Summary
Cis -regulatory elements such as transcription factor (TF) binding sites can be identified genome-wide, but it remains far more challenging to pinpoint genetic variants affecting TF binding. Here we introduce a pooling-based approach to mapping quantitative trait loci (QTLs) for molecular-level traits. Applying this to five TFs and a histone modification, we mapped thousands of cis-acting QTLs, with over 25-fold lower cost compared to standard QTL mapping. We found that single genetic variants frequently affect binding of multiple TFs, and that CTCF can recruit all five TFs to its binding sites. These QTLs often affect local chromatin and transcription, but can also influence long-range chromosomal contacts, demonstrating a role for natural genetic variation in chromosomal architecture. Thousands of these QTLs have been implicated in genome-wide association studies, providing candidate molecular mechanisms for many disease risk loci, and suggesting that TF binding variation may underlie a large fraction of human phenotypic variation.
Introduction
In the past decade, genome-wide association studies (GWAS) have transformed our view of common diseases: thousands of loci have been found to associate with common disease risk, compared to just a handful before the GWAS era. However despite their success, GWAS have suffered from several major limitations (Edwards et al., 2013). First, they generally do not reveal the causal variants underlying the associations, due to linkage disequilibrium (LD) between variants. Second, they give no information about the molecular mechanisms of how these variants affect disease risk; this generally requires laborious follow-up studies of individual loci. As a result, GWAS have made little contribution to our understanding of disease etiologies.
Perhaps the clearest trend to emerge from most GWAS is that the vast majority of genetic variants affecting complex traits resides outside of protein-coding regions (Hindorff et al., 2009; Schaub et al., 2012; Edwards et al., 2013); recent evolutionary adaptations follow a similar pattern (Fraser 2013; Enard et al., 2014). These trait-associated noncoding variants are highly enriched in transcriptional enhancers (Gusev et al., 2014; Farh et al., 2015; Roadmap Epigenomics Consortium 2015), but whether they affect enhancer function—and if so, how—remains largely unknown. One approach to annotate candidate functions for GWAS hits is to intersect them with quantitative trait loci (QTLs) mapped for molecular-level traits such as gene expression (eQTLs). This can be informative, but only in a small fraction of cases (e.g. 5.9% of GWAS hits were in LD with an eQTL from the multi-tissue GTEx analysis (GTEx Consortium, 2015)), and still does not indicate how these variants affect gene expression.
A complementary approach for annotating noncoding variants is to determine which of them are associated with transcription factor (TF) binding. This may be quite powerful, since TF binding 1) is mostly determined by highly local sequence (White et al., 2013; Levo and Segal, 2014), reducing the search space for QTLs; 2) is causally upstream of transcription and thus may be more strongly linked to genetic variants (c.f. Kaplow et al., 2015); and 3) involves many more traits (TF binding sites), and thus potential QTLs, than the number of genes.
Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in many genotyped human samples could allow the genetic mapping of variants affecting TF binding, but in practice, this is quite challenging due to both experimental variation between samples (e.g. batch effects) (Bonhoure et al., 2014), and the prohibitive cost of such an experiment. As a result, previous studies comparing TF binding across individuals—which have revealed extensive inter-individual variation—have been limited to relatively small sample sizes (Kasowski et al., 2010; Kasowski et al., 2013; Kilpinen et al., 2013; Ding et al., 2014; Waszak et al., 2015). In this work, we developed a more efficient and cost-effective approach to mapping QTLs for molecular-level traits, which allowed us to map thousands of QTLs affecting TF binding or histone modification. Their extensive overlap with variants previously implicated by GWAS provides candidate molecular mechanisms underlying disease etiologies, while more broadly suggesting a major role for TF binding in complex disease risk.
Results
Pooled ChIP-seq for high-throughput QTL mapping
We devised a simple modification to existing methods for mapping QTLs controlling molecular-level traits that reduces both experimental variation and cost. The central idea is that samples from any number of individuals can be combined into a single pool, in which ChIP-seq is performed only once. For genetic variants that affect binding of a TF in cis, the high-affinity allele will be present at a higher frequency after the ChIP, as compared to before (Figure 1A). In contrast, variants with no effect on binding should have no significant shift in allele frequencies. Post-ChIP allele frequencies are estimated based on the observed counts of each allele in the ChIP-seq reads. One potential complication is that samples with greater abundance or activity of a TF will have more bound sites, and thus contribute more to the post-ChIP pool; therefore we estimated pre-ChIP allele frequencies with a regression-based approach that accounts for any differences in TF activities between samples (see Supplemental Experimental Procedures). Our approach is limited to variation in local sequence (covered by the ChIP-seq reads), which is the primary determinant of TF binding (White et al., 2013; Levo and Segal, 2014).
