Abstract
BCL-XL is a dominant inhibitor of apoptosis and a significant anti-cancer drug target. Endogenous BCL-XL is integral to the mitochondrial outer membrane (MOM). Indeed, BCL-XL reconstituted in detergent-free lipid bilayer nanodiscs is anchored to the nanodisc lipid bilayer membrane by tight association of its C-terminal tail, while the N-terminal head retains the canonical structure determined for water-soluble, tail-truncated BCL-XL, with the surface groove solvent-exposed and available for BH3 ligand binding. To better understand the conformation and dynamics of this key region of BCL-XL we have developed methods for isolating the membrane-embedded C-terminal tail from its N-terminal head and for preparing protein suitable for structural and biochemical studies. Here, we outline the methods for sample preparation and characterization and describe previously unreported structural and dynamics features. We show that the C-terminal tail of BCL-XL forms a transmembrane α-helix that retains a significant degree of conformational dynamics. We also show that the presence of the intact C-terminus destabilizes the soluble state of the protein, and that the small fraction of soluble recombinant protein produced in E. coli is susceptible to proteolytic degradation of C-terminal residues beyond M218. This finding impacts the numerous previous studies where recombinant soluble BCL-XL was presumed to be full-length. Notably, however, the majority of recombinant BCL-XL produced in E. coli is insoluble and protected from proteolysis. This protein retains the complete C-terminal tail and can be reconstituted in lipid bilayers in a folded and active state.
Keywords: BCL-2, apoptosis, membrane, nanodisc, structure, NMR
1. INTRODUCTION
The BCL-2 family proteins are the principal regulators of the mitochondrial pathway to apoptosis, a major homeostatic process of programmed cell death whose deregulation is central to many human diseases [1, 2]. Their pro- and anti-apoptotic activities are regulated by a range of intermolecular interactions that converge at the mitochondrial outer membrane (MOM) and result in life or death decisions for the cell. As a dominant inhibitor of apoptosis, the BCL-2 family member BCL-XL is a significant anti-cancer drug target [3] and the subject of numerous studies aimed at understanding its molecular mechanism of action. BCL-XL is the major product of bcl-x, a gene that is highly conserved in vertebrate evolution, expressed in a wide variety of tissues and cell types, and overexpressed in many tumors where it acts to promote tumor cell survival, tumor formation and tumor resistance to chemotherapy [4]. The majority of BCL-XL localizes to intracellular membranes, primarily the MOM, but some protein is also found in the cytosol of some cell types [5–8].
The 233-residue sequence of BCL-XL encompasses the four, signature BCL-2 homology (BH) motifs that define the protein family, forming an N-terminal head domain followed by a C-terminal, hydrophobic tail that is required for membrane association and cytoprotective activity [9]. The structure of the head domain [10] defines the canonical globular fold of the BCL-2 family proteins [11], with six amphipathic α-helices folded around two central α-helices to form a surface-exposed groove that engages the BH3 motifs of its BCL-2 protein partners. Three alternative splice variants of bcl-x have been identified [5–7]. One form (bcl-x3) lacks the thirty C-terminal residues and expresses as a fully soluble rather than membrane-bound protein, a second form (bcl-xβ) possesses a shorter and completely different C-terminal sequence, and a third form (bcl-xs) expresses as a much shorter protein, missing residues 126–188 in helices 4–6 of the globular domain, and has cytotoxic rather than cytoprotective activity.
Most structural studies of BCL-2 proteins have required complete or partial deletion of the hydrophobic C-terminal tail to promote protein solubility [2, 11]. As a result, only limited molecular data are available for their active membrane-associated states. Recently, we described the conformation of membrane-associated BCL-XL, derived by NMR spectroscopy after reconstitution of recombinant, full-length protein in detergent-free lipid bilayer nanodiscs [12]. In this state, the protein is anchored to the membrane by tight association of the C-terminal tail with the lipid bilayer, and the N-terminal head retains the canonical structure determined for water-soluble, tail-truncated BCL-XL, with the surface groove solvent-exposed and competent for BH3 ligand binding. Notably, membrane-associated BCL-XL has enhanced BH3 binding affinity compared to its tail-truncated, soluble counterpart, indicating that BCL-2 protein-protein interactions may experience additional levels of regulation at the membrane-water interface, and underscoring the importance of understanding their structures and functions in this critical environment.
To this end, we have set out to characterize the structure of the membrane-anchored C-terminal tail of BCL-XL in atomic detail. Here we describe methods for generating samples of the tail of BCL-XL embedded in detergent-free lipid nanodiscs and in mixed lipid-DePC. Using NMR, we show that the tail forms a membrane-embedded α-helix with backbone dynamics that reflect the presence of varying degrees of conformational exchange in the N-terminal region linking it to the N-terminal head. These features of the tail help explain how cytoprotective BCL-XL can insert in the MOM post-translationally, after synthesis in the nucleus.
2. MATERIALS AND METHODS
Protein expression and purifications
Four DNA sequences, based on the gene encoding human BCL-XL (Genbank NM_001191) were cloned into the NcoI and HindIII restriction sites of the pET-28a plasmid, and expressed in E. coli BL21 cells. The expressed protein sequences all include an N-terminal His-tag (Fig. 1). To obtain isotopically labeled proteins, bacterial cells were grown at 37°C, in M9 minimal media containing (15NH4)2SO4 and/or 13C-glucose (Cambridge Isotope Laboratories). Protein expression was induced by adding 1 mM isopropyl-1-thio-β-D-galactopyranoside to the culture when the cell density reached OD600 = 0.6. After growing the cells for an additional 3 hrs, they were harvested by centrifugation (6,000 × g, 4°C, 15 min) and stored at −80°C overnight.
