (diluted 1:2000; #111-036-003; Jackson ImmunoResearch Laboratories, West Grove, PA), and 2) detection by 3,3′-diaminobenzidine (DAB) staining (DAB kit, Vector Laboratories, Burlingame, CA). Antibodies against branched chain alpha-keto acid dehydrogenase E1 component alpha (BCKDHA) were diluted 1:2500. Antibodies against dihydrolipoamide branched chain transacylase E2 (DBT) were diluted 1:5000. The antibodies were used as described earlier [46, 47]. Lipid staining in mouse tissue with Oil Red O was conducted as described earlier [29]. Lipids in human tissue samples were stained with HCS LipidTOX Deep Red Neutral Lipid Stain (H34477, ThermoFisher) using manufacturer’s recommendations.
2.9. Gene expression profiling
The human and mouse TP tissue samples were sent for gene expression profiling to the UT Southwestern Genomics and Microarray core facility. The analyses were performed using Affymetrix (Santa Clara, CA) reagents, protocols, and microarray according to the manufacturer’s recommendations [48]. The samples were stored at −80°C before the analyses. Each of the human TP tissue samples was ~50 mg or more, and was analyzed individually. Unlike human tarsal plates, the tarsal plates of mice are small. Thus, to maximize the signal-to-noise ratio and equalize the possible inter-donor variations, samples from five animals were pooled. After homogenization of the tissues samples, the mRNA was extracted and quantitated. Individual human samples produced about 40 and 290 ng/μL, while the pooled mouse one – 21 ng/μL. Their quality was checked electrophoretically using an Agilent 2100 Bioanalyzer and Nano chips by measuring the 28s/18s tRNA ratios, which were 2.7 and 2.4 for human samples, and 2.6 for the mouse one. The RNA integrity numbers (RIN) for the two individual human and the pooled mouse samples were high 8.7, 9.1 and 8.3, correspondingly. The human tarsal plate mRNA was analyzed using a GeneChip® Human Transcriptome Array 2.0 that covers 44,699 protein-coding genes, while the mouse mRNA was profiled using the GeneChip® Mouse Transcriptome Assay 1.0 (covering >23,000 protein-coding genes). Datasets #GDS1832 [49], GDS1361, GDS1009, GDS3113, and GDS3334, among others, available through NIH GEO database, were used to compare our results with those reported earlier.
To characterize the relative expression of genes in the TP and other tissue samples of humans and mice, we adhered to the following semi-quantitative scale: 1) extremely high expression [15<log2(SST-RMA gene full signal)<20; with log2 value of 20 being the maximum, i.e. 100%; Rank Order between 75% and 100%]; 2) high, 10<log2(RMA)<15, Rank Order 50 to 75%; moderate expression 5<log2(RMA)<10, Rank Order 25 to 50%; low expression – 2<log2(MRA)<5, Rank Order between 10 and 25%. A level of expression of log2(MRA)=2 (the cutoff value) and less was considered a background noise. All the genes reported in this paper fall into the category of highly to moderately expressed genes. The NIH GEO Profiles used in this study were already available in normalized form, with values varying from 1% (low expression) to 100% (high expression). When necessary, full soft dataset files downloaded from NIH GEO database were used instead.
2.10. Chromatographic and mass spectrometric analyses of lipid secretions
GC-MS analyses of meibum, sebum, and authentic lipid standards (such as WE, FFA, SQL, CHL and other sterols) were conducted as follows. A Trace Ultra gas chromatograph and an ITQ 1100 mass spectrometric detector (both from Thermo Electron) were used. High temperature separation of the analytes was performed as described before [31] using a TG-5MS capillary column (length 30 m, internal diameter 0.25 mm, film coating 0.25 μm from Thermo Electron). The ITQ1100 detector was used in the electron impact (EI) mode. The following parameters were used: electron energy (EE) (−30) V; ionization current 250 μA; m/z range 50 to 900; 3×25ms microscans; isolation width 1.5 mass units; ion source temperature 250 °C; carrier gas – helium; gas flow rate of 1 mL/min; the injector temperature 300 °C; the MS-transfer line was maintained at 325 °C. To remove traces of oxygen and water from helium, a Triple Trap (Thermo Scientific) was used. All lipid samples were dissolved in either HIP solvent mixture, or in deuterochloroform. The majority of the samples were dissolved in the HIP solvent mixture, while those destined for NMR were analyzed as solutions CDCl3. Splitless injections of 0.2 to 2 μL of samples (depending on their concentrations) were performed using an AI3000 autoinjector (Thermo Scientific). The analytes were eluted using a temperature gradient as follows. The starting temperature of 100°C was maintained for the first 5 min. Then, the column temperature rose linearly at a 5°C/min rate for 50 minutes. After the final temperature of 350 °C was achieved, it was held steady for 10 minutes followed by a cooling cycle from 350°C to 100°C during the next 10 minutes, and re-equilibration time of 5 min. The total duration of the run was ~85 min.
Calibration curves for individual lipid standards were generated. Lipids were dissolved in the same solvents as meibum and sebum. Under the conditions of the EI GC-MS analysis, most analytes were detected as (M+) molecular ions. CHL was detected as (M)+ and (M – H2O)+ ions due to a spontaneous loss of H2O.
For HPLC-MS analyses, a tandem of a Waters Alliance 2695 gradient system (Waters Corp., Milford, MA) and a LCQ Fleet ion trap mass spectrometer (Thermo Electron) was used. The spectrometer was alternately used with either an electrospray ionization (ESI) ion source (for amphiphilic lipids, such as FFA, OAHFA, PL, and SM) or an atmospheric pressure chemical ionization (APCI) ion source (for non-polar lipids, such as WE, CE, CHL-OAHFA, SQL, CHL, and others). Both ion sources were from Thermo Electron. The MS instrument was operated under the Xcalibur software. Authentic oleic-acid based homologous WE standards ranging from lauryl oleate (C12:0-C18:1) to lignoceryl oleate (C24:0-C18:1) were used as positive controls to identify and quantitate meibomian WE as described earlier [32–34]. Authentic homologous CE standards ranged from cholesteryl laurate to cholesteryl lignocerate. Tested authentic homologous TAG ranged from trialaurin to trilignocerin. Various authentic free fatty acids, cholesterol, dehydrocholesterol, and squalene were used as standards to confirm the nature of corresponding meibomian lipids. Synthetic DiAD, OAHFA, and CHL-OAHFA were of the types described earlier [30, 34, 50].
Normal-phase HPLC-MS (NP HPLC-MS)
A Lichrosphere Diol HPLC column (3.2 × 125 mm; particles 5μm) was used for isocratic normal phase HPLC separation of lipids [30, 32, 33, 42, 43]. The column was equilibrated in a n-hexane:iso-propanol:glacial acetic acid (HIPAA, 950:50:1, vol/vol) solvent mixture at a flow rate of 0.3 mL/min and 30°C. The meibum samples were dissolved in a small volume of the eluent to make ~0.1 mg/mL sample stock solution. Between 0.5 and 7 μL of the stock solutions were injected. The analytes were eluted isocratically for up to 1 hr. Then, the column was washed with iso-propanol for 10 min at 0.3mL/min, and re-equlibrated with the HIPAA eluent to make it ready for the next run. Using these conditions, the major and most nonpolar lipids of meibum – WE, CE, TAG, DiAD and CHL-OAHFA – eluted between 3 and 7 minutes, while more polar lipids – CHL and CER – eluted later and were clearly separated based on their polarity [42, 43]. The elution was monitored using the APCI ion source operating in the positive ion mode with the following analyte-optimized settings: source voltage ~3.75 kV; vaporizer temperature between 250 and 350 °C; sheath gas (nitrogen) – 20 to 25 arbitrary units; sweep gas – 5 units; capillary voltage – between 5 and 25 V; capillary temperature −300 °C. In these conditions, WE and SQL were visible as (M + H)+ adducts, CHL and CER – as (M + H – H2O)+ ions, DiAD – as ions, CE as (M + H – FA)+ ions, while CHL-OAHFA – as ions (M + H)+ and (M + H – CHL)+. The loss of FA and CHL residues from DiAD, CE, and CHL-OAHFA was due to spontaneous in-source fragmentation of these complex lipid molecules. FFA and OAHFA were detected in the negative ion mode (NIM) as either (M – H)– ions, or (M – H + acetic acid]– adducts. The MS parameters were similar, except for the source voltage (–1 to –2 kV) and the capillary voltage (−10 to −15 V).
Reversed phase HPLC-MS (RP HPLC-MS)
The gradient reversed phase HPLC experiments were conducted on a C18 Hypersil Gold column (2.1 × 150 mm; particles 5μm) using a ternary gradient of 5 mM ammonium formate in water, acetonitrile, and iso-propanol [34, 39]. Neutral and cationic analytes were detected in the positive ion mode, while the anionic lipids (such as FFA, OAHFA, and acidic PL) were monitored in the negative ion mode. Using APCI, most of the neutral lipids were detected as (M + H)+ and (M + H – H2O)+ adducts, while ESI produced a large percentage of (M + K)+ adducts. The distribution patterns of these adducts were specific for each lipid class and eluent, and must have been evaluated for each condition separately.
To determine the molar ratios of major ML, mixtures of authentic lipid standards composed of WE, TAG, CE, CHL, and SQL were prepared and analyzed alongside meibum samples. For tested lipids, the low limit of detection (LLoD), linearity of response, and reproducibility of injections were evaluated as illustrated in Fig. S1 (Supplemental Materials).
2.11. NMR analyses of human meibum
1H- and 13C-NMR experiments were conducted on an Agilent 400 MHz-MR DD2 NMR spectrometer equipped with an Agilent 5mm PFG OneNMR probe, operating at 399.677 MHz (for 1H) and 100.510 MHz (for 13C) frequencies. All experiments were carried out at an ambient temperature of about 22 °C. The number of scans and other parameters varied from sample to sample and were chosen based on the optimal balance between the signal-to-noise ratios, resolution, and the duration of the experiments.
1H spectra were acquired with a spectral width of 3396.7 Hz, 8682 complex data collection points, a 45 degree RF pulse, 1 relaxation delay, and 16 to 64 scans. 13C spectra were acquired with a spectral width of either 19531.2 or 24038.5 Hz, 32768 complex data points, a 45 degree RF pulse, a 45 degree RF pulse, 0.3 sec relaxation delay, broadband 1H decoupling during the relaxation delay and acquisition periods, and numbers of scans varying between 512 and 18812. gHSQCAD data were acquired with a spectral width of 3396.7 Hz and 510 complex data points on F2 and a spectral width of between 4522.8 and 14571.9 Hz and 96 to 256 t1 increment on F1 (all sample-specific), a 1 sec relaxation delay and between 2 and 16 scans per increment. All study samples were dissolved in CDCl3 to make up to ~20 mg/0.6 mL solutions of lipid standards, and about 3 and 5mg/0.6 mL for two pooled human meibum samples.