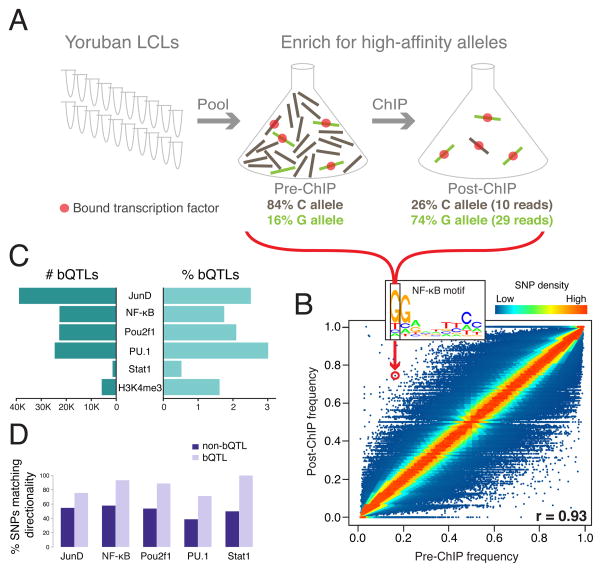
Figure 1. Outline and results of pooled ChIP-seq.
A. Performing ChIP in a pool of individuals selects only the DNA molecules bound by a TF (or with a particular histone modification), thus enriching for high-affinity alleles. In this example, the G allele has a low pre-ChIP frequency but a high post-ChIP frequency, due to its higher affinity. The allele frequencies and read counts are for the SNP highlighted in panel B. B. For NF-κB, plotting pre-ChIP vs. post-ChIP allele frequencies shows that the vast majority of allele frequencies change very little, as expected. One SNP (rs10263017) significantly off the diagonal is highlighted. This SNP occurs in the boxed position of an NF-κB binding site motif, where the G allele has higher predicted affinity, consistent with the allele frequency shift. C. The number of independent bQTLs (after removing those in LD) (left), and the percent of all bound SNPs called as bQTLs (right). D. Directionality agreement between predicted effects of SNPs in TF binding motifs, and their observed associations. SNPs that are bound but have no allele frequency shift (p > 0.5) show ~50% agreement, as expected by chance; bQTL (p < 0.005) agreement is significantly higher. H3K4me3 has no motif, so is not included. All comparisons are significant at Fisher’s exact test p < 0.005 except for Stat1, due to its small number of bQTLs. See also Figure S1 and Table S1.
We performed ChIP-seq for five TFs critical for immune cell development and function (NF-κB, PU.1/Spi1, Stat1, JunD, and Pou2f1/Oct1) in a pool of 60 Yoruban lymphoblastoid cell lines (LCLs), and obtained ~300 million reads per TF (except for Stat1 with ~126 million reads) with excellent agreement between biological replicates (Figure S1A). We also produced ~804 million reads for a histone modification associated with active transcription (H3K4me3) in a pool of 71 Yoruban LCLs. In all experiments, we found that the vast majority of SNPs had similar pre-ChIP and post-ChIP allele frequencies (Figure 1B and Figure S1B), as expected. However many SNPs had significantly shifted frequencies, assessed by a modified binomial test that accounts for uncertainty in our allele frequency estimates (see Supplemental Experimental Procedures). We refer to these SNPs as binding QTLs (bQTLs). We found between 1,533–38,944 independent bQTLs per TF at a nominal p < 0.005, which represented 0.5–3% of all bound SNPs (Figure 1C, Figure S1C, Table S1; false discovery rates discussed below).
For example, the G allele of rs10263017 was strongly over-represented in the NF-κB post-ChIP sample (Figure 1B; p = 1.8×10−15), consistent with its being located in an NF-κB binding motif where the G allele has a higher predicted affinity. Similarly, among all of our bQTLs predicted to affect the corresponding TF’s motif (see Supplemental Experimental Procedures), we found that most acted in the expected direction (Figure 1D). For all five TFs, <0.9% of bQTLs were located in predicted motifs, and <1.3% of SNPs in predicted motifs were bQTLs (Figure S1D). This minor role of motifs suggests that many bQTLs may involve recruitment of one TF by another, which can occur in the absence of a binding motif for the recruited TF; and/or that sequences flanking the binding motif play a major role, as has been observed before (White et al., 2013; Dror et al., 2015; Levo et al., 2015).