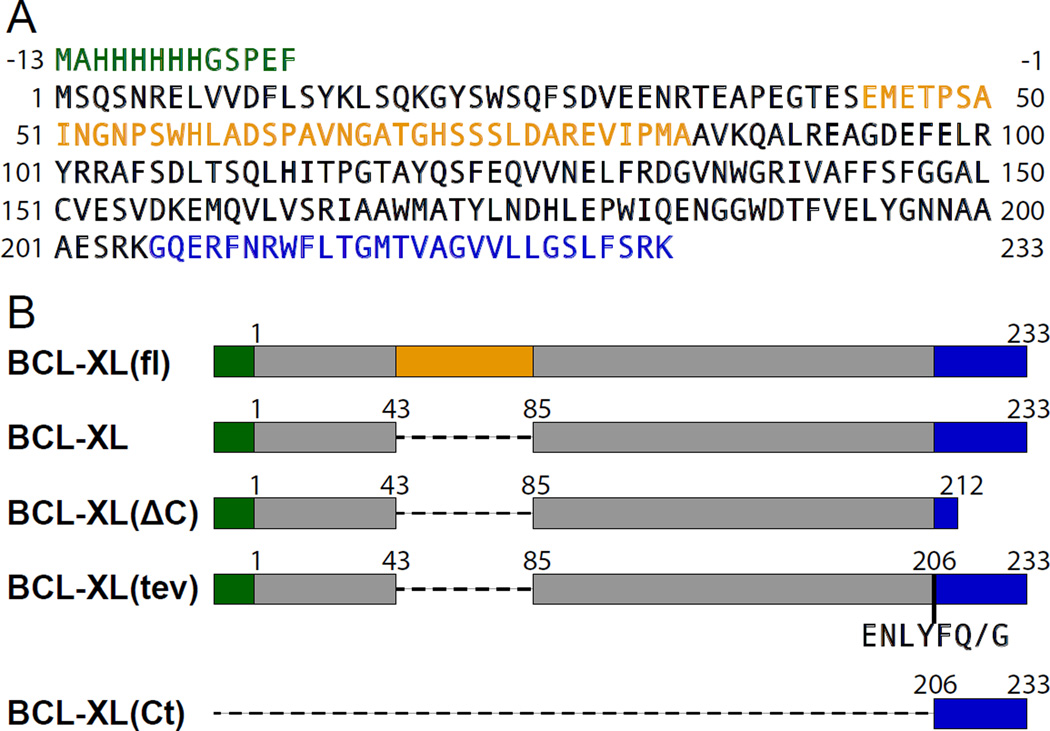
Figure 1. Amino acid sequence of human BCL-XL and recombinant polypeptides prepared in this study.
(A) Sequence of human BCL-XL color-coded to show: additional N-terminal residues encoding the His-tag (green), the flexible loop (gold) and the membrane-targeting C-terminus (blue). (B) Expressed sequences. BCL-XL(fl) is the full-length protein. BCL-XL lacks residues 44–84 in the flexible loop. BCL-XL(ΔC) lacks both loop residues 44–84 and C-terminal residues 213–233. BCL-XL(tev) lacks loop residues 44–84 and contains the TEVp recognition sequence (ENLYFQ/G) inserted before residue 206; TEVp cleaves between Q and G. BCL-XL(Ct) encompasses only the C-terminal tail.
BCL-XL(ΔC) (Fig. 1B) was purified as described previously [13]. BCL-XL and BCL-XL(fl) were purified in two fractions, as follows. Cells harvested from 1 L of culture were suspended in 30 mL of buffer A (25 mM Tris-Cl, pH 8, 100 mM NaCl), supplemented with protease inhibitors (cOmplete Mini EDTA-free cocktail; Roche), and lysed using a French Press. The soluble fraction was isolated as the supernatant from centrifugation after cell lysis, and purified first by Ni affinity chromatography (HisTrap FF 5 mL column, GE Healthcare) and then by ion exchange chromatography (HiPrep 16/10 Q FF column, GE Healthcare) with a linear gradient of NaCl in buffer B (20 mM Tris-Cl, pH 8, 2 mM DTT, 1 mM EDTA). Purified protein was stored at 4°C. The insoluble fraction, resulting from centrifugation of the cell lysate, was dissolved in 6 M urea, in buffer B, and then purified by size exclusion chromatography (HiPrep 16/60 Sephacryl S-200 column, GE Healthcare) followed by ion exchange chromatography (HiPrep 16/10 Q FF column, GE Healthcare) with a linear gradient of NaCl. Purified protein was precipitated by dialysis against water, lyophilized, and stored at −20°C.
Protein incorporation in lipid nanodiscs
BCL-XL and BCL-XL(tev) were incorporated in nanodiscs, prepared with the phospholipids (Avanti) dimyristoyl-phosphatidyl-choline (DMPC) and dimyristoyl-phosphatidyl-glycerol (DMPG), in 3/1 molar mixture, as described [12]. MSP1D1Δh5, the short form of membrane scaffold protein (MSP) used to prepare the nanodiscs, was prepared as described [14].
Briefly, three solutions were prepared and combined, as follows. Pure lyophilized BCL-XL protein (2–5 mg) was dissolved in 0.5 mL of nanodisc buffer (20 mM Tris-Cl, pH 7.5, 2 mM DTT, 1 mM EDTA) containing 100 mM n-decyl-phosphocholine (DePC; Anatrace). The lipids DMPC (13.4 mg) and DMPG (1.6 mg) were separately co-dissolved in 1 mL of nanodisc buffer containing Na-cholate (Anatrace) to obtain a final 1/2 molar ratio of lipid to cholate. Finally, purified MSP1D1Δh5 (1/50 molar ratio to total lipid) was separately dissolved in 700 µL of nanodisc buffer. The three solutions, each containing BCL-XL, lipid, or MSP1D1Δh5, were combined to obtain a final volume of 2.2 mL. After incubation at room temperature for 1 hr, 2 g of Biobeads (Biorad SM-2), prewashed in nanodisc buffer, were added, and the mixture was further incubated at room temperature, with gentle shaking overnight. The Biobeads were removed by centrifugation and the resulting nanodiscs were washed twice with one sample volume of nanodisc buffer. The nanodisc solution was concentrated using a 10 kD cutoff Vivaspin concentrator (Viva Products) and stored at 4°C.
Preparation of isolated membrane-associated C-terminal tail
The BCL-XL(Ct) peptide corresponding to C-terminal residues 206–233 was prepared by Tobacco Etch Virus protease (TEVp) cleavage of recombinant BCL-XL(tev). TEVp was expressed, purified and used as described [15]. To remove the N-terminal head and isolate pure, nanodisc-incorporated tail peptide, BCL-XL(tev) was first incorporated in nanodiscs and then incubated with TEVp, (8/1 molar ratio of TEVp to substrate) in cleavage buffer (20 mM Tris-Cl pH 7.5, 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA), for 12 hrs, at room temperature. The cleavage reaction was then transferred to buffer C (25 mM Tris-Cl pH 8, 150 mM NaCl) and mixed with 4 mL of Ni-NTA resin for 2 hrs, to isolate nanodiscs containing BCL-XL(Ct) in the unbound flow-through fraction, and remove the His-tagged N-terminus which binds to the resin. Pure, chemically synthesized, 15N Leu labeled BCL-XL(Ct) tail peptide was also obtained commercially (GenScript).