3. RESULTS
3.1. Comparison of meibum and sebum lipids of humans
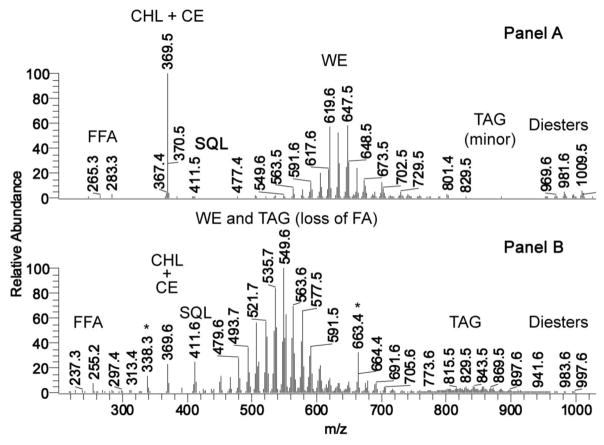
Human meibum and sebum samples were compared with each other using the NP HPLC-APCI MS protocol described in Materials and Methods. Resulting integrated mass spectra of the samples, taken in the positive ion mode, revealed marked differences between the secretions (Fig. 1). Major lipid classes detected in meibum (Fig. 1, Panel A) were: free fatty acids [mainly, oleic acid, m/z 283 (M+H)+ and 265 (M+H-H2O)+]; CHL and extremely long chain CE (who share a common analytical ion m/z 369); SQL [ion m/z 411, (M+H)+]; various very, and extremely long chain WE [ions m/z 477–673, (M+H)+]; very minor amounts of TAG [major component – triolein, m/z 885, (M+H)+]; and a group of complex, extremely long chain lipids called “diesters” (DiAD, CHL-OAHFA, and others; ions with m/z values between 900 and 1000) [30; 50, 51]. A representative APCI mass spectrum of a human sebum sample taken in the positive ion mode is shown in Fig. 1, Panel B. Major lipid classes detected in sebum were: free fatty acids [mainly, palmitoleic acid, m/z 255 (M+H)+ and 237 (M+H-H2O)+]; CHL and CE (common analytical ion m/z 369); SQL [ion m/z 411, (M+H)+]; various WE and fragments of TAGs, formed via the loss of one of their FA residues due to spontaneous in-source fragmentation of TAG [ions m/z 450–600, (M+H)+]; noticeable amounts of regular TAG [major components – C16-C18FA-based TAF with m/z values of 843, 857, and 869 (M+H)+]; and a group of complex lipids called “diesters” (DiAD, CHL-OAHFA, and others) [30; 50, 51]. Signals m/z 338 and 663 (Irgafos, [36]), labeled with asterisk, are contaminations. Notable was a high enrichment of meibum with CHL/CE (peak with m/z 369), and a very low intensity of the SQL signal m/z 411, compared to other detected components of meibum, while the opposite was true for sebum – the signal of CHL/CE was markedly lowered, while SQL signal increased more than ten-fold. Also, very, and extremely long chain WE were substantially reduced in sebum, but highly represented in meibum. On average, the decrease in molecular masses of WE of sebum was about 7 carbons, compared to meibomian WE. Though the comprehensive comparison of meibum and sebum goes beyond the scope of this manuscript, the presented data, taken together with independently published results, clearly outlined marked differences between two secretions.
Figure 1. Mass-spectrometric comparison of human meibum and sebum.
Panel A. A representative APCI mass spectrum of a human meibum sample taken in the positive ion mode. Major lipid classes detected in meibum were: free fatty acids [mainly, oleic acid, m/z 283 (M+H)+ and 265 (M+H-H2O)+]; CHL and CE (common analytical ion m/z 369); SQL [ion m/z 411, (M+H)+]; various WE [ions m/z 477–673, (M+H)+]; minor amounts of TAG [major component – triolein, m/z 885, (M+H)+]; and a group of complex lipids called “diesters” (DiAD, CHL-OAHFA, and others; ions with m/z values between 900 and 1000) [30, 50, 51].
Panel B. A representative APCI mass spectrum of a human sebum sample taken in the positive ion mode. Major lipid classes detected in sebum were: free fatty acids [mainly, palmitoleic acid, m/z 255 (M+H)+ and 237 (M+H-H2O)+]; CHL and CE (common analytical ion m/z 369); SQL [ion m/z 411, (M+H)+]; various WE and fragments of TAGs, formed via the loss of one of their FA residues due to spontaneous in-source fragmentation of TAG [ions m/z 450–600, (M+H)+]; noticeable amounts of TAG [major components – C16-C18FA-based TAF with m/z values of 843, 857, and 869 (M+H)+]; and a group of complex lipids called “diesters” (DiAD, CHL-OAHFA, and others)[30, 50, 51]. Signals m/z 338 and 663 (Irgafos, [36]), labeled with asterisk, are contaminations.
Note a high enrichment of meibum with CHL/CE, and a very weak signal of SQL, and a reverse situation for sebum, where the CHL/CE signal is diminished, and SQL in increased.
3.2. Comparison of meibum of humans and mice
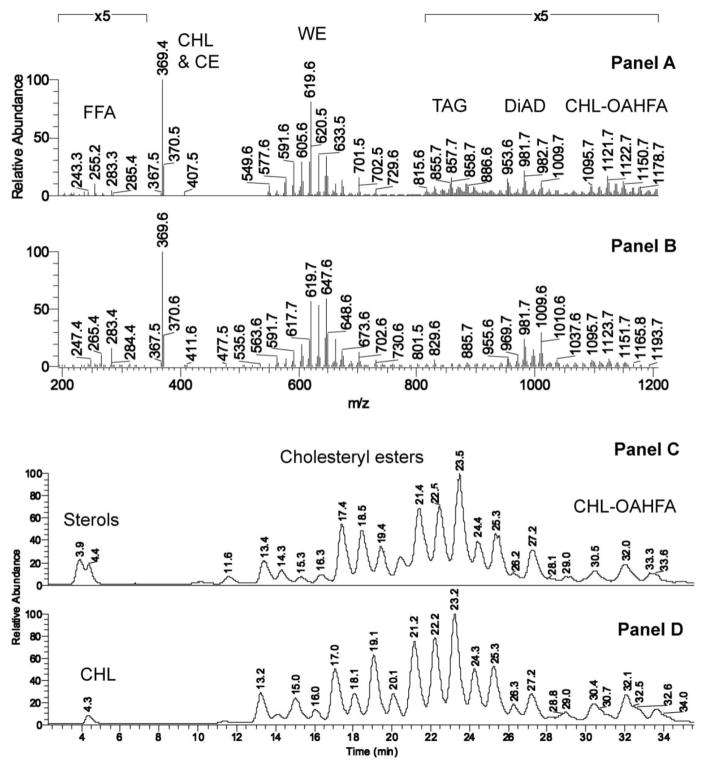
Key features of meibum of the two species were compared in NP HPLC-MS and GC-MS experiments (Figs. 2, S2–S5). The following groups of lipids were found to be similar: 1) FFA (with C18:1 and C16:1 as major components, detected as ions with m/z values of, respectively, 283/265 and 255/237); 2) CHL and CE (both groups were detected using a common analytical ion m/z 369); 3) WE (detected as ions with m/z values between 500 and 740; major homologous waxes with m/z 533/535, 547/549, 561/563, 575/577, 589/591, 603/605, 617/619, 631/633, 645/647, and 659/661, differing by numbers of double bonds in their structures and the nature of their fatty acid residues; most mouse WE were based on palmitoleic acid, while the human WE were mostly based on oleic acid); 4) TAG (m/z values between 800 and 900; major components m/z 853/855/857 and 883/885/887); 5) CHL-OAHFA (ions with m/z values between 900 and ~1200; major components m/z 1093/1095, 1107/1109, 1121/1123, 1135/1137, and 1149/1151, differing in the number of double bonds); and 6) a very diverse group of “di- and tri-esters” [30; 50, 51], such as DiAD (ions with m/z values between 900 and ~1000; major ions 953/955/957, 979/981/983, and 1007/1009) and more complex compounds whose structural analyses has not been accomplished yet. Importantly, the elution profile of mouse CE – one of the two largest classes of meibomian lipids (WE and CE) – was virtually a carbon copy of the human one (Fig. 2, Panels C and D). The FA residues of major CE observed in the mouse meibum as (M + H)+ and (M + K)+ ions were of C18:n to C36:n nature with the total number of double bond n=0–4, and major FA and FAL residues being saturated and monounsaturated ones.
Figure 2. HPLC-MS analysis of mouse and human meibum.
Panel A. Mass spectrum of mouse meibum in the m/z range of 200 to 1200. NP HPLC-APCI MS, positive ion mode. Averaged spectrum of meibomian lipids is shown.
Panel B. Mass spectrum of human meibum. Same conditions as in Panel A.
Panel C. Extracted ion chromatogram of ion m/z 369.4 detected in mouse meibum in RP HPLC-APCI MS experiments in positive ion mode. Notes: Ion m/z 369.4 is an analytical ion generated from CHL (M – H2O + H+) and CE (M – FA + H+) due to spontaneous in-source fragmentation of CHL and CE. Structural assignments of major human and mouse WE were made as follows: CHL, dehydro-CHL [peaks with retention times (RT) 3.9–4.4 min]; C18:1-CHL (11.6 min); C20:1-CHL (13.4 min); unknown(14.3 min); C20:0- and C22:1-CHL (15.3 min); C21:0-CHL (16.3 min); C22:0- and C24:1-CHL (17.4 min); C23:0-CHL (18.5 min); C24:0- and C26:1-CHL (19.4 min): C26:0- and C28:1-CHL (21.4 min); C27:0-CHL (22.5 min); C28:0- and C30:1-CHL (23.5 min); C29:0-CHL (24.2 min); C30:0- and C32:0-CHL (25.3 min); C34:1-CHL (27.2 min); C36:1-CHL (29:0 min); peaks with RT of 30.5 min, 32 min, and 33.6 min are a complex mixture of CHL-OAHFA [30, 50].
Panel D. Extracted ion chromatogram of ion m/z 369.4 detected in human meibum. Note very close similarities in the elution profiles of human and mouse CHL, CE, and CHL-OAHFA. The analytes detected in eluted peaks are the same as in Panel C.
Saturated WE, on the other hand, were almost undetectable in the HPLC-MS experiments because of their poor ionizability, and their analyses required the use of high temperature GC-MS (Figs. S2–S5), which was conducted according to the published protocol [31]. Importantly, when tested in an equimolar ratio, the intensities of ions of saturated WE were about 15 times stronger than those of their unsaturated counterparts, which had to be taken into account when quantifying both types of lipids. As was with unsaturated WE, their saturated counterparts were very similar in mice and humans (Fig. S3). The major saturated FA in this group of mouse lipids were C16:0, C17:0, and C18:0, in the ratio of about 10:2:1, respectively. In human meibum, the C16:0 to C17:0 to C18:0 FA ratio was about 2:4:1.
3.3. Establishing key features of meibomian lipids
3.3.1. Assessment of the degree of unsaturation of meibomian lipids
The major group of meibomian lipids - unsaturated WE – were detected in NP HPLC-MS experiments almost exclusively as (M+H)+ adducts (Fig. 2, Panels A and B), while in RP HPLC-MS experiments WE appeared as a mix of (M+H)+ and (M+K)+ ions (Fig. S2). No Na+ adducts were observed in either experiments. WE of both tested species produced signals with almost identical distributions of molecular masses across the species ranging from as low as 506 Da to as high as 732 Da. The major WE were present as families of homologous compounds differing in their degree of unsaturation in the following order: mono-unsaturated≫di-unsaturated≫poly-unsaturated.
The nature of the unsaturated FA residues (UFA) in mouse WE and other ML was identified in: 1) source induced dissociation MS experiments which were designed to cause mild in-source fragmentation of intact complex lipids (mostly, various esters in nature) to release their fatty acid fragments as (FA+H)+ product ions, and 2) in MS/MS experiments. The vast majority of mouse ML were found to have C16:1-C16:3 and C18:1-C18:3 mono-, di- and, occasionally, tri-unsaturated FA in their structures. The major UFA fragments detected in mouse meibum were identified as those of palmitoleic acid (C16:1), while the second largest ion was that of oleic acid (C18:1), with the overall C16:1/C18:1 molar ratio being about 1.7:1. Relatively weaker signals of linoleic acid (C18:2) were also regularly observed. In humans, the major UFA component of ML was oleic acid with palmitoleic acid being the second largest unsaturated FA, present in a C18:1/C16:1 molar ratio of 5.5:1. Again, linoleic acid was the third most abundant unsaturated FA in mouse meibum at about 5–10% of oleic acid levels, while linolenic – an exceedingly small 1% or less. These observations recapitulated our previous reports on human meibum [22–25, 30–38].