To investigate the bQTLs falling outside motifs, we tested the idea that the GC content surrounding motifs plays an important role in TF binding (White et al., 2013; Dror et al., 2015). Specifically, it has been observed that high local GC content promotes the binding of some TFs, whereas high AT content is ideal for others (Dror et al., 2015). To test this in our data, we asked whether the high-affinity bQTL alleles showed a skewed GC content, particularly when close to a binding motif. For NF-κB bQTLs, we observed a significant enrichment for high-affinity G/C alleles specifically within 25 bp of the motif (Fisher’s exact p = 0.005; Figure S1E). In contrast, Stat1 showed the opposite pattern, with a strong preference for high-affinity A/T alleles near the motif (Fisher’s exact p = 0.003). None of the bQTLs showed any significant GC content bias at distances further than 25 bp from the motif (Figure S1E). Therefore our results support the idea that for some TFs, GC content surrounding binding motifs can play an important role in determining affinity.
Comparing pooled bQTLs with ChIP-seq in individual samples
To validate our pooled QTL mapping approach, we asked how well our bQTLs can predict allele-specific binding in individual LCLs heterozygous for those bQTLs. Specifically, we compared our PU.1 bQTLs with PU.1 ChIP-seq data from 47 individual LCLs (Waszak et al., 2015); these samples were from another population (European/CEU), so were independent of those used in our experiments. We found that out of 592 of our PU.1 bQTLs (covered by at least 20 reads in the 47 CEU samples when summing across all heterozygous individuals), 518 (88%) showed an allelic bias in the direction predicted by our bQTLs, compared to 50% expected by chance (Figure 2A). However since the allelic bias for most of these was of a small magnitude and not statistically significant, we then asked how well our bQTLs predict the individual-level allele-specific binding when we restrict the comparison to the 998 PU.1 bQTLs reported by Waszak et al. (2015). In this case, the agreement is much stronger (88/89 SNPs, 99%; Figure 2B), and notably the one discordant SNP was also the one with the fewest reads (summed across both data sets).
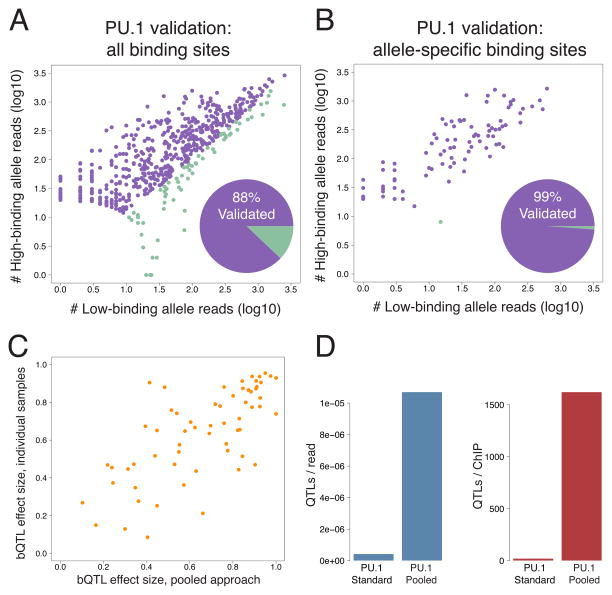
Figure 2. Comparing pooled bQTLs with ChIP-seq in individual samples.
A. Examining allele-specific binding of PU.1 (measured in individual heterozygous samples) at 592 PU.1 bQTLs (Waszak et al., 2015), we found 88% directionality agreement. B. Comparing our PU.1 bQTLs to allele-specific binding at 89 SNPs previously reported as PU.1 bQTLs (Waszak et al., 2015), we found 99% directionality agreement. C. Effect sizes of our bQTLs compared to the strength of allelic bias summed across individual heterozygous samples. D. Number of PU.1 bQTLs per sequence read and per ChIP, at equivalent significance cutoffs for each data set. See also Figure S2.
As a second test of validation, we asked whether the quantitative effect sizes inferred in our pooled data were also in agreement with the strength of allele-specific binding in individual heterozygotes. Among the 88 bQTLs identified above, the effect sizes were well correlated (Pearson r = 0.575; p = 5×10−9), and the agreement was higher still for those bQTLs covered by more reads (e.g. for the 61 bQTLs with at least 100 reads in our data, Pearson r = 0.705; p = 2×10−10; Figure 2C). Altogether, these results demonstrate a high degree of concordance between our pooling-based bQTLs and individual-level data.