NMR experiments
Samples for NMR studies were transferred to NMR buffer (20 mM Na-phosphate pH 6.5, 2 mM DTT, 1 mM EDTA). For studies of BCL-XL(Ct) in mixed lipid-DePC, the purified BCL-XL(Ct) nanodiscs were supplemented with 170 mM DePC in NMR buffer. NMR samples of BCL-XL or BCL-XL(Ct) in pure detergent micelles were prepared by dissolving purified protein directly in 170 mM DePC, in NMR buffer.
Solution NMR experiments were performed on a Bruker AVANCE 600 MHz spectrometer equipped with a Bruker 1H/15N/13C triple-resonance cryoprobe. Backbone 1H and 15N chemical shifts were resolved and assigned using two-dimensional 1H/15N HSQC experiments and three-dimensional 1H/15N NOESY-HSQC experiments. The NMR pulse sequences are described in detail in the literature [16–21]. The NMR data were processed and analyzed using NMRPipe [22], NMRViewJ [23] and Sparky [24]. Solvent paramagnetic relaxation enhancement (PRE) experiments were performed by acquiring one- or two-dimensional 1H/15N NMR spectra after incremental additions of either lipid-soluble 16-doxyl-stearic acid (16-DSA; Sigma), or water-soluble Mn-EDTA. Additions were made from stock solutions of 100 mM 16-DSA in methanol or 200 mM Mn-EDTA in water.
Protein analysis
Analytical size exclusion chromatography was performed in NMR buffer with a Superdex 75 10/300 GL column (GE Healthcare) and a Breeze HPLC system (Waters). SDS polyacrylamide gel electrophoresis (PAGE) was performed with 4–12% Bis–Tris gels stained with Coomassie brilliant blue. Western immuno-blots were performed by loading protein solutions directly onto nitrocellulose, and visualized using antibody-conjugated alkaline phosphatase (Biorad), with 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt substrate, and nitro-blue tetrazolium chloride developer. His-tagged protein was probed with mouse anti-His monoclonal antibody (Qiagen; 1/5000 dilution). Nanodisc MSP1D1Δh5 was probed with goat anti-ApoA1 polyclonal antibody (Millipore; 1/1000 dilution).
Mass spectra were obtained using a Bruker Daltonics Autoflex II Matrix Assisted Laser Desorption/Ionization (MALDI) time of flight (TOF) spectrometer. Samples for MALDI were prepared by mixing 2 uL of protein solution (0.2 mg/mL of lyophilized protein in 25 mM Tris pH 8.0, 2 mM DTT, 1 mM EDTA, 200 mM NaCl) with MALDI matrix solution (20 mg/mL of Sinapic acid in aqueous 50% acetonitrile, 0.1% trifluoroacetic acid), loading the mixture onto a MALDI plate and allowing the solvents to evaporate for 15 min at room temperature.
3. RESULTS AND DISCUSSION
Preparation of soluble and membrane-associated BCL-XL
Upon gene expression in E. coli, recombinant BCL-XL accumulates predominantly (90% of total protein) in the insoluble cellular fraction and produces only a small amount (10%) of water-soluble protein (Fig. 2A). The soluble fraction can be readily purified to obtain folded protein with its C-terminal tail residues bound to the surface-exposed BH3-binding groove [12]. The insoluble fraction, on the other hand, can be purified to homogeneity under denaturing condition, and then refolded in the presence of lipids to yield a membrane-associated form of BCL-XL in which the globular head adopts the canonical BCL-2 tertiary fold, the BH3-binding groove is ligand-accessible and the C-terminus is tightly associated with the lipid bilayer membrane [12].
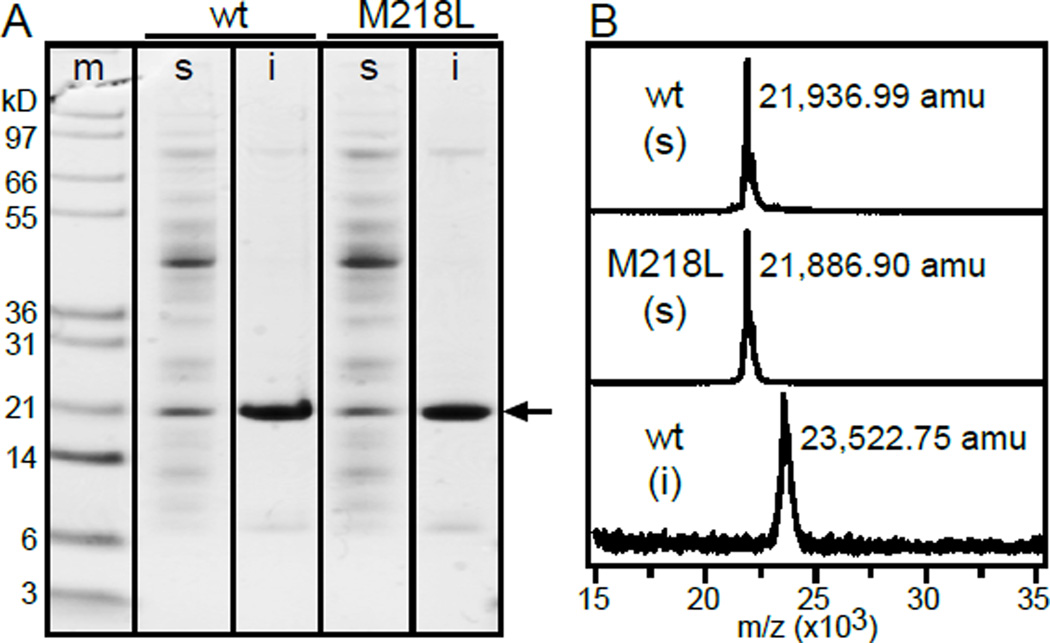
Figure 2. The soluble and insoluble fractions of recombinant BCL-XL produced in E. coli have different molecular masses.
(A) SDS-PAGE showing expression of wild-type (wt) or Met218 mutant (M218L) BCL-XL (arrow) in the soluble (s) and insoluble (i) E. coli cell fractions. Gels were stained with Coomassie blue. Molecular weight markers are shown in left lane (m). (B) MALDI mass spectra of 15N-labeled proteins purified from soluble (s) or insoluble (i) cell fractions. Molecular mass is given in atomic mass units (amu) derived from mass to charge (m/z) ratio.
Notably, mass spectrometry analysis reveals that the proteins purified from the soluble and insoluble fractions have different masses (Fig. 2B). The mass (21,936.99 Da) observed for 15N-labeld soluble protein is smaller than the theoretical mass (23,535.4 Da) expected for the expressed sequence of 15N BCL-XL after the common proteolytic removal of the N-terminal Met, but matches that of a sequence that is processed after both the initiating Met and M218. By contrast, the mass of BCL-XL purified from the insoluble cell fraction corresponds exactly to that of the expressed sequence, including the intact C-terminus, after removal of the initiating Met. Retention of the His-tag in both proteins is confirmed by Western-blotting with anti-His antibody.