The GC-MS analyses of major WE of mice (Figs. S3 and S4) corroborated the results of HPLC-MS experiments discussed above. For example, a group of M+ ions m/z 642.6, 644.6, 646.7 and 648.6 (Fig. S4) was found to belong to a family of C44:n wax esters, with a total number of double bonds per molecule (n) of 3, 2, 1, and 0, correspondingly. Note that many isobaric WE coexisted in meibum of both species (Fig. S4, Panels C–F).
3.3.2. Evaluation of the presence of odd-chain fatty acid and alcohol moieties in meibomian lipids
Considering the noted variations in the lengths of FA residues, the apparent equality of the molecular weights (MW) of intact human and mouse WE (Fig. 2) could have been achieved only by corresponding increases in the lengths of their FAL moieties. For major C16:1-FA based WE with MW 604.6, 618.6, 632.6, 646.7, and 660.6, this means that their FAL moieties are C25:0, C26:0, C27:0, C28:0 and C29:0, while for C18:1-FA based WE the FAL moieties are two methylene groups shorter, i.e. C23:0, C24:0, C25:0, C26:0, and C27:0. These structural assignments were confirmed by analyzing FAL fragments of parent WE as described earlier [31]. Characteristically, as discussed in the previous section, a large number of ML had C17:0 residues FA in their structures. Also found were odd-chain FFA (C19-C33, both of saturated and mono-unsaturated nature), WE with C15:0 and C19:0 FA, large amounts of CE with odd-chain C21:0-C31:0 FA, and odd-chain extremely long chain FA moieties in OAHFA and DiAD (up to C29-C33). Thus, we concluded that meibum of both mice and humans had a substantial presence of odd carbon chain length FA and FAL, a rather rare feature in organs and tissues other than meibomian and sebaceous glands.
3.3.3. Establishing cis, trans-geometry of the double bonds in meibomian lipids
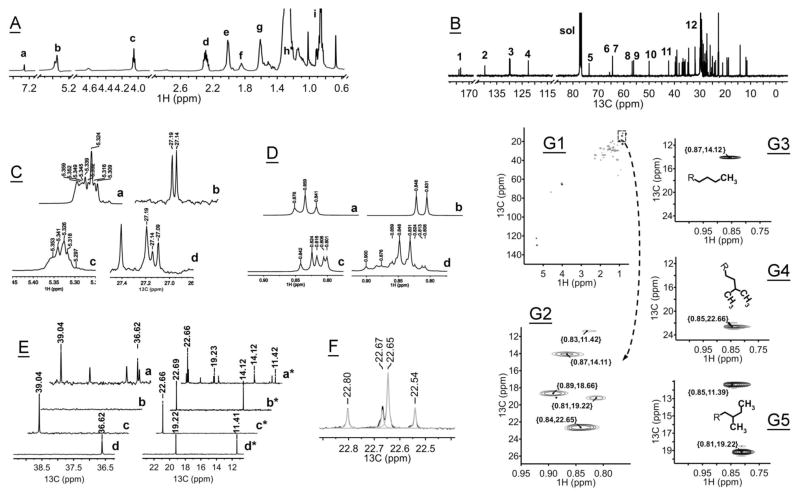
To determine the predominant stereochemistry of the double bonds in ML, 13C- and 1H-NMR experiments were conducted with human meibum and a few lipid standards (Fig. 3 and Table S1). A typical 13C-NMR analysis requires multi-milligram quantities of meibum to be accomplished in a reasonable amount of time. Therefore, two pooled human samples each from 4 different donors were analyzed, totaling 3 and 5 mg each. Also, this approach was to minimize possible inter-donor variations in the chemical composition of ML, though such variations typically do not exceed ±15% of the average [36, 38, 39]. The NMR spectra of two pooled samples were virtually identical. Typical full 1H- and 13C-NMR spectra are shown in Fig. 3, Panels A and B. The 1H-NMR spectra (Fig. 3, Panels A and C, trace “c”) clearly revealed strong characteristic resonances of cis-double bonds –CH=CH– with chemical shifts δ 5.30–5.38 ppm (also evident in the spectrum of cholesteryl oleate, Panel C, trace “a”), and lacked those of trans-double bonds (which would be visible as chemical shifts δ 5.38–5.52 ppm [52]). Similarly, 13C-NMR spectra of meibum (Fig. 3, Panels B and C, trace “d”) did show a resonance at δ 27.19 ppm (allylic carbons adjacent to cis-monoenes and dienes –CH2-CH=CH–, also visible in the spectrum of cholesteryl oleate, Panel C, trace “b”), but not at δ 32.5 ppm, which would indicate the presence of trans-mono- and dienes. Thus, we concluded that double bonds in ML are predominantly in cis-configuration.
Figure 3. NMR spectra of human and mouse meibum.
Panel A. Full 1H-NMR spectrum of human meibum. For experimental details, see Material and Methods. The following assignments were made: resonance “a” – δ 7.25 ppm, residual signals of CHCl3; resonance “b” – δ 5.33 ppm, olefinic double bonds in cis-configuration, -CH2-CH=CH-CH2- (for more details see Panel C, this Figure); resonance “c” – δ 4.03 ppm, -CH2-O-C(O)- of wax esters and analogs; resonance “d” – δ 2.27 ppm, -O-C(O)-CH2- of fatty acid esters; resonance “e” – δ 1.98–1.99 ppm, -CH2-CH=CH-CH2 – of unsaturated aliphatic chains; resonance “f” – δ 1.7–1.9 ppm, -O-C(O)-CH2-CH2- of fatty acid esters; resonance “g” – δ 1.59 ppm, -CH2-CH2-O-C(O)- of wax esters and analogs; resonance “h” – δ 1.3 ppm, -CH2-(CH2)n-CH2- of the “methylene pocket” of aliphatic chains; resonance “i” – δ 0.8–0.9 ppm, terminal CH3 groups of aliphatic chains (straight, iso-, and anteiso-branched).
Panel B. Full 13C-NMR spectrum of human meibum. Peak assignments are shown in Table S1.
Panel C. Partial 1H-NMR spectra (traces “a” and “c”) and 13C-NMR spectra (traces “b” and “d”) of cholesteryl oleate (traces “a” and “b”) and human meibum (traces “c” and “d”) in the vinyl region. The spectra demonstrate that double bonds of unsaturated meibomian lipids exist predominantly in cis-configuration. Resonances which would point toward trans-double bonds (δ 32.5 ppm, 13C, and δ ~5.4 ppm, 1H) were not observed.
Panel D. Partial 1H-NMR spectra of human meibum and three fatty acid methyl ester standards in the ω-terminal region of the spectrum. Trace “a” – 1H-NMR spectrum of methyl stearate (straight chain); trace “b” – 1H-NMR spectrum of methyl (13-methyl)myristate (iso-branched); trace “c” – 1H-NMR spectrum of methyl (12-methyl)myristate (anteiso-branched); trace “d” – 1H-NMR spectrum of human meibum. The spectra demonstrate an unexpectedly high abundance of iso-branched lipids (characteristic resonances δ 0.848 and 0.831 ppm) compared to straight chain lipids (triplet δ 0.876, 0.859, and 0.841 ppm) in human meibum.
Panel E. Partial 13C-NMR spectrum of human meibum and three fatty acid methyl ester standards. Traces “a” and “a*” – 13C-NMR spectrum of human meibum; traces “b” and “b*” – methyl stearate (straight chain); traces “c” and “c*” – methyl (13-methyl)myristate (iso-branched); traces “d” and “d*” – ethyl (12-methyl)-myristate (anteiso-branched).
Panel F. Experimental (black) and deconvoluted (grey) 13C-NMR spectra of human meibum. Experimental notes: Resonance δ 22.66 ppm confirms the enrichment of meibum with iso-branched lipids, while δ 22.69 ppm – straight chain lipids. Note the relatively higher intensity of the resonance with δ 22.66, which might indicate a larger presence of branched lipids in meibum. The ratio of the peak areas of the two deconvoluted resonances δ 22.66/δ 22.69 was 2.7/1.0.
Panel G. 1H,13C-gHSQCAD NMR spectra of of human meibum (A, B) and standards (C–E).
Panel G1: full 2-D spectrum of human meibum; Panel G2: expanded portion of spectrum G1; Panel G3: signal of a terminal ω3-CH3 group (straight chain methyl stearate); Panel G4: signal of terminal -(CH3)2 [iso-branched methyl (13-methyl)myristate]; Panel G5: signal of ω3-CH3 group [anteiso-branched methyl (12-methyl)myristate].
We attempted to measure the levels of poly-unsaturated fatty chains in ML by monitoring resonances δ 25.2 ppm (methylene-interrupted cis-pentadienes) and δ 35.5 ppm (their trans-counterparts). However, no such signals were detected. It appears that the levels of lipids with polyunsaturated fatty chains in meibum were below LLoD of our current 1H- and 13C-NMR approaches. Also not found in meibum were conjugated dienes, which should have produced characteristic resonances in the region of the spectrum between δ 6.1 ppm and 5.4 ppm, but were not visible in the spectra of human meibum (not shown).
3.3.4. Studying extensive branching of meibomian lipids
Our earlier experiments [31] indicated that, unlike most other organs and tissues which make and/or accumulate mostly straight chain lipids, MG produced a mixture of WE with straight chain and branched FA moieties. In this paper, we corroborated this conclusion using 1-D 13C-, 1-D 1H-, and 2-D 1H,13C-gHSQCAD NMR approaches (Table S1 and Fig. 3, Panels D-G) as well GC-MS (Fig. S5) as follows.
To evaluate the degree of branching of meibomian lipids, 1H-NMR spectra of straight chain and branched lipid standards [specifically, methyl esters of stearic acid, (13-methyl)myristic acid, and (12-methyl)myristic acid] were recorded. The spectra showed distinctive differences in the resonances of their ω-CH3 groups (Fig. 3, Panel D). While comparing them with the corresponding regions of human meibum samples, we concluded that meibum is highly enriched with iso-branched lipids (δ 0.848 and 0.831 ppm), whose signals dominated resonances of other geometrical isomers – anteiso-branched (δ 0.842, 0824, 0.816, 0.806, and 0.801 ppm) and straight chain lipids (δ 0.876 and 0.831 ppm). If one assumes that the relative intensity of the proton resonances is proportional to the molar ratio of lipids in meibum, than the overall order of different structural isomers in human meibum should be as follows: iso-branched> straight chain>anteiso-branched, a rather unexpected finding.
Next, we recorded 13C-NMR spectra of human meibum and the same three standard fatty acid methyl esters (Fig. 3, Panel E), and compared them with each other and with published data [53, 54]. Two 13C resonances – δ 39.04 ppm and 22.66 ppm – were observed only in ML (Fig. 3, trace “a”) and the iso-branched lipid standard (trace “c”), and were assigned to their ω3 and ω1 carbons, correspondingly. No such signals were detected in the spectra of straight chain (trace “b”) and anteiso-branched (trace “d”) isomers. At the same time, a trio of resonances δ 36.62, 19.23 and 11.42 ppm detected in meibum corresponded well with resonances δ 36.62, 19.22 and 11.41 ppm of authentic anteiso-branched alkyl chains. An expected presence of straight chain lipids in meibum led to the raise of resonances δ 22.69 and 14.12 ppm, which were identical to those of a straight chain lipid standard (methyl stearate, trace “b”). Deconvolution of the 13C-NMR peaks of meibum in the region of 23 to 22 ppm (Fig. 3, Panel F) confirmed the presence of a large peak δ 22.66 (produced by terminal methyl groups of iso-branched lipids), and a smaller peak δ 22.69 ppm of ω2 methylene groups of straight chain lipids. Again, the apparent signals of iso-branched lipids were much stronger than those of straight chain ones, corroborating our 1H-NMR data.