Cleavage after M218 in the soluble protein is consistent with our previous observations [12] that several signals from the C-terminus could not be detected in the NMR spectra of soluble BCL-XL, that the NMR peak intensities of observable C-terminal signals reflect varying degrees of dynamic conformational exchange, and that the signal from M218 is very intense, with chemical shifts consistent with the highly dynamic conformations typically observed for C-terminal extremities. The mass spectra indicate that M218 is indeed the C-terminus of the purified soluble polypeptide. Similar NMR spectral features were also observed for the soluble fraction of full-length protein BCL-XL(fl), which, like its loop-deleted analog, is only slightly soluble in water and accumulates primarily in the insoluble cell fraction upon expression in E. coli [12].
The C-terminal tail of BCL-W, a related pro-survival protein that shares 44% sequence identity with BCL-XL, is also partially disordered relative to the well-defined helical conformation of the N-terminal head. NMR structures of BCL-W [25, 26] determined for soluble forms of the protein that were stabilized by point mutations and deletion of as few as ten tail residues, reveal that residues E157-L183 fold into the BH3-binding groove of the globular head. However, the C-terminus of the BCL-W loses helical order at residue S169 and appears to be fully disordered at A177, the sequence position that aligns with that of M218 in BCL-XL.
Attempts to inhibit C-terminal cleavage of BCL-XL, by adding high concentrations of protease inhibitors before and during cell lysis, resulted in similarly processed protein with the same, reduced mass, suggesting that C-terminal cleavage occurs in the bacterial cell, before any manipulations aimed at protein purification. Indeed, this appears to be what allows a small portion of BCL-XL to remain in solution, while BCL-XL that accumulates in the insoluble fraction is protected from proteolysis.
The active protease or lytic mechanism does not appear to be highly sequence-specific, since mutating M218 to Leu had no affect on cleavage and resulted in a polypeptide with the same reduced mass as wild-type (Fig. 2A, B). Cleavage at sequence position 218 is likely to be facilitated by the greater extent of dynamic conformational exchange observed in this region of the C-terminus by NMR [12], leading to the question whether similar processing might also occur endogenously in mammalian cells. In mammalian cells, protein [27–29] and/or lipid [30, 31] cofactors can help promote protein stability, prevent C-terminal cleavage and chaperone BCL-XL during its transition from cytosolic to membrane-associated protein. However, it is also interesting to note early reports [7] where Western blotting of tissue homogenates identified BCL-XL as two bands with molecular weights of about 29 and 31 kDa, that were not derived from splice variants.
As the cleavage site appears to be dictated more by the dynamic conformation of the C-terminus than sequence specificity, BCL-XL sequences expressed with other N-terminal tags may also be expected to undergo similar processing. The finding that soluble BCL-XL produced in bacteria is cleaved at the C-terminus impacts the many biochemical studies performed with recombinant, N-terminal-tagged protein, which was presumed to include all C-terminal residues. Thus, effects attributed to the C-terminal tail may need to be reinterpreted in light of C-terminal truncation.
The C-terminal tail of BCL-XL associates tightly with lipid bilayer nanodiscs
In most mammalian cells, BCL-XL is tightly associated with intracellular membranes, mostly the MOM. In vitro, the N-terminus of membrane-bound BCL-XL adopts the canonical BCL-2 globular fold and is anchored to the membrane through its C-terminus, while water-soluble protein that is cleaved after M218 fails to associate with lipid bilayers [12]. Structure determination of the C-terminal tail has been hindered by severe line broadening in the NMR spectra of BCL-XL bound to lipid bilayer nanodiscs. These nanodiscs were prepared with a short form of MSP, developed for solution NMR studies of membrane proteins, and have been shown to have an average diameter of 8 nm by electron microscopy [14]. Despite the relatively small size of the nanodiscs, and although the spectra of nanodisc-integrated BCL-XL display well-resolved signals from the N-terminal globular head of the protein, most signals from the C-terminus remain undetected [12].
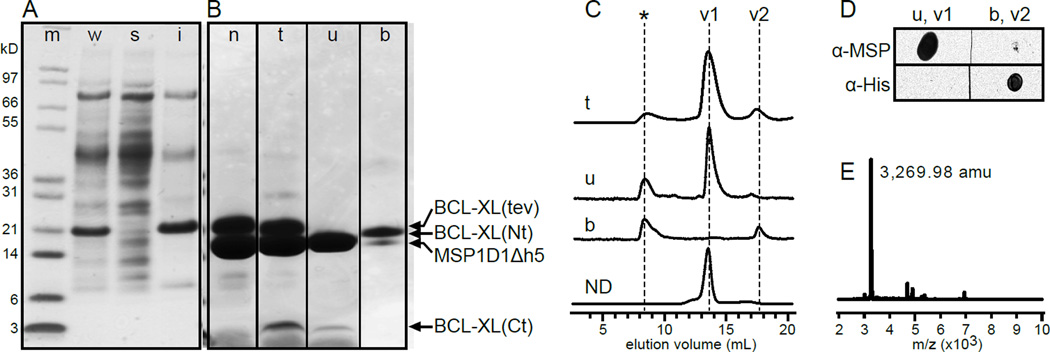
To simplify NMR spectral analysis and characterize the structure of the C-terminal tail we isolated it from the N-terminal head by inserting a TEVp recognition sequence between the two domains and separating them after treatment of the nanodisc-bound protein with TEVp. As observed for BCL-XL, the cleavable protein, BCL-XL(tev), accumulates predominantly in the insoluble E. coli cell fraction (Fig. 3A) and can be reconstituted into nanodiscs (Fig. 3B). Incubation with TEVp followed by Ni-NTA affinity purification removes the His-tagged N-terminal head and yields pure nanodisc-bound BCL-XL(Ct) encompassing tail residues G206-K233 (Fig. 3B). Analysis of the cleavage reaction and its Ni-NTA fractions, by SDS-PAGE (Fig. 3B), size exclusion chromatography (Fig. 3C), Western blot (Fig. 3D) and MALDI mass spectrometry (Fig. 3E), shows that the liberated nanodisc-anchored tail is present in the unbound Ni-NTA fraction (u), and elutes with a size exclusion volume (v1) similar to empty nanodiscs (ND). This fraction is devoid of His-tagged head but contains MSP1D1Δh5, as expected for the presence of nanodiscs (Fig. 3D), and its major polypeptide component has the exact mass (Fig. 3E) expected for the sequence of 15N-labeld BCL-XL(Ct). By contrast, the severed His-tagged head is found in the Ni-NTA bound fraction (b) and elutes later, with a size exclusion volume (v2) similar to that of soluble BCL-XL(ΔC) [12].
Figure 3. The C-terminus of BCL-XL associates tightly with lipid bilayer nanodiscs.
(A, B) SDS-PAGE analysis. Gels were stained with Coomassie blue. Molecular weight markers are shown in left lane (m). Arrows mark BCL-XL(tev), its cleavage products and nanodisc MSP1D1Δh5. (A) Expression of BCL-XL(tev) in E. coli whole cells (w), and in the soluble (s) and insoluble (i) cell fractions. (B) Purified BCL-XL(tev) reconstituted in nanodiscs, before treatment with TEVp (n), after cleavage by TEVp (t), and after cleavage by TEVp followed by incubation with Ni-NTA to obtain a flow-through unbound fraction (u) and a bound fraction (b) that elutes with imidazole. (C) Size exclusion chromatography analysis of Ni-NTA fractions t, u and b. Empty nanodiscs (ND) elute at volume v1. BCL-XL(ΔC) elutes at volume v2. Aggregated protein elutes early (*). (D) Western dot blots of the two major volume fractions (v1, v2) from size exclusion chromatography of Ni-NTA fractions u and b. Blots were probed with anti-MSP antibody to detect the nanodiscs, or anti-His antibody to detect the His-tagged N-terminus of BCL-XL. (E) MALDI mass spectrum of Ni-NTA unbound fraction (u). Molecular mass is given in atomic mass units (amu), derived from the mass to charge (m/z) ratio.
NMR Characterization of the C-terminal tail of BCL-XL
Isolated nanodisc-anchored BCL-XL(Ct) yields a 1H/15N correlation NMR spectrum (Fig. 4A, blue) that clearly shows signals from several backbone sites, including three Gly residues, the N-containing side chains of Q207, N210 and W213, and the K233 C-terminus. Removal of the N-terminal head yields a spectrum that is significantly simplified compared to that obtained for BCL-XL in nanodiscs (Fig. 4A, red). Nevertheless, the NMR lines are significantly broader than expected for a peptide of this size, even when considering the slower tumbling rate of the peptide-nanodisc assembly. Furthermore, the resonance line widths are highly heterogeneous, with narrower, strong peaks observed for residues in the C-terminus of the tail peptide, and broader, weak peaks observed for the N-terminus. Doubling is observed for some peaks, most prominently for the signal from the W213 sidechain (Fig. 4A, inset). These spectral features indicate that several sites in the isolated C-terminal tail experience varying degrees of dynamic conformational exchange.
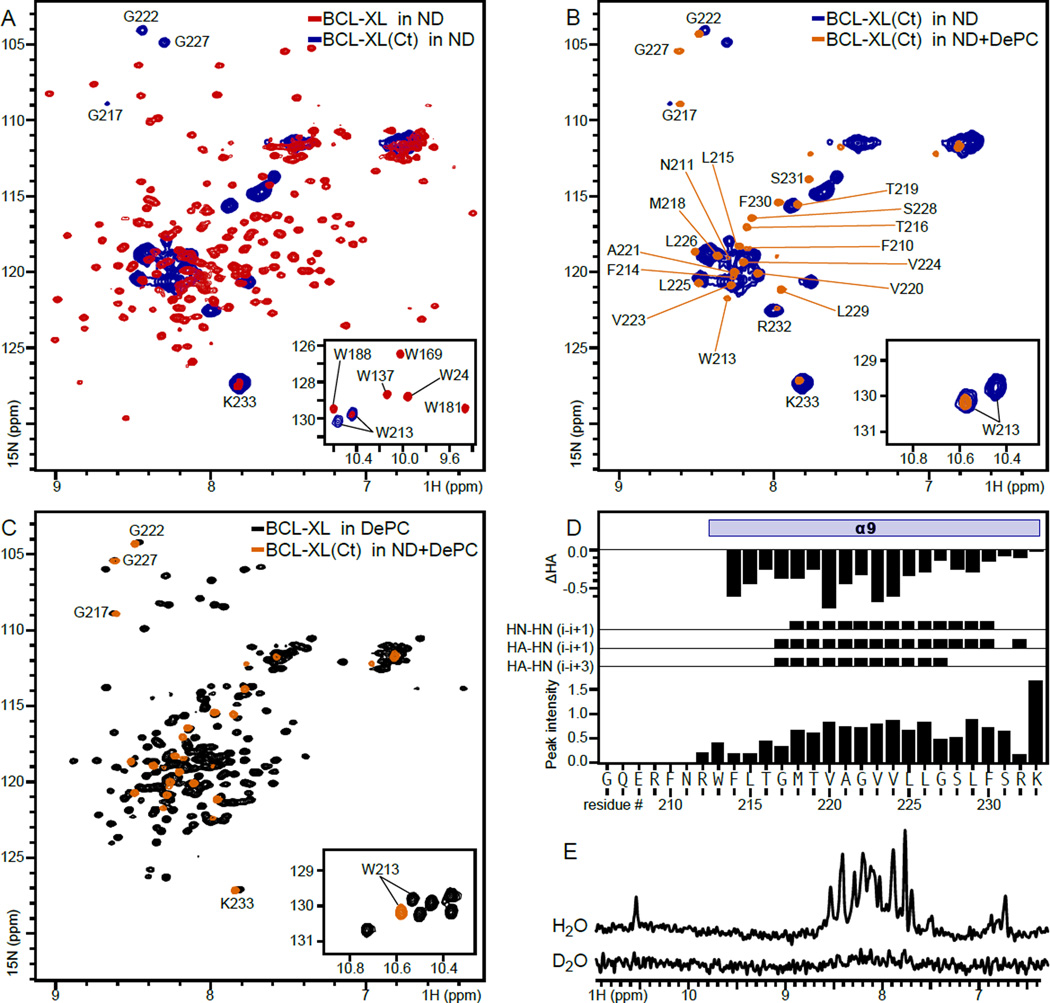
Figure 4. NMR spectra of BCL-XL and BCL-XL(Ct) in lipid nanodiscs, mixed lipid-DePC, or DePC micelles.
(A–C) Solution NMR 1H/15N correlation spectra of BCL-XL in nanodiscs (red), BCL-XL in 170 mM DePC (black), BCL-XL(Ct) in nanodiscs (blue) or BCL-XL(Ct) in nanodiscs supplemented with 170 mM DePC (mixed lipid-DePC; orange). (D) Secondary structure and dynamics features of BCL-XL(Ct) in mixed lipid-DePC. The HA chemical shift difference (ΔHA) was calculated by subtracting the random coil values from the experimental chemical shifts. Experimental 1H-1H NOE data were obtained between atomic sites: HN(i) and HN(i+1), HA(i) and HN(i+1), or HA(i) and HN(i+3). HN Peak intensities (Io) were derived from the 1H/15N correlation spectra. (E) One-dimensional 1H spectra of 15N labeled BCL-XL(Ct) in mixed lipid-DePC obtained in H2O or 2H2O.