The structural assignments of tested lipids were verified in 1H,13C-HSQCAD NMR experiments (Fig. 3, Panels G1–G5). An expanded portion (Panel G2) of the entire spectrum of human meibum (Panel G1) showed characteristic resonances of the same trio of structural isomers – straight chain lipids [δ {0.87,14.07} ppm], iso-branched chains [δ {0.84,22.65} ppm] and anteiso-branched [δ {0.83,11.37} and {0.81,19.19} ppm], which were close to those of standards, and to the values obtained in 13C NMR experiments described above. The peak δ 22.66 ppm indicated the presence of large amounts of iso-branched lipids in human meibum. Similar comparative analysis of spectral characteristics of methyl (12-methyl)myristate and ML confirmed the presence of anteiso-branched lipids in the secretion.
Independent confirmation of the presence of large amounts of branched FA in ML was obtained in GC-MS experiments. As an illustration, mouse WE with m/z 648.6 (M+) has been shown to be a complex mixture of straight chain and iso-branched C16:0 and C18:0 FA-based WE, whose structural assignments were made using previously described MS/MS approaches [31]. An almost exclusive anteiso-branching was observed for mouse C17:0 FA-based WE. For example, a major isoform of compound with m/z 634.6 (M+, Fig. S5) produced the following fragmentation pattern: 634→270→(241 + 213; ΔM=30 and 58, typical of anteiso-FA [31]). Still, a small portion of the compound produced a different ion with m/z 256.2 – a straight chain C16 fatty acid (not shown). Another commonly found product ion m/z 285.2 that originated in MS/MS experiments from a precursor ion m/z 648.6, fragmented to produce analytical ions with m/z values of 241 (i.e. being iso-branched saturated C18 FA [31]), and a set of ions that were characteristic of its straight chain isomer (not shown). Thus, MS and NMR experiments established that a large part of ML is iso- and anteiso-branched. It remains to be seen whether branching affects only FA residues, or FAL as well. The overall prevalence of the NMR signals of branched lipids implies that FAL might be branched as well.
3.3.5. Verification of the extreme lengths of meibomian lipids
Very and extremely long chain FA (VLCFA and ELCFA, respectively) have been observed in HPLC-MS/MS and GC-MS/MS fragmentation experiments as principal species in the following groups of ML of mice and humans: FFA, CE, OAHFA and CHL-OAHFA. The carbon chains of their FA residues approached C34 in all three classes of lipids. Omega-fatty acid moieties of OAHFA and CHL-OAHFA were in the C26 to C36 range. Very, and extremely, long chain FAL (VLCFAL and ELCFAL, respectively) moieties were observed as components of WE and DiAD. In WE, which had molecular masses between 534 and 661, and were based largely on C16 to C18 fatty acids, major FAL ranged from about C16 to C34, while α,ω-diols in DiAD – from C26 to C34.
3.3.6. Detecting ω-hydroxylated fatty acid and fatty α,ω-diols-based lipids
Though free ω-hydroxylated fatty acids (OHFA) have not been targeted in our experiments, their (O)-acylated products – OAHFA and CHL-OAHFA – were observed in meibum of humans and mice, and so were DiAD – products of di-(O)-acylation of fatty α,ω-diols. Both OHFA and α,ω-diols found in ML were of ELC nature, reaching C32-C34 in length. As levels of any FA with C>22 in blood are low, and decrease progressively with the increase in their carbon chain length [55, 56], we expect OHFA, OAHFA, and DiAD to be made in situ in MG.
3.3.7. Measuring enrichment of meibum with cholesteryl esters
The CHL and CE content of human and mouse meibum samples was determined using NP and RP HPLC-MS as described in Section 2.10 and in earlier publications [32, 35, 39]. The RP HPLC elution profiles of CE of both species were very close (Fig. 2, Panels C and D). In both species, the major CE were based on C18–C32 FA. The most abundant CE were those with C26-C28 FA residues, though FA residues of CE extended to at least C34. Similarly to WE, a large portion of saturated CE in mouse meibum were also based on FA with an odd number of carbons (C17:0 to C31:0). CHL-OAHFA, eluting with retention times (RT) between 29 and 35 min, were also similar between the species, both structurally and quantitatively. Their OHFA fragments ranged from C28 to at least C34 before (O)-acylation with C16 to C18 FA and esterification with CHL. In fresh human meibum samples, the amounts of free CHL varied (depending on the sample) from 0.05 to 0.5% of all lipids (w/w), while in mouse meibum it could approach 1.5–2% (w/w).
The MS analyses revealed the presence of CHL, VLC and ELC CE, and CHL-OAHFA as major components of meibum in both species. Note the highly similar natures of both secretions. The CE fraction contained compounds with their FA carbon chains ranging from C16 (minor peak with RT of around 11.6 min) to C32 (RT of 27.2 min), with the majority of FA moieties being in the C24 to C30 range.
3.3.8. Minor lipid components
SQL was rarely detected in mouse and human samples, and only in miniscule quantities. Two most sensitive analytical approaches – GC-MS and HPLC-MS – were utilized in an attempt to detect the compound. The amounts of detected SQL in the meibum of humans and mice varied substantially, but never exceeded those of CHL. Typically, the CHL:SQL ratio varied from 10:1 to virtually undetectable 100:1. This made SQL a very minor component of meibum (≪0.05%, w/w), whose presence could have been further exaggerated by inadvertent contamination of the study samples with skin lipids originated from eyelid margins or hands of the experimenters. Characteristically, no traces of SQL, which would produce 1H resonances δ 5.12, 5.14, 5.24, 5.27, and 5.46 ppm, were observed in 1H-NMR experiments with human meibum, either. Other hydrocarbons (both saturated and non-saturated ones) have never been found in meibum neither by MS, nor NMR.
Meibum was also checked for the presence of TAG, diacylglycerols (DAG) and monoacylglycerols. A standard TAG tripalmitin was used as a reference lipid. NMR spectra of TAG can be found at the Lipid Library website (http://lipidlibrary/aocs.org). In our hands, tripalmitin produced a characteristic signal of an sn-2 proton (δ 5.25 ppm, a triplet of triplets), and a pair of doublets of doublets (δ 4.27 and 4.17 ppm), indicative of protons sn-1 and sn-3. The 1H- and 13C-NMR data, obtained for human meibum, were consistent with the presence of small, but noticeable, amounts of TAG. The latter manifested themselves as distinctive proton resonances with chemical shifts δ 4.27, 4.25, 4.15, 4.13, 4.12, and 4.10 ppm. The absence of visible resonances δ 2.48 ppm (an sn-2 hydroxyl group of 1,3-DAG) and 3.97 and 4.2 ppm (sn-1 and sn-3 protons in 1,3-DAG) ruled out the existence of 1,3-DAG in the samples. No characteristic signals of monoacylglycerols (δ 4.18, 4.25, 3.97, 3.73, 3.64 ppm) were observed in the human samples, either. Thus, the only clearly observed type of acyl glycerols in the samples was TAG, mostly triolein. Finally, large amounts of various CER had been detected in skin lipid samples of mice [42, 43, 57], but not in the samples of mouse meibum (current experiments).
3.4. Gene expression analysis of major lipid-synthesizing enzymes in tarsal plates of mice and humans
To evaluate the nature of the major lipid-metabolizing enzymes in the TP of humans and mice, we conducted a genome-wide mRNA microarray analysis of their TP tissue samples.
Our data indicated that the most highly expressed genes in TP of mice were those related to the biosynthesis of ATP (Atp8), mitochondrial electron transport via NADH (Nd4/Nd5), Krebs cycle (Gm23935, encoding dihydrolipoamide succinyltransferase), and desaturation of fatty acids (Scd1), all ranking at 100%, followed by Gm10222 (or mt-Nd4l, encoding NADH dehydrogenase 4L; Rank 99.5%), and the elongase of very long chain fatty acids Elovl4 (Rank 98.6%).
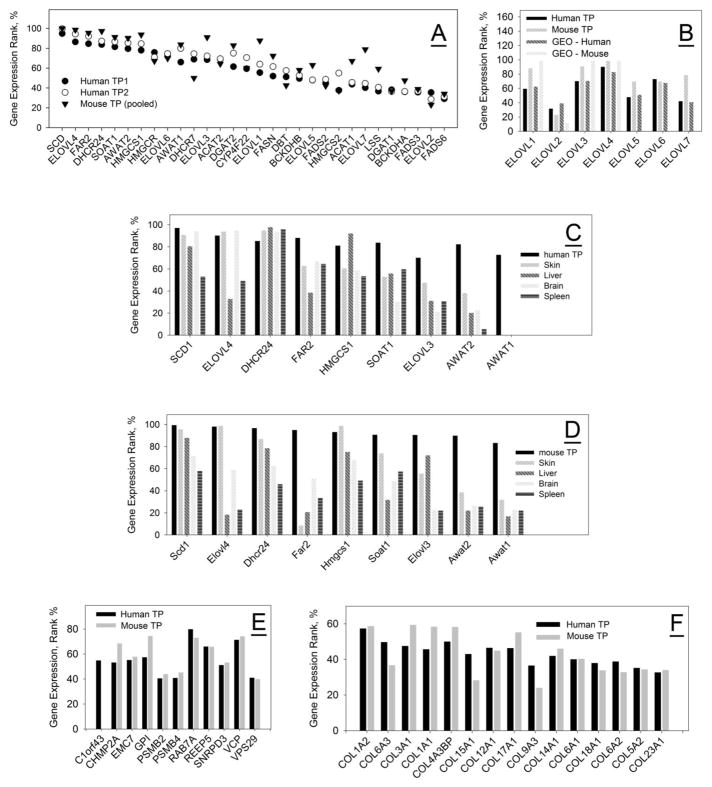
A list of highly expressed genes encoding major lipid-synthesizing enzymes in TP of humans and mice is shown in Table 3, and illustrated in Fig. 4, Panel A. Almost all entries shown fall into the category of highly expressed, identified genes, with Scd1, Elovl4, Fads2p1, Dhcr24, and Far2 topping the entire list with log2 values between 19 and 20 (the highest possible). Note a very tight pattern of expressed genes for human samples TP1 and TP2. Importantly, the expression levels of ELOVL1-ELOVL7 for humans and mice measured in our experiments were quite similar, and were close to the data that were retrieved from the NIH GEO dataset #GSE17822 (Fig. 4, Panel B).
Table 3.
Expression patterns of genes encoding major lipid-synthesizing enzymes in the tarsal plates of mice and humans
| Gene Symbol | Enzyme (EC number) | Expression Level (Rank Order, %) | |||
|---|---|---|---|---|---|
| Mice, NIH GEO Data (accession ## GDS 1009, 1136, 1832) | Mice, our data (average of five animals) | Humans, NIH GEO Data (accession # GSE 17822) | Humans, our data (average of two samples) | ||
| Acat1,2/Soat1,2 | Acetyl-CoA/sterol O-acyltransferases (2.3.1.9/26) | Moderate (15–30) | High (65/91) | Mod. to High (44, 55/58, 39) | High (69/84) |
| Awat1/Awat2 | Acyl-CoA wax alcohol acyltransferase (2.3.1.75) | No info | High (84/90) | No info | High (73/82) |
| Bckd (various) | Branched chain fatty acid synthase multienzyme complex | No info | High (≥50) | Mod. to High (44–65) | Mod. to High (37–53) |
| Cyp (various) | Cytochrome oxidases (1.14.-.-) | Mod. to High (35–100) | High (74–86) | High (74–86) | |
| Dgat1/Dgat2 | Diglyceride acyltransferase (2.3.1.20) | High (>70) | Mod./High (37/83) | High (77/77) | Mod./High (37/68) |
| Dbt | Dihydrolipoamide branched chain transacylase | Mod (42) | High (54) | ||
| Dhcr7/Dhcr24 | Dehydrocholesterol reductases (1.3.1.21/72) | High (>80) | High (50/97) | High (63/65) | High (72/85) |
| Elovl1 | Elongase of very long chain fatty acids (2.3.1.199) | High (100) | High (88) | High (63) | High (60) |
| Elovl2 | same as above | Low (<10) | Low to Mod. (23) | Mod. (39) | Mod. (32) |
| Elovl3 | same as above | High (100) | High (91) | High (71) | High (70) |
| Elovl4 | same as above | High (100) | High (99) | High (83) | High (90) |
| Elovl5 | same as above | No info | High (63) | High (51) | Mod. (48) |
| Elovl6 | same as above | No info | High (70) | High (68–73) | High (73) |
| Elovl7 | same as above | No info | High (79) | Mod. (41) | Mod. (42) |
| Fads1/Fads2 | Fatty acid desaturase (1.14.19.-) | Moderate (20–50) | Mod. (39/41) | Mod. to High (51/40) | Mod. (39/47) |
| Far1/Far2 | Fatty acyl-CoA reductase (1.2.1.50/84) | No info | High (82/96) | No info | High (52/88) |
| Fasn | Fatty acid synthase (2.3.1.85) | High (80–95) | High (72) | High (83) | High (57) |
| Hmgcr | 3-Hydroxy-3-methylglutaryl-CoA reductase (1.1.1.34/38) | High (95) | High (67) | High (71) | High (73) |
| Hmgcs1/Hmgcs2 | 3-Hydroxy-3-methylglutaryl-CoA synthase (2.3.3.10) | High (80) | High/Mod. (94/37) | High (85/54) | High (81/55) |
| Lss | Lanosterol synthase (5.4.99.7) | No info | High (59) | Mod. (39) | Mod. (41) |
| Scd (various) | Stearoyl-CoA desaturases (1.14.19.1) | High (90) | High (88–100) | High (93) | High (97) |
Figure 4. Analysis of gene expression patterns in the tarsal plates of humans and mice.