The most prominent peaks in the spectrum of BCL-XL in nanodiscs come from the N-terminal head of the protein, which is anchored to the nanodisc by the tail but otherwise free to undergo additional motions. Comparison with the spectrum of BCL-XL(Ct) in nanodiscs shows overlap in a number of peaks, most prominently for K233. Notably, the W213 indole sidechain yields a single signal in the spectrum of BCL-XL compared to the split signal that is resolved in the spectrum of the isolated tail. This observation suggests that additional regions of the protein upstream of G206 may be needed to stabilize the membrane-associated conformation of the C-terminal tail.
Addition of 170 mM DePC to nanodisc-embedded BCL-XL(Ct) significantly reduces the NMR linewidths (Fig. 4B) and enables assignments of backbone and sidechain resonances, as well as measurements of NOE contacts for structural analysis. The signals resolved in the presence of detergent have significant overlap with those observed in detergent-free nanodiscs, thus enabling derivation of assignments by spectral comparison. The NMR signals resolved for BCL-XL(Ct) in mixed lipid-DePC also have nearly complete overlap with those from BCL-XL in pure DePC (Fig. 4C). These results indicate that the tail adopts a similar conformation in lipid nanodiscs, mixed lipid DePC and pure DePC, and that the tail is independently structured from the N-terminal head of the protein.
The precise structure of the mixed lipid-DePC aggregate, generated by adding DePC to the nanodiscs, is not known. The environment is likely very similar to that of pure DePC micelles, given the very high degree of NMR signal overlap noted in Fig. 4C, and hence we deduce that it must be a micellar or bicellar colloidal aggregate. The most prominent difference is observed for the signals of the W213 indole nitrogen (Fig. 4C, inset). This sidechain is sensitive to the lipid environment and the shift in 1H/15N signal indicates that it may sense the phospholipids present from the nanodisc preparation.
Resonance assignments were obtained for N, HN, HA and HB atomic sites in residues R212-K233 of BCL-XL(Ct) in mixed lipid-DePC. By contrast, signals from residues G206-N211 at the N terminus could not be detected, probably due to 1H exchange with solvent. The 1H and 15N chemical shifts together with the NOE measurements indicate that the C terminal tail of BCL-XL adopts α-helical structure from W213 to K233 in the mixed lipid-detergent environment. This is consistent with the profile of 1H/15N peak intensities, which reveals relatively uniform backbone dynamics for residues M218-S231, higher mobility and/or conformational exchange at the N-terminus, and a highly mobile C-terminus (Fig. 4D).
The presence of conformational exchange is also borne out by 1H/2H exchange experiments showing that all amide 1H sites of BCL-XL(Ct) in lipid-detergent are readily exchanged for 2H (Fig. 4E). Typically, transmembrane α-helices have shorter, stronger intra-molecular hydrogen bonds to effectively shield polar backbone atoms from the low dielectric environment of the membrane or micelle interior, and their amide protons are highly resistant to exchange with the surrounding water. By contrast, the rapid 1H/2H exchange observed for BCL-XL(Ct) in micelles is consistent with a relatively loose hydrogen bond network across the entire C-terminal tail.
In light of this result we assessed the membrane-associated topology of BCL-XL(Ct) by means of solvent-induced PRE. The paramagnetic electrons of nitroxyl groups and Mn2+ induce distance-dependent broadening of NMR signals. Signals from protein sites in the hydrophobic interior of the micelle, such as those in a transmembrane helix, are affected by paramagnetic electrons embedded in the micelle interior, but not by water-soluble paramagnetic agents. Conversely, signals from solvent exposed protein sites are sensitive to water-soluble but not lipid-soluble paramagnetic molecules.
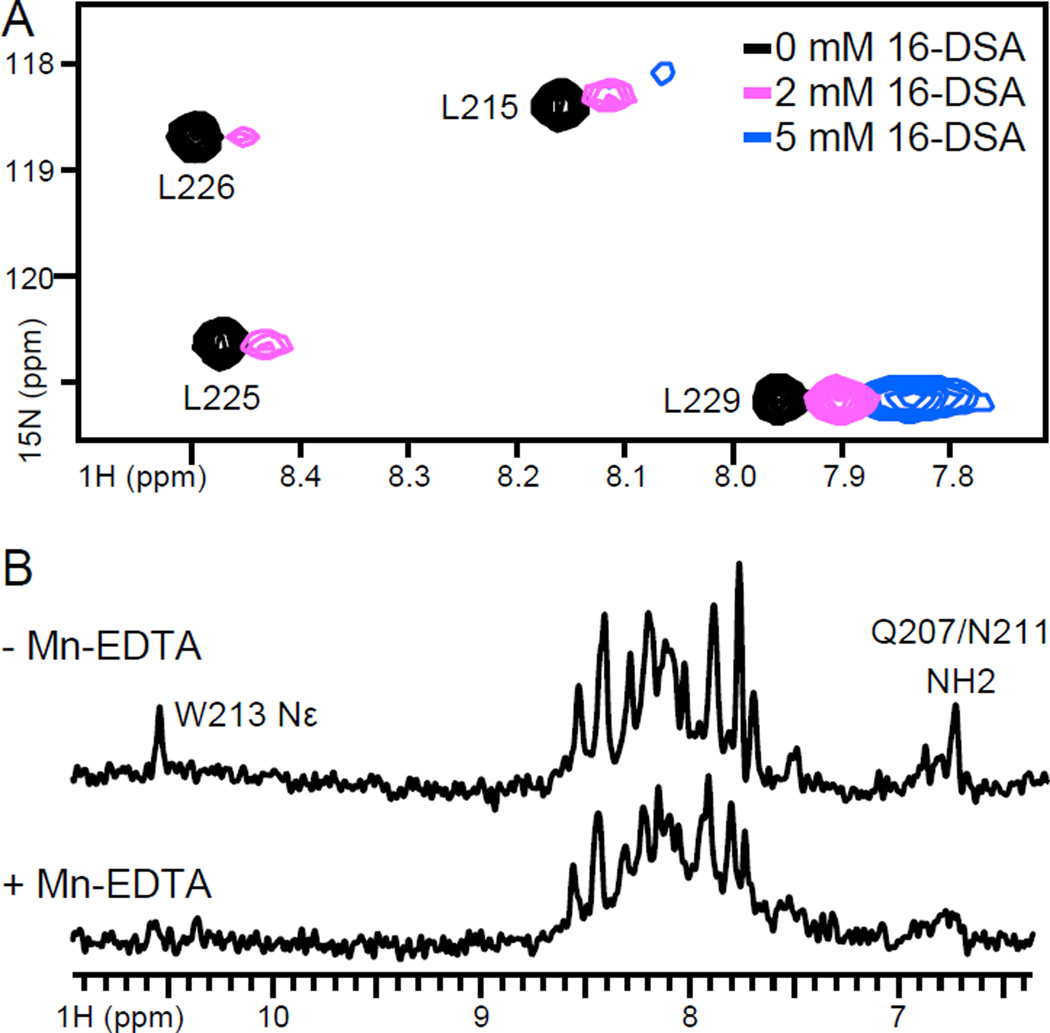
Addition of the micelle-soluble paramagnetic lipid 16-DSA, to 15N-Leu labeled BCL-XL(Ct) in micelles induced selective broadening of the 1H/15N signals (Fig. 5A), with significantly greater effects at L225 and L226 near the center of the hydrophobic tail sequence, and much less broadening at L215 and L229 near the termini. As 16-DSA bears the paramagnetic moiety at the very end of its hydrophobic 16-carbon acyl chain, we conclude that L225 and L266 must also be situated at the hydrophobic core of the micelle, while L115 and L229 are further removed from the center, as expected for sites near the helix termini. Conversely, addition of water-soluble Mn- EDTA to 15N-Leu labeled BCL-XL(Ct) in mixed lipid-DePC caused significant line broadening and disappearance of peaks from the sidechain atoms of Q207, N211 and W213, while the majority of peaks from visible backbone sites remained largely unaffected (Fig. 5B). These results demonstrate that BCL-XL(Ct) is deeply embedded in the mixed lipid-DePC micelle, consistent with 1H/15N solid-state NMR spectra of BCL-XL in lipid bilayer macrodiscs [12].