Panel A. Direct comparison of expression profiles of lipid metabolism-related genes in tarsal plates of humans (TP1 and TP2, circles) and mice (pooled samples; triangles). For clarity, only human genes are labeled.
Panel B. Expression profiles of ELOVL enzymes in human (average of two samples) and mouse tarsal tissues. Note that GEO datasets GDS 1832, 1361, and 1009, unlike our data, do not report any presence of ElovL5, Elovl6, and Elovl7 in MG of mice.
Panel C. Comparison of expression profiles of major lipid-synthesizing enzymes in selected human tissues (tarsal plates – our data; other tissues – from GEO dataset GDS3113)
Panel D. Comparison of expression profiles of major lipid-synthesizing enzymes in selected mouse tissues (tarsal plates – our data; other tissues – from GEO dataset GDS3334)
Panel E. Average expression profiles of reference house-keeping genes in tarsal plates of humans and mice.
Panel F. Direct comparison of average expression profiles of collagen-encoding genes in tarsal plates of humans and mice.
Importantly, the gene expression patterns in human and mouse TP were not replicated in other tissues, such as brain, liver, spleen, and skin (evaluated using sample datasets from Affymetrix and GEO datasets GDS3113 and 3334): the expression levels of these genes in different tissues varied from 2 to 1,000-fold (Fig. 4, Panels C and D). In case of some genes (e.g. AWAT/Awat, FAR1/Far1 and FAR/Far2), the expression levels in mouse brain, liver and spleen were only slightly above the background (Log2<5), indicating a low likelihood of corresponding enzymes playing a significant role in those tissues, but a high one – in TP. The skin was found to be the closest tissue to TP in terms of its gene expression pattern, but still featuring noticeably different levels of, for example, FAR2/Far2, SOAT1/Soat1, ELOVL3/Elovl3, AWAT2/Awat2 and AWAT1/Awat1, SCD4/Scd4, and other genes.
A side-by-side comparison of the TP expression patterns of lipid-metabolism related enzymes in mice and humans (Fig. 4, Panel A) revealed that not only were the lipid profiles of human and mouse meibum very close, but the major enzymes that we had predicted to be involved in meibogenesis, were found to be expressed in both species at similar levels as well. The Spearman rank order correlation coefficient between the gene expression patterns in humans and mice was calculated using the SigmaStat® statistical software package and found to be a high 0.8 (P-value of 2×10−7) and a normal distribution of data within both datasets. As the majority of ML are of VLC and ELC nature, we anticipated to detect high levels of expression of ELOVL1, 3, 4, 5, and 6, and much lower levels of ELOVL2 and 7 (Fig. 4, Panel B).
To validate the results presented above, we also measured the expression profiles of standard reference housekeeping genes (such as C1orf43, CHMP2A, EMC2, GPI, PSMB2, PSMB4, RAB7A, REEP5, SNRPD3, VCP, and VPS29 [58]) in TP of humans and mice (Fig. 4, Panel E). The expression data for humans and mice were distributed normally as confirmed by Kolmogorov-Smirnov tests using SigmaStat software. With the exception of C1orf43, which was not detected in mice, the rest of the reference genes in human and mouse TP were expressed almost identically with a Pearson product moment correlation coefficient of 0.9 (P-value <0.001) and a Spearman rank order correlation coefficient of 0.9 (P-value 0). Both results were indicative of a very strong correlation between the expression patterns of the standard reference genes in the two species.
To further validate our findings, we compared the gene expression profiles of structural proteins that contribute to cartilage and connective tissues, such as collagens, aggrecan (encoded in humans and mice by ACAN and Acan, correspondingly), tenascin (TNN and Tnn), and versican (VCAN and Vcan). These proteins were chosen as they had been previously identified in human TP using immunohistochemical approaches [5]. First, the collagen expression was evaluated. Humans are known to express dozens of different types of collagens [59, 60].
Fifteen most highly expressed collagen variants in TP of mice and humans were compared (Fig. 4, Panel F). The Kolmogorov-Smirnov test indicated a normal distribution of the data. The Pearson coefficient was a high 0.6 (P-value <0.015), while the Spearman coefficient was also a high 0.6 (P-value <0.01) – all indicative of a strong correlation between the gene expression profiles observed in TP of humans and mice. Next, we compared the expression levels of ACAN/Acan, TNN/Tnn, and VCAN/Vcan. Again, all three genes were expressed in both species with Rank Orders of 31% and 23% (ACAN and Acan, respectively), 31% and 26% (TNN and Tnn), and 44% and 38% (VCAN and Vcan).
3.5. Histochemical and immunohistochemical staining of human and mouse tarsal plate tissue samples
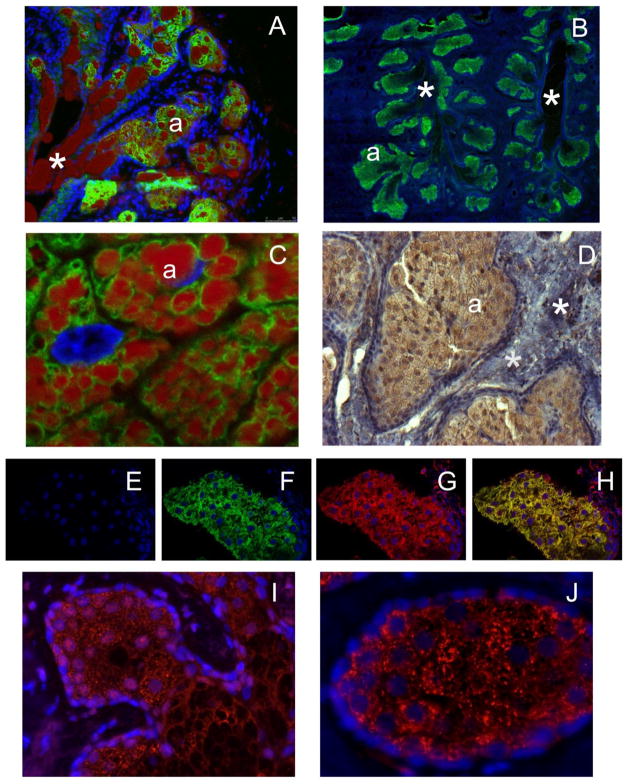
Our next step was to verify the gene profiling experiments by IHC. As all-inclusive IHC verification of the mRNA microarray data discussed above goes beyond the scope of this manuscript, targeted IHC analyses of selected proteins from Table 3 and Fig. 4 (specifically, ELOVL3, ELOVL4, BCKDHA, and DBT) was conducted instead. Previously, we reported the abundant expression of ELOVL4 protein in mouse TP [29]. A triply stained cryosection of a mouse TP, with a clearly visible central duct, is shown in Fig. 5, Panel A. Therefore, we anticipated that ELOVL4 and other enzymes of the ELOVL family would be highly expressed in human TP as well. Indeed, our IHC experiments demonstrated, for the first time, the abundant expression of ELOVL3 and ELOVL4 proteins in human TP (Fig. 5, Panels B–D). Note similarities in the distribution patterns of the anti-ELOVL4 staining, which occurred in the ascini, but not within the ducts, of mice and humans (Panels A and B). Also noticeable was accumulation of large quantities of lipid droplets in the meibocytes, but not within the connective tissue, in the immediate areas next to ELOVL4 (Panels A and C). These results fully corroborated our gene expression analysis data (above), are in agreement with our predictions about their involvement in meibogenesis (see below, Discussion), and support our hypothesis about similarities in meibogenesis in mice and humans. Three types of negative controls were tested. First, negative controls with no primary Ab showed no staining of the TP tissues of humans and mice. Second, the tarsal plates themselves, in the areas with no developed MG, did not show any ELOVL4-specific staining. Third, we used HEK293T cells without and with overexpressed human ELOVL4 (to be published elsewhere), and triply stained both types of cells with antibodies against human ELOVL4 and calnexin, as well as DAPI to locate the nuclei (Fig. S6). Cells with no overexpressed human ELOVL4 showed no specific staining, while the Ab clearly stained the ELOVL4-positive cells. Note that ELOVL4 staining clearly co-localized with a typical staining pattern of calnexin – a protein marker of endoplasmic reticulum (Fig. S6, Panel E), which replicates the staining pattern in the TP tissues. ELOVL4 was detected in the meibocytes that concentrated in the ascini of MG, but not within their ducts. The presence of ELOVL4 in the mouse TP was verified by Western blot experiments using published protocols [44]. The only protein band detected with the anti-mouse ELOVL4 antibody had a molecular mass of 32–33 KDa identical to the size reported before [44]. Anti-ELOVL3 staining pattern of the ascini was essentially similar to the anti-ELOVL4 staining (Fig. 5, Panel D), which became even more obvious when a tissue section of human TP was triply-stained for ELOV3, ELOVL4, and DAPI (Fig. 5, Panels E–H).
Figure 5. Immunohistochemical staining of tarsal plates of mice and humans.
Panel A. A cryosection of a mouse TP triply stained with anti-ELOVL4 antibodies (green; same antibodies as in [29]), lipid stain Oil Red O (red), and DAPI (blue). Cryosectioning was needed to preserve meibum droplets in the sample. Central duct section is shown. Labels: (*) – duct; (a) – ascinus. Image size: 690×550 μm
Panel B. A section of a human TP stained with anti-human ELOVL4 antibodies (green) and DAPI (blue). Labels: (*) – duct; (a) – ascinus. Image size: 90×90 μm.
Panel C. A cryosection of a human TP stained with anti-human ELOVL4 antibodies (green), LipidTOX (red), and DAPI (blue). Labels: (*) – duct; (a) – ascinus. Image size: 90×90 μm.
Panel D. A cryosection of a human TP stained with anti-human ELOVL3 antibodies (brown) and hematoxylin (purple-blue). Labels: (*) – connective tissue; (a) – ascinus. Image size: 444×332 μm.
Panels E–H. Triple-staining of human TP with DAPI (Panel E), anti-ELOVL4 Ab (Panel F, green), andti-ELOVL3 Ab (Panel G, red), and a merged image (Panel H). Confocal microscope. Image size: 185×127 μm.
Panel I. Anti-DBT (red) and DAPI (blue) staining of human TP. Image size: 221×166 μm.
Panel J. Anti-BCKDHA (red) and DAPI (blue) staining of human TP. Image size: 138×104 μm.