Figure 5. Effect of solvent PRE on the spectra of BCL-XL(Ct) in lipid-DePC or DePC.
(A) Solution NMR 1H/15N correlation spectra of 15N Leu labeled BCL-XL(Ct) synthetic peptide, in 170 mM DePC, obtained before (black) and after addition of 2 mM (blue) and 5 mM (pink) 16-DSA. The 1H chemical shift scale of the spectra obtained with 16-DSA are shifted by 0.05 ppm to display the PRE effect. The spectrum measured in the presence of 5 mM 16-DSA is displayed with a contour level four times lower than the others to display residual signal intensity. (B) One-dimensional 1H spectra of 15N labeled recombinant BCL-XL(Ct), in mixed lipid-DePC, obtained without or with 10 mM Mn-EDTA.
Taken together, the NMR data for BCL-XL(Ct) in mixed lipid-DePC show that the NH sites undergo rapid 1H/2H exchange, are largely protected from interactions with water-soluble Mn-EDTA, and come into contact with 16-DSA to varying degrees. These apparently conflicting observations can be reconciled by recent data [32] showing that water molecules penetrate the interior of micelles to a very significant extent. This is very different from the case of detergent-free lipid bilayers where water is largely excluded from the hydrophobic interior. Given the dynamic conformational properties of BCL-XL(Ct), its weakly hydrogen-bonded amide hydrogens would be expected to exchange with the penetrating water molecules. By contrast, large ionic species, too polar to penetrate the micelle, would be expected to remain sufficiently distant from micelle-dissolved protein, consistent with the lack of PRE broadening from Mn-EDTA. Finally, PRE broadening from 16-DSA reflects the proximity of BCL-XL(Ct) to the lipid hydrocarbon chains in the lipid-DePC aggregate.
4. CONCLUSIONS
We have shown that the C-terminal tail of BCL-XL forms a membrane-embedded α-helix that anchors the protein's globular head to the lipid bilayer membrane, yet retains a significant degree of conformational dynamics. The conformational dynamics of the C-terminal tail may facilitate its proteolytic cleavage in aqueous solution, as the small fraction of recombinant BCL-XL produced in the soluble bacterial cell fraction only attains solubility after lytic removal of C-terminal residues T219-K233. By contrast, the majority of recombinant BCL-XL produced in the insoluble cell fraction is protected from proteolysis, retains the full C-terminal tail and can be reconstituted in lipid bilayers for structure and activity studies.
Endogenous BCL-XL has been shown to be integral, rather than peripheral, to the MOM [33]. The C-terminal tail is essential for membrane integration and amino acids upstream of its hydrophobic sequence are important for determining specific localization to the MOM instead of other intracellular membranes. Whether MOM insertion of BCL-2 family proteins occurs spontaneously or requires initial recognition of the mitochondrial Translocase of the outer membrane (TOM) complex is still unclear [34–38]. However, specific, post-translational targeting of tail-anchored proteins to the MOM does depend on specific features of the C-terminal tail sequence.
As in most tail-anchored proteins that target and insert post-translationally in the MOM, the C-terminal sequences of the BCL-2 family proteins are not highly conserved, but share a set of common features that, combined, constitute the membrane targeting signal [33, 39]. The BCL-2 tail sequences are moderately hydrophobic, relatively short, and are flanked by positively charged residues.
Notably, they are also enriched in Gly and Ala, residues that enhance helix flexibility, as noted for the MOM-targeting sequences of many other proteins [39]. Indeed, the NMR data do show that the C-terminal tail of BCL-XL is highly flexible and forms a relatively loose hydrogen bond network compared to other transmembrane helices. A certain level of conformational dynamics may be important for facilitating insertion of the tail across the MOM and/or for mediating intra-membrane interactions with other MOM-localized partners during the apoptosis process. Additional factors may influence the flexibility of the membrane-inserted C-terminal tail of BCL-XL. It is possible that the conformation and dynamics of the tail are susceptible to the specific lipid composition of the membrane, as suggested by a study on the tail of BCL-2 [15]. Furthermore, the observation of peak doubling for the signal from W213 in the spectrum of nanodisc-inserted BCL-XL(Ct) but not BCL-XL suggests that additional residues upstream of G206 may be important for stabilizing the membrane-inserted conformation. This would be consistent with the ability of helix α8 (G196–G206) to reorient upon BH3 ligand binding by BCL-XL [10] and also with our previous observation that this is the site of major NMR perturbations detected upon membrane association of BCL-XL [12]. Additional studies aimed at resolving the structure and interactions of membrane-bound BCL-XL will be needed to understand the full extent of functionality of this important class of proteins.
HIGHLIGHTS.
Soluble BCL-XL is readily proteolysed after C-terminal residue M218.