Finally, enzymes of the BKDH complex – branched chain alpha-keto acid dehydrogenase E1 component alpha (encoded by BCKDHA gene), and dihydrolipoamide branched chain transacylase E2 (encoded by DBT gene) were detected in the ascini of human MG, but not in the MG-free tissues of TP (Fig. 5, Panels I and K), and demonstrated very similar distribution patterns.
4. DISCUSSION
To establish plausible mechanisms of meibogenesis, we conducted an integrated evaluation of expression patterns of major lipid-synthesizing enzymes in TP, and correlated them with immunohistochemical and lipidomic data, expecting that this approach would help us in uncovering and mapping major biosynthetic pathways that are responsible for meibogenesis. Comprehensive evaluation of meibum, regardless of its source, has been a challenging task because of the diversity of lipids produced by MG. However, in this paper we will isolate and summarize key features of the meibomian lipidomes of mice and humans that differentiate meibum from other known lipid secretions and lipid-enriched tissues and organs. Our first working hypothesis was that the rather unique combination of features that characterize ML requires a very particular (and different from other tissues) combination of enzymes that are specific to TP. Our second hypothesis was that similarities in the composition of meibum of mice and humans might be strong indicators of similarities in their corresponding biosynthetic pathways.
As is evident from the facts presented and discussed above, meibum is a complex and rather unusual secretion formed of lipids that are extremely long, often branched, and rarely found in other cells and tissues. For example, the closest related secretion – sebum, produced by sebocytes in the epidermis (Fig. 1 and [26, 27] – differs markedly from meibum. Unlike meibum, human sebum is enriched with TAG, SQL, and FFA, and depleted of CE, while the mouse one is enriched with lanosterol and its derivatives, and is much lower in CE. Also, in our hands lipids of sebum were found to be considerably shorter than meibomian lipids (Fig. 1, Table 1). This corresponds well with published data of Lanz et al. [61], who demonstrated that the longest detected FA in sebaceous lipids was C26, and a paper by Pappas et al. [62], where FA from C12 to up to C22 were reported instead of C34 FA moieties in meibomian lipids (Table 2). Lastly, a detailed analysis of human sebaceous lipids conducted by Camera et al [63] demonstrated that FA residues of CE found in sebum ranged from C14 to C18, DAG were mostly of C30 to C35 nature (two FA residues combined), TAG barely reached C57 (three FA residues combined), while WE were based on C16:1 FA esterified to C12 to C26 FAL. Thus, FA and FAL found in meibum are, on average, substantially longer than those found in sebum, and the chemical diversity of ML seems to be greater (Table 2). These differences between sebum and meibum imply either 1) tissue-specific alterations in the expression patterns and the substrate/product specificity of known enzymes (or their isoforms), or 2) the expression and metabolic activity of still unknown enzymatic systems in MG. Let’s discuss the main features of ML.
Table 2.
Extremely long chain and branched lipids are major components of human and mouse meibum (this study and references [22–24]). Very and extremely long chain lipids are shown in bold text
| Lipid classes | Humans | Mice | ||
|---|---|---|---|---|
| WE | Major FA | Major FAL | Major FA | Major FAL |
| C16-C20; branched and straight; saturated and unsaturated | C22-C28 mostly saturated | C15-C20; branched and straight; saturated and unsaturated | C22-C28 mostly saturated | |
| CE | Major FA | none | Major FA | none |
| C18-C28 | C16-C28 | |||
| OAHFA (FA:OHFA) | Major FA | Major OHFA | Major FA | Major FA2 |
| C16, C18 | C28-C34 | C16, C18 | C28-C36 | |
| Free FA | C18-C32; straight and branched; saturated and unsaturated | |||
| DiAD (FA1:α,ω-Diol:FA2) | Major FA1, FA2 | Major α, ω-Diols | Major FA1, FA2 | Major α,ω-Diols |
| C16-C20 | C28-C34 | C16-C20 | C28-C36 | |
| CHL-OAHFA (FA:OHFA:CHL) | Major FA | Major OHFA | Major FA | Major FA2 |
| C16, C18 | C28-C34 | C16, C18 | C28-C36 | |
There are six main characteristics of ML, a combination of which differentiates them from lipids found in other human cells, tissues, and secretions, including sebum: 1) the extreme lengths of their FA and/or FAL residues [up to C34 and C36 (Table 2)]; 2) the abundance of branched lipids, especially iso-branched ones; 3) the high presence of odd-numbered FA (C15 to, at least, C31) and FAL (C19 to, at least, C33); 4) the high presence of ω-hydroxylated FA and α,ω-diols (both ELC in nature; up to C34 in length), primarily in the form of OAHFA, CHL-OAHFA, and DiAD; 5) the high content of WE, CE and CHL-OAHFA, and a low content of PL and TAG; and 6) the relatively low degree of unsaturation (mostly, one or two cis-double bonds per FA/FAL residue; all classes of lipids). These features are summarized in Table 2.
Importantly, experimental observations suggest that: 1) ELCFA are found in blood only in small quantities, and their presence drop with the increase in the chain length [55, 56, 64, 65], and 2) enzymes that are responsible for the biosynthesis of ELCFA and ELCFAL (e.g. various ELOVL and FAR1/FAR2) are abundantly expressed in human and mouse TP (see gene expression and IHC data, above). These facts imply that the vast majority of ML should be biosynthesized in MG de novo and/or in situ, and thus would require appropriate enzymes to be expressed and working in a concerted manner. Notably, only fragmentary information on the molecular enzymology of meibogenesis is currently available [66, 67].
It is also noteworthy that: 1) the gene expression profiles of major lipid-synthesizing enzymes of mice and humans are similar (Fig. 4), and 2) genes that encode enzymes that are needed for making compounds with the features listed in Table 4, are prominently expressed in human and mouse TP (Table 3 and Fig. 4). These similarities are especially striking considering that human and mouse samples were analyzed on different species-specific microarrays. Though not a guarantee of the identity of the molecular mechanisms involved (various post-translational modifications of expressed proteins, variability of the intracellular levels of substrates, cofactors, and effectors, and many other factors might influence the activity of enzymes of interest in a complex way), the similarities still make it possible to predict that the general biosynthetic reactions that define meibogenesis in humans and mice might be very similar. Supporting this conclusion are the facts that normal housekeeping genes are also expressed in TP of both species at similar levels (Fig. 4, Panels C and D), which makes comparison of the expression patterns of lipid-metabolism related genes even more relevant.
Table 4.
Major features of meibomian lipids and key enzymes responsible for their appearance. Predictions about sub-cellular localization of enzymes are based on previous studies of other organs and tissues.
| Feature | Relevant Enzymes And Their Predicted Localization in Meibocytes |
|---|---|
| Low degree of unsaturation of fatty chains (mostly, n-1 to n-3) | Δ9-SCD1, Δ5-FADS2 and Δ6-FADS1 (ER, NU) |
| Branching (“iso” and “anteiso”) | BCKA (MC), FASN (CP, MC) |
| High levels of odd-numbered FA in lipids | unknown |
| ω-Hydroxylation of FA and FAL | Various CYP (ER, MS) |
| High levels of VLC and ELC FA in lipids | ELOVL1-7 (ER) |
| High levels of ELC FAL in WE and DiAD | FAR1/FAR2 (PS) |
| High levels of sterols and their esters | HMGR (ER, PS), LSS (ER), Δ7-DHCR (ER, NU) ACAT (MC), LCAT (SEC), SOAT (ER, PM, MC) |
| High levels of WE | AWAT2/WES (ER, GA) |
| High levels of OAHFA and CHL-OAHFA | unknown |
CP – cytoplasm; GA – Golgi apparatus; ER – endoplasmic reticulum; MC – mitochondrium; MS – microsome; NU – nucleus; PS – peroxisome; SEC – secreted (e.g., blood plasma)
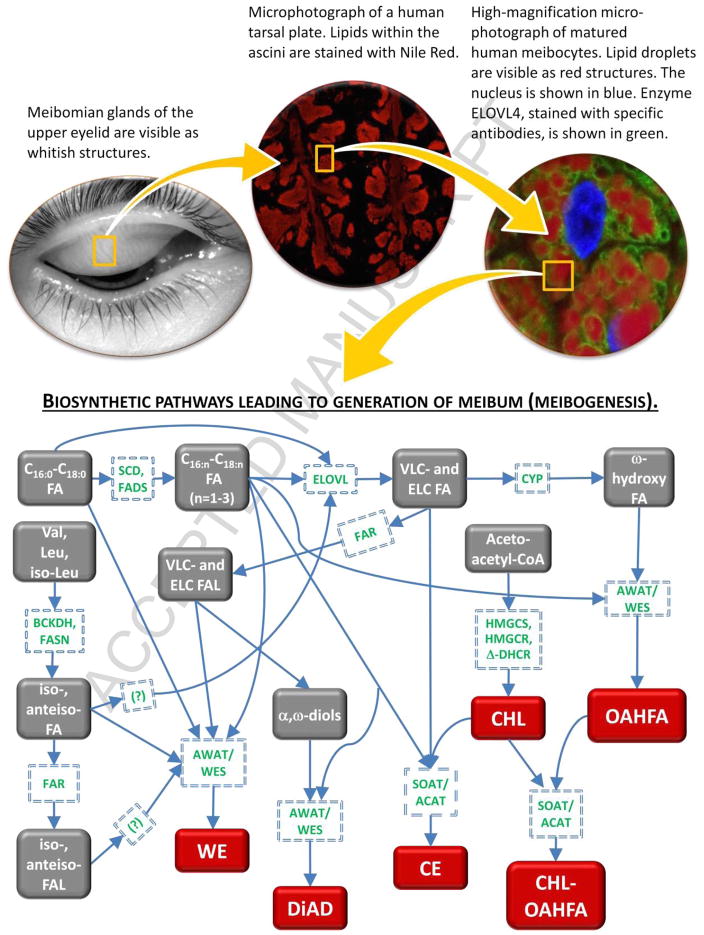
Thus, by combining results of the lipidomic and gene expression analyses described above, immunohisto-chemical data, structures and quantities of lipids found in meibum, and available literature data [4, 22–25, 29–39, 50, 51, 67–72], we expect the following major subsets of enzymatic reactions to be involved in meibogenesis (Fig. 6 and Table 4). Each subset of reactions shown in Fig. 6 leads to formation of either a structural element (such as ELCFA, branched FA, OHFA, CHL, etc.) of a more complex ML, or a final product – a complex lipid (WE, CE, OAHFA, CHL-OAHFA, DiAD, etc.). Each individual step in those reactions is catalyzed by an enzyme, or a group of enzymes, whose nature has been predicted based on: 1) gene expression patterns in the tarsus of mice and humans discussed above; and 2) similar studies conducted with other organs and tissues of humans, mice and other animals. Note that the proposed pathway addresses the existence of every class of lipids positively identified in our lipidomic experiments, with a possibility of adding new steps to describe compounds that are not included in this basic scheme.
Figure 6. Meibogenesis – a network of metabolic pathways in meibomian glands of humans and mice.
Unknown and yet unconfirmed hypothetical metabolic steps are marked with question marks. Major final products of meibogenesis (WE, DiAD, CE, CHL, OAHFA, CHL-OAHFA) are shown in red.
It seems that one of the most critical steps in meibogenesis is elongation of C16-C18 FA by the ELOVL family of enzymes. The major reason for this conclusion is the established fact that most of the tested lipid classes have moieties (either FA, or FAL) of very and extremely long chain nature (Table 2). Thus, they must be elongated before incorporation into complex lipids such as WE, CE, DiAD, OAHFA, etc. This makes elongation a critically important, key step in meibogenesis. Another critical step that may or may not be related to the elongation reactions per se is biosynthesis of branched FA. So far, we confirmed iso- and anteiso-branching for medium-length saturated FA (such as C17 FA, which is the most abundant anteiso-branched FA in meibum). We hypothesize that longer chain FA and FAL are also branched, which the apparent domination of the NMR signals of branched lipids (Fig. 3, Panel F) favors.
If this is the case, then the branching step should precede the elongation step, as our Fig. 6 illustrates. Finally, the very existence of a pool of major ω-hydroxylated FA and α,ω-diols – structural elements of OAHFA, CHL-OAHFA, DiAD, and, possibly, even more complex lipids – makes enzymes that are involved in their synthesis interesting, important, and totally unexplored subjects.
It is not known at this juncture if the enzymes that are involved in meibogenesis (such as those shown in Tables 2–4 and Figs. 4 and 6) differ in any way from similar enzymes in other cells and tissues. What is important is the uniqueness of their substrates and products, more specifically, their lengths and branching. Human ML are longer than even already highly elongated lipids of sebum (Fig. 1 and references [61–63]), which might be caused by either alterations in the substrate and/or product specificity of known enzymes through effectors or post-translational modifications, or the existence of isozymes with altered structures and catalytic activities. The same can be argued for the branched lipids, as we do not know how branching of FA and FAL impacts specificity of enzymes that elongate FA or produce FAL, WE, CE, OAHFA, CHL-OAHFA, DiAD and other lipids found in meibum. We are confident that future studies will answer these questions. There is no doubt that careful examination of meibum using ever more sensitive techniques will result in detection of new compounds, especially those with regulatory and signaling functions. Nevertheless, the compounds listed above form the backbone of meibum, and are likely to be responsible for its physiological roles.
Finally, in this manuscript, we were not concerned with regulation of meibogenesis, differentiation of meibocytes, or developmental aspects of MG themselves (which were subjects of a few recent papers, for example [3, 73–76]). However, there have been no reports so far that would demonstrate that MG-derived cell cultures in vitro were able to produce lipids that would be identical (or even similar) to the mature MG secretions described in this paper. Indeed, the reported lipidomes of the cell cultures were much closer to the regular lipid profiles of non-differentiated epithelial cells that are enriched with PL and TAG, but lack WE, CE and other typical components of meibum [74–76]. Thus, it remains unknown what regulatory factors are responsible for triggering meibogenesis, which our current mechanistic approach (Fig. 6) may help with by identifying possible targets for stimulating proper biosynthetic steps.
Supplementary Material
HIGHLIGHTS.
Biosynthesis of lipid-rich Meibomian gland secretions (meibum) has been elucidated.
Chemical composition of meibum is vastly different from other lipid-rich secretions.
Meibum is synthesized in meibocytes via a complex network of reactions which we call meibogenesis.
We propose a unified view of meibogenesis in humans and mice.
Acknowledgments
The authors would like to thank Dr. Robert E. Anderson (University of Oklahoma Health Sciences Center) for providing us with anti-mouse ELOVL4 antibodies, Dr. Robert S. Molday (University of British Columbia) for anti-human ELOVL4 antibodies used in our immunohistochemical studies, and Dr. David Chuang (UT Southwestern Medical Center) for anti-BCKDHA and anti-DBT antibodies. We gratefully acknowledge help of Ms. Janie Burroughs and Mr. Mike Molai – our clinical research coordinators – in gathering human meibum samples. The gene expression patterns in human and mouse tarsal plates were analyzed in the Genomics and Microarray Core facility at the UT Southwestern Medical Center with a help of Drs. Indu Raman and George Adams.
The project has been supported in part by an NIH grant R01EY019480 (to IAB), a Core grant for Vision Research P30EY020799, and an unrestricted grant from Research to Prevent Blindness (New York, NY).
IAB designed research, performed chromatographic and mass spectrometric experiments, analyzed NMR and gene expression data, and wrote the manuscript; AMcM collected mouse meibum and tarsal plate samples and performed immunohistochemical experiments; JCW collected human and mouse meibum samples and analyzed the data; FL performed NMR experiments; RM and KI collected human tarsal plate tissue. All authors discussed the data, participated in editing and proof-read the manuscript.
ABBREVIATIONS
- APCI
atmospheric pressure chemical ionization
- CE
cholesteryl ester
- CER
ceramide
- CHL
cholesterol
- CHL-OAHFA
cholesteryl ester of (O-acyl)-ω-hydroxy fatty acid
- DAG
diacylglycerols
- DE
dry eye
- DiAD
diacylated fatty α,ω-diol
- EI
electron impact
- ESI
electrospray ionization
- ELC
extremely long chain
- FAL
fatty alcohol
- FFA
free fatty acid
- MG
Meibomian gland
- ML
meibomian lipids
- OAHFA
(O-acyl)-ω-hydroxy fatty acid
- OHFA
ω-hydroxy fatty acid
- PL
phospholipid
- SM
sphingomyelin
- SQL
squalene
- STE
sterol
- TAG
triacylglycerol
- TF
tear film
- TFLL
tear film lipid layer
- TP
tarsal plate
- UFA
unsaturated fatty acid
- VLC
very long chain
- WE
wax ester
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meibom H. De vasis palpebrarum novis epistolae. H. Muller; Helmstadt: 1666. [Google Scholar]
- 2.Knop E, Knop N. Meibomian glands. Part I: anatomy, embryology and histology of the Meibomian glands. Ophthalmologe. 2009;106:872–873. doi: 10.1007/s00347-009-2006-1. [DOI] [PubMed] [Google Scholar]
- 3.Nien CJ, Massei S, Lin G, Liu H, Paugh JR, Liu CY, Kao WW, Brown DJ, Jester JV. The development of meibomian glands in mice. Mol Vis. 2010;16:1132–1140. [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolaides N, Kaitaranta JK, Rawdah TN, Macy JI, Boswell FM, 3rd, Smith RE. Meibomian gland studies: comparison of steer and human lipids. Invest Ophthalmol Vis Sci. 1981;20:522–536. [PubMed] [Google Scholar]
- 5.Milz S, Neufang J, Higashiyama I, Putz R, Benjamin M. An immunohistochemical study of the extracellular matrix of the tarsal plate in the upper eyelid in human beings. J Anat. 2005;206:37–45. doi: 10.1111/j.0021-8782.2005.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bron AJ, Tiffany JM, Gouveia SM, Yokoi N, Voon LW. Functional aspects of the tear film lipid layer. Exp Eye Res. 2004;78:347–360. doi: 10.1016/j.exer.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Olson MC, Korb DR, Greiner JV. Increase in tear film lipid layer thickness following treatment with warm compresses in patients with meibomian gland dysfunction. Eye Contact Lens. 2003;29:96–99. doi: 10.1097/01.ICL.0000060998.20142.8D. [DOI] [PubMed] [Google Scholar]
- 8.Foulks GN. The correlation between the tear film lipid layer and dry eye disease. Surv Ophthalmol. 2007;52:369–374. doi: 10.1016/j.survophthal.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Paananen RO, Rantamäki AH, Holopainen JM. Anti-evaporative mechanism of wax esters: implications for the function of tear fluid. Langmuir. 2014;30:5897–5902. doi: 10.1021/la501678t. [DOI] [PubMed] [Google Scholar]
- 10.Mudgil P. Antimicrobial role of human meibomian lipids at the ocular surface. Invest Ophthalmol Vis Sci. 2014;55:7272–7277. doi: 10.1167/iovs.14-15512. [DOI] [PubMed] [Google Scholar]
- 11.Cher I. Fluids of the ocular surface: concepts, functions and physics. Clin Experiment Ophthalmol. 2012;40:634–643. doi: 10.1111/j.1442-9071.2012.02758.x. [DOI] [PubMed] [Google Scholar]
- 12.Montes-Mico R. Role of tear film in the optical quality of the human eye. J Cataract Refract Surg. 2007;33:1631–1635. doi: 10.1016/j.jcrs.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Knop E, Knop N, Schirra F. Meibomian glands. Part II: physiology, characteristics, distribution and function of meibomian oil. Ophthalmologe. 2009;106:884–892. doi: 10.1007/s00347-009-2019-9. [DOI] [PubMed] [Google Scholar]
- 14.Green-Church KB, Butovich IA, Willcox M, Borchman D, Paulsen F, Barabino S, Glasgow BJ. The international workshop on meibomian gland dysfunction: report of the subcommittee on tear film lipids and lipid-protein interactions in health and disease. Invest Ophthalmol Vis Sci. 2011;52:1979–1993. doi: 10.1167/iovs.10-6997d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolaides N, Flores A, Santos EC, Robin JB, Smith RE. The lipids of chalazia. Invest Ophthalmol Vis Sci. 1988;29:482–484. [PubMed] [Google Scholar]
- 16.Wojtowicz JC, Butovich IA, McMahon A, Hogan RN, Itani KM, Mancini R, Molai M, Linsenbardt E. Time-dependent degenerative transformations in the lipidome of chalazia. Exp Eye Res. 2014;127:261–269. doi: 10.1016/j.exer.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knop E, Knop N. Meibomian glands. Part IV: Functional interactions in the pathogenesis of meibomian gland dysfunction. Ophthalmologe. 2009;106:980–987. doi: 10.1007/s00347-009-2044-8. [DOI] [PubMed] [Google Scholar]
- 18.Galor A, Feuer W, Lee DJ, Florez H, Carter D, Pouyeh B, Prunty WJ, Perez VL. Prevalence and risk factors of dry eye syndrome in a United States veterans affairs population. Am J Ophthalmol. 2011;152:377–384. doi: 10.1016/j.ajo.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gayton JL. Etiology, prevalence, and treatment of dry eye disease. Clin Ophthalmol. 2009;3:405–412. doi: 10.2147/opth.s5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry HD, Donnenfeld ED. Topical 0.05% cyclosporin in the treatment of dry eye. Expert Opin Pharmacother. 2004;5:2099–2107. doi: 10.1517/14656566.5.10.2099. [DOI] [PubMed] [Google Scholar]
- 21.Lane SS, DuBiner HB, Epestein RJ, Ernest PH, Greiner JV, Hardten DR, Holland EJ, Lemp MA, McDonald JE, 2nd, Silberd DJ, Blackie CA, Stevens CA, Bedi R. A new system, the LipiFlow, for the treatment of meibomian gland dysfunction. Cornea. 2012;31:396–404. doi: 10.1097/ICO.0b013e318239aaea. [DOI] [PubMed] [Google Scholar]
- 22.Butovich IA. Tear film lipids. Exp Eye Res. 2013;117:4–27. doi: 10.1016/j.exer.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butovich IA. Lipidomics of human meibomian gland secretions: Chemistry, biophysics, and physiological role of meibomian lipids. Prog Lipid Res. 2011:278–301. doi: 10.1016/j.plipres.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butovich IA. The Meibomian puzzle: combining pieces together. Prog Retin Eye Res. 2009;28:483–498. doi: 10.1016/j.preteyeres.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butovich IA, Millar TJ, Ham BM. Understanding and analyzing meibomian lipids – a review. Curr Eye Res. 2008;33:405–420. doi: 10.1080/02713680802018419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinde E, Haslam IS, Schneider MR, Langan EA, Kloepper JE, Schramm C, Zouboulis CC, Paus R. A practical guide for the study of human and murine sebaceous glands in situ. Exp Dermatol. 2013;22:631–637. doi: 10.1111/exd.12207. [DOI] [PubMed] [Google Scholar]
- 27.Smith KR, Thiboutot DM. Thematic review series: skin lipids. Sebaceous gland lipids: friend or foe? J Lipid Res. 2008;49:271–281. doi: 10.1194/jlr.R700015-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Mouse Genome Informatix database. http://www.informatics.jax.org (downloaded 02/15/2016)