BCL-XL is reconstituted in detergent-free lipid nanodiscs.
The N-terminal head retains the canonical structure determined for water-soluble, tail-truncated BCL-XL.
The C-terminal tail anchors the protein to the nanodisc membrane.
The C-terminal tail forms a transmembrane α-helix that has significant conformational dynamics.
Acknowledgments
This work was supported by grants (R01CA179087, R01GM100265, P41EB002031, P30CA030199) from the National Institutes of Health.
ABBREVIATIONS
- 16-DSA
16-doxyl-stearic acid
- BH
BCL-2 homology
- DMPC
dimyristoyl-phosphatidyl-choline
- DMPG
dimyristoyl-phosphatidyl-glycerol
- MALDI
Matrix Assisted Laser Desorption/Ionization
- MOM
mitochondrial outer membrane
- MSP
membrane scaffold protein
- PAGE
polyacrylamide gel electrophoresis
- TEVp
Tobacco Etch Virus protease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi X, Kale J, Leber B, Andrews DW. Regulating cell death at, on, and in membranes. Biochim Biophys Acta. 2014;1843:2100–2113. doi: 10.1016/j.bbamcr.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 4.Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Garcia M, Perez-Ballestero R, Ding L, Duan L, Boise LH, Thompson CB, Nunez G. bcl-XL is the major bcl-x mRNA form expressed during murine development and its product localizes to mitochondria. Development. 1994;120:3033–3042. doi: 10.1242/dev.120.10.3033. [DOI] [PubMed] [Google Scholar]
- 6.Fang W, Rivard JJ, Mueller DL, Behrens TW. Cloning and molecular characterization of mouse bcl-x in B and T lymphocytes. J Immunol. 1994;153:4388–4398. [PubMed] [Google Scholar]
- 7.Mizuguchi M, Sohma O, Takashima S, Ikeda K, Yamada M, Shiraiwa N, Ohta S. Immunochemical and immunohistochemical localization of Bcl-x protein in the rat central nervous system. Brain Res. 1996;712:281–286. doi: 10.1016/0006-8993(95)01453-5. [DOI] [PubMed] [Google Scholar]
- 8.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todt F, Cakir Z, Reichenbach F, Youle RJ, Edlich F. The C-terminal helix of Bcl-x(L) mediates Bax retrotranslocation from the mitochondria. Cell Death Differ. 2013;20:333–342. doi: 10.1038/cdd.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 11.Kvansakul M, Hinds MG. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell death & disease. 2013;4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y, Fujimoto LM, Hirshman N, Bobkov AA, Antignani A, Youle RJ, Marassi FM. Conformation of BCL-XL upon Membrane Integration. J Mol Biol. 2015;427:2262–2270. doi: 10.1016/j.jmb.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzin CM, Choi J, Zhai D, Reed JC, Marassi FM. Structural studies of apoptosis and ion transport regulatory proteins in membranes. Magn Reson Chem. 2004;42:172–179. doi: 10.1002/mrc.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torrecillas A, Martinez-Senac MM, Goormaghtigh E, de Godos A, Corbalan-Garcia S, Gomez-Fernandez JC. Modulation of the membrane orientation and secondary structure of the C-terminal domains of Bak and Bcl-2 by lipids. Biochemistry. 2005;44:10796–10809. doi: 10.1021/bi0503192. [DOI] [PubMed] [Google Scholar]
- 16.Fesik SW, Zuiderweg ER. Heteronuclear three-dimensional NMR spectroscopy of isotopically labelled biological macromolecules. Q Rev Biophys. 1990;23:97–131. doi: 10.1017/s0033583500005515. [DOI] [PubMed] [Google Scholar]
- 17.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 18.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR spectroscopy : principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- 19.Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Curr Opin Chem Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 20.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 21.Kay LE. Nuclear magnetic resonance methods for high molecular weight proteins: a study involving a complex of maltose binding protein and beta-cyclodextrin. Methods Enzymol. 2001;339:174–203. doi: 10.1016/s0076-6879(01)39314-x. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. Journal of Biomolcular NMR. 1995;5:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 24.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2004. [Google Scholar]
- 25.Denisov AY, Madiraju MS, Chen G, Khadir A, Beauparlant P, Attardo G, Shore GC, Gehring K. Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix. J Biol Chem. 2003;278:21124–21128. doi: 10.1074/jbc.M301798200. [DOI] [PubMed] [Google Scholar]
- 26.Hinds MG, Lackmann M, Skea GL, Harrison PJ, Huang DC, Day CL. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 2003;22:1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millar DG, Shore GC. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J Biol Chem. 1996;271:25823–25829. doi: 10.1074/jbc.271.42.25823. [DOI] [PubMed] [Google Scholar]
- 28.Egan B, Beilharz T, George R, Isenmann S, Gratzer S, Wattenberg B, Lithgow T. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 1999;451:243–248. doi: 10.1016/s0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- 29.Beddoe T, Lithgow T. Delivery of nascent polypeptides to the mitochondrial surface. Biochim Biophys Acta. 2002;1592:35–39. doi: 10.1016/s0167-4889(02)00262-8. [DOI] [PubMed] [Google Scholar]
- 30.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 31.Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta. 2009;1788:2022–2031. doi: 10.1016/j.bbamem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Long JA, Rankin BM, Ben-Amotz D. Micelle Structure and Hydrophobic Hydration. J Am Chem Soc. 2015;137:10809–10815. doi: 10.1021/jacs.5b06655. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann T, Schlipf S, Sanz J, Neubert K, Stein R, Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janiak F, Leber B, Andrews DW. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J Biol Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 35.Nakai M, Takeda A, Cleary ML, Endo T. The bcl-2 protein is inserted into the outer membrane but not into the inner membrane of rat liver mitochondria in vitro. Biochem Biophys Res Commun. 1993;196:233–239. doi: 10.1006/bbrc.1993.2239. [DOI] [PubMed] [Google Scholar]
- 36.Motz C, Martin H, Krimmer T, Rassow J. Bcl-2 and porin follow different pathways of TOM-dependent insertion into the mitochondrial outer membrane. J Mol Biol. 2002;323:729–738. doi: 10.1016/s0022-2836(02)00995-6. [DOI] [PubMed] [Google Scholar]
- 37.Schleiff E, Shore GC, Goping IS. Human mitochondrial import receptor, Tom20p. Use of glutathione to reveal specific interactions between Tom20-glutathione S-transferase and mitochondrial precursor proteins. FEBS Lett. 1997;404:314–318. doi: 10.1016/s0014-5793(97)00145-2. [DOI] [PubMed] [Google Scholar]
- 38.Schleiff E, Shore GC, Goping IS. Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J Biol Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 39.Rapaport D. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]