- 29.McMahon A, Lu H, Butovich IA. A role for ELOVL4 in the mouse meibomian gland and sebocyte cell biology. Invest Ophthalmol Vis Sci. 2014;55:2832–2840. doi: 10.1167/iovs.13-13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butovich IA, Lu H, McMahon A, Eule JC. Toward an animal model of the human tear film. Biochemical comparison of the mouse, canine, rabbit and human meibomian lipidomes. Invest Ophthalmol Vis Sci. 2012;53:6881–6896. doi: 10.1167/iovs.12-10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butovich IA, Arciniega JC, Lu H, Molai M. Evaluation and quantitation of intact wax esters of human meibum by gas-liquid chromatography-ion trap mass spectrometry. Invest Ophthalmol Vis Sci. 2012;53:3766–3781. doi: 10.1167/iovs.11-9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butovich IA, Uchiyama E, McCulley JP. Lipids of human meibum: Mass-spectrometric analysis and structural elucidation. J Lipid Res. 2007;48:2220–2235. doi: 10.1194/jlr.M700237-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Butovich IA. Lipidomic Analysis of Human Meibum Using HPLC-MSn. Methods Mol Biol. 2009;579:221–246. doi: 10.1007/978-1-60761-322-0_11. [DOI] [PubMed] [Google Scholar]
- 34.Butovich IA, Wojtowicz JC, Molai M. Human tear film and meibum. 1. Very long chain wax esters and (O-acyl)-omega-hydroxy fatty acids of meibum. J Lipid Res. 2009;50:2471–2485. doi: 10.1194/jlr.M900252-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butovich IA. Cholesteryl esters as a depot for very long chain fatty acids in human meibum. J Lipid Res. 2009;50:501–513. doi: 10.1194/jlr.M800426-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butovich IA. On the lipid composition of human meibum and tears: Comparative analysis of nonpolar lipids. Invest Ophthalmol Vis Sci. 2008;49:3779–3789. doi: 10.1167/iovs.08-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wojtowicz JC, Butovich IA, Uchiyama E, Aronowicz J, Agee S, McCulley JP. Pilot, prospective, randomized, double-masked, placebo-controlled clinical trial of an omega-3 supplement for dry eye. Cornea. 2011;30:308–314. doi: 10.1097/ICO.0b013e3181f22e03. [DOI] [PubMed] [Google Scholar]
- 38.Butovich IA, Uchiyama E, DiPascuale M, McCulley JP. Liquid chromatography-mass spectrometric analysis of lipids present in human meibomian gland secretions. Lipids. 2007;42:765–776. doi: 10.1007/s11745-007-3080-2. [DOI] [PubMed] [Google Scholar]
- 39.Butovich IA. Fatty acid composition of cholesteryl esters of human meibomian gland secretions. Steroids. 2010;75:726–733. doi: 10.1016/j.steroids.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarys P, Barel A. Quantitative evaluation of skin surface lipids. Clin Dermatol. 1995;13:307–321. doi: 10.1016/0738-081x(95)00079-u. [DOI] [PubMed] [Google Scholar]
- 41.Ashraf Z, Pasha U, Greenstone V, Akbar J, Apenbrinck E, Foulks GN, Borchman D. Quantitation of human sebum on skin and human meibum on the eye lid margin using Sebutape®, spectroscopy and chemical analysis. Curr Eye Res. 2011;36:553–562. doi: 10.3109/02713683.2011.574331. [DOI] [PubMed] [Google Scholar]
- 42.McMahon A, Butovich IA, Mata NL, Klein M, Ritter R, 3rd, Richardson J, Birch DG, Edwards AO, Kedzierski W. Retinal pathology and skin barrier defect in mice carrying a Stargardt disease-3 mutation in elongase of very long chain fatty acids-4. Mol Vis. 2007;13:258–272. [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon A, Butovich IA, Kedzierski W. Epidermal expression of an Elovl4 transgene rescues neonatal lethality of homozygous Stargardt disease-3 mice. J Lipid Res. 2011;52:1128–1138. doi: 10.1194/jlr.M014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agbaga MP, Brush RS, Mandal MN, Henry K, Elliott MH, Anderson RE. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc Natl Acad Sci U S A. 2008;105:12843–12848. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grayson C, Molday RS. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J Biol Chem. 2005;280:32521–32530. doi: 10.1074/jbc.M503411200. [DOI] [PubMed] [Google Scholar]
- 46.Fisher CW, Chuang JL, Griffin TA, lau KS, Cox RP, Chuang DT. Molecular phenotypes in cultured maple syrup urine disease cells. Complete E1 alpha cDNA sequence and mRNA and subunit contents of the human branched chain alpha-keto acid dehydrogenase complex. J Biol Chem. 1989;264:3448–3453. [PubMed] [Google Scholar]
- 47.Lau KS, Griffin TA, Hu CW, Chuang DT. Conservation of primary structure in the lipoyl-bearing and dihydrolipoyl dehydrogenase binding domains of mammalian branched-chain alpha-keto acid dehydrogenase complex: molecular cloning of human and bovine transacylase (E2) cDNAs. Biochemistry. 1988;27:1972–1981. doi: 10.1021/bi00406a025. [DOI] [PubMed] [Google Scholar]
- 48.User Guide. GeneChip® WT PLUS Reagent Kit. P/N 703147. Rev. 4 (from: http://www.affymetrix.com)
- 49.Liu S, Richards SM, Lo K, Hatton M, Fay A, Sullivan DA. Changes in gene expression in human meibomian gland dysfunction. Invest Ophthalmol Vis Sci. 2011;52:2727–2740. doi: 10.1167/iovs.10-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butovich IA, Borowiak AM, Eule JC. Comparative HPLC-MS analysis of canine and human meibomian lipidomes: many similarities, a few differences. Sci Rep. 2011;1:24. doi: 10.1038/srep00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicolaides N, Santos EC. The di- and triesters of the lipids of steer and human meibomian glands. Lipids. 1985;20:454–467. doi: 10.1007/BF02534237. [DOI] [PubMed] [Google Scholar]
- 52.Butovich IA, Reddy CC. Enzyme-catalyzed and enzyme-triggered pathways in dioxygenation of 1-monolinoleoyl-rac-glycerol by potato tuber lipoxygenase. Biochim Biophys Acta. 2001;1546:379–398. doi: 10.1016/s0167-4838(01)00162-5. [DOI] [PubMed] [Google Scholar]
- 53.Knothe G. Branched chain and cyclic fatty acids. 1H-NMR spectroscopy of fatty acids and their derivatives. ( http://lipidlibrary.aocs.org/Analysis/content.cfm?ItemNumber=40259)
- 54.Gunstone FD. Branched chain acids. 13C-NMR spectroscopy. ( http://lipidlibrary.aocs.org/Analysis/content.cfm?ItemNumber=40273)
- 55.Hsu WY, Lin WD, Hwu WL, Lai CC, Tsai FJ. Screening assay for very long chain fatty acids in human plasma with multiwalled carbon nanotube-based surface-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2010;82:6814–6820. doi: 10.1021/ac100772j. [DOI] [PubMed] [Google Scholar]
- 56.Johnson DW, Trinh MU, Oe T. Measurement of plasma pristanic, phytanic and very long chain fatty acids by liquid chromatography-electrospray tandem mass spectrometry for the diagnosis of peroxysomal disorders. J Chromat B Analyt Technol Biomed Life Sci. 2003;798:159–162. doi: 10.1016/j.jchromb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 57.Ebel P, Imgrund S, Vom Dorp K, Hofmann K, Maier H, Drake H, Degen H, Dormann P, Echhardt M, Franz T, Willecke K. Ceramide synthase 4 deficiency in mice causes lipid alterations in sebum and results in alopecia. Biochem J. 2014;461:147–158. doi: 10.1042/BJ20131242. [DOI] [PubMed] [Google Scholar]
- 58.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 59.Prockop DJ, Kivirikko KI. Collagens: molecular biology, diseases, and potential therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 60.Mienaltowski MJ, Birk DF. Structure, physiology, and biochemistry of collagens. Adv Exp Med Biol. 2014;802:5–29. doi: 10.1007/978-94-007-7893-1_2. [DOI] [PubMed] [Google Scholar]
- 61.Lanz C, Ledermann M, Slavik J, Idle Jeffrey R. The production and composition of rat sebum is unaffected by 3 Gy gamma radiation. Int J Radiat Biol. 2011;87:360–371. doi: 10.3109/09553002.2010.537432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pappas A, Johnsen S, Liu JC, Eisinger M. Sebum analysis of individuals with and without acne. Dermato-Endocrinology. 2009;1:157–161. doi: 10.4161/derm.1.3.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camera E, Ludovici M, Galante M, Sinagra JL, Picardo M. Comprehensive analysis of the major lipid classes in sebum by rapid resolution high-performance liquid chromatography and electrospray mass spectrometry. J Lipid Res. 2010;51:3377–3388. doi: 10.1194/jlr.D008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Dirbashi OY, Santa T, Rashed MS, Al-Hassnan Z, Shimozawa N, Chedrawi A, Jacob M, Al-Mokhadab M. Rapid UPLC-MS/MS method for routine analysis of plasma pristanic, phytanic, and very long chain markers of peroxisomal disorders. J Lipid Res. 2008;49:1855–1862. doi: 10.1194/jlr.D800019-JLR200. [DOI] [PubMed] [Google Scholar]
- 65.Lam C, Wong D, Cederbaum S, Lim B, Qu Y. Peanut consumption increases levels of plasma very long chain fatty acids in humans. Mol Genet Metab. 2012;107:620–622. doi: 10.1016/j.ymgme.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 66.Miyazaki M, Man WC, Ntambi JM. Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J Nutr. 2001;131:2260–2268. doi: 10.1093/jn/131.9.2260. [DOI] [PubMed] [Google Scholar]
- 67.Cheng JB, Russell DW. Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem. 2004;279:37789–37797. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lam SM, Tong L, Yong SS, Li B, Chaurasia SS, Shui G, Wenk MR. Meibum lipid composition in Asians with dry eye disease. PLoS One. 2011;6:e24339. doi: 10.1371/journal.pone.0024339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mori N, Fukano Y, Arita R, Shirakawa R, Kawazu K, Nakamura M, Amano S. Rapid identification of fatty acids and (O-acyl)-ω-hydroxy fatty acids in human meibum by liquid chromatography/high-resolution mass spectrometry. J Chromatogr A. 2013;1347:129–136. doi: 10.1016/j.chroma.2014.04.082. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, Green-Church KB, Nichols KK. Shotgun lipidomic analysis of human meibomian gland secretions with electrospray ionization tandem mass spectrometry. Invest Ophthalmol Vis Sci. 2010;51:6220–6231. doi: 10.1167/iovs.10-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicolaides N, Ruth EC. Unusual fatty acids in the lipids of steer and human meibomian gland excreta. Curr Eye Res. 1982–1983;2:93–98. doi: 10.3109/02713688208997682. [DOI] [PubMed] [Google Scholar]
- 72.Kolattukudy PE, Rogers LM, Nicolaides N. Biosynthesis of lipids by bovine meibomian glands. Lipids. 1985;20:468–474. doi: 10.1007/BF02534238. [DOI] [PubMed] [Google Scholar]
- 73.Nien CJ, Massei S, Lin G, Nabavi C, Tao J, Brown DJ, Paugh JR, Jester JV. Effects of age and dysfunction on human meibomian glands. Arch Ophthalmol. 2011;129:462–469. doi: 10.1001/archophthalmol.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sullivan DA, Liu Y, Kam WR, Ding J, Green KM, Shaffer SA, Hatton MP, Liu S. Serum-induced differentiation of human meibomian gland epithelial cells. Invest Ophthalmol Vis Sci. 2014;55:3866–3877. doi: 10.1167/iovs.13-13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S, Kam WR, Ding J, Hatton MP, Sullivan DA. Effect of growth factors on the proliferation and gene expression of human meibomian gland epithelial cells. Invest Ophthalmol Vis Sci. 2013;54:2541–2550. doi: 10.1167/iovs.12-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hampel U, Schroder A, Mitchell T, Brown S, Snikeris P, Garreis P, Kunnen C, Willcox M, Paulsen F. Serum-induced keratinization processes in an immortalized human meibomian gland epithelial cell line. PLoS One. 2015;10:e0128096. doi: 10.1371/journalpone.0128096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.