Another important question is how the cost and effort of our pooled-QTL approach compares with standard QTL mapping. To investigate this, we again compared our PU.1 bQTLs with those previously mapped in 47 individual LCLs (Waszak et al., 2015). To ensure a fair comparison, we sought a p-value cutoff for our PU.1 bQTLs that yielded similar enrichments for an independent set of QTLs, suggesting that the two lists are of comparable quality. We therefore intersected both bQTL lists with published QTLs for three histone modifications (hQTLs) (Grubert et al., 2015). At a p < 5×10−5 cutoff for our bQTLs, we found comparable levels of enrichment for the two PU.1 bQTL lists (Figure S2; slightly higher enrichments for our bQTLs in 2/3 comparisons). At this cutoff, we mapped 3,246 bQTLs, compared to 998 for the standard, non-pooled approach. We achieved this despite generating 7.6-fold fewer ChIP-seq reads and performing 23.5-fold fewer ChIPs (Figure 3D). Therefore in terms of cost per QTL, we estimate that for PU.1 our method is over 25-fold more effective than the standard approach. This difference will likely increase with the number of samples studied, since pooling provides the greatest savings for large sample sizes.
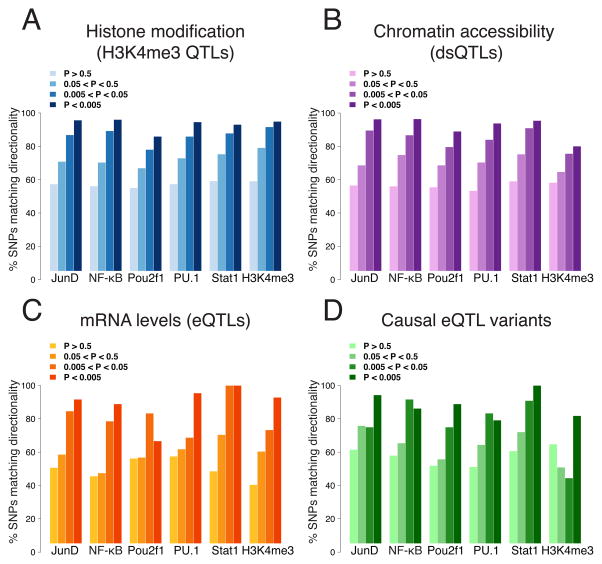
Figure 3. Comparisons with other molecular-level QTLs.
For each type of previously published QTL, we calculated the degree of allelic concordance with our bQTLs. A. H3K4me3 QTLs (Grubert et al., 2015). B. dsQTLs (Degner et al., 2012). C. eQTLs (Lappalainen et al., 2013). D. Causal SNPs underlying eQTLs (R. Tewhey and P. Sabeti, pers. comm.). See also Figure S3.
We hypothesize that these improvements are due to two key aspects of our approach. First, by performing ChIP with all samples pooled together, we reduce the inevitable experimental variability between samples. Second, our approach takes full advantage of the allele-specific information in each read, whereas traditional QTL mapping—by focusing on the mean of both alleles in each sample—ignores an important source of information in heterozygous samples (Kaplow et al., 2015; van de Geijn et al., 2015). In the extreme case of a cohort with only heterozygotes at a particular SNP, traditional QTL mapping would have no power, in contrast to our pooling approach. However we note that pooling is not necessary to utilize this extra information, since it is also present in individual heterozygotes (van de Geijn et al., 2015).
Comparing bQTLs with other molecular QTLs
Many other types of molecular-level QTLs have been mapped in LCLs, including QTLs for mRNA levels (eQTLs), chromatin accessibility (dsQTLs), DNA methylation (mQTLs), and histone modifications (Fraser and Xie 2009; Degner et al., 2012; Fraser et al., 2012; Kasowski et al., 2013; Lappalainen et al., 2013; McVicker et al., 2013; Kilpinen et al., 2013; Gutierrez-Arcelus et al., 2013; Ding et al., 2014; Banovich et al., 2014; Waszak et al., 2015; Grubert et al., 2015). To explore the relationships between TF binding and these other regulatory levels, we intersected our bQTLs with each of these other QTL types, and calculated the fold-enrichment compared to SNPs bound by our TFs but not classified as bQTLs (to control for any general characteristics of each TF’s binding sites, such as proximity to promoter regions) (see Supplemental Experimental Procedures). We found dsQTLs to have the highest overall enrichment for bQTLs (9.2-fold median enrichment; Figure S3A), as expected since these are the most directly linked to TF binding (Degner et al., 2012). mQTLs had the lowest levels of enrichment (Figure S3A), suggesting that binding of these TFs is largely independent of DNA methylation.
Because our TFs are all primarily transcriptional activators, we expected that bQTLs would be biased in their direction of effects on other molecular traits—alleles with higher binding should typically have more accessible chromatin, greater local H3K4me3, and higher mRNA levels. To investigate this, we calculated the directionality agreement between bQTLs and these other QTL types, for SNPs found as QTLs for both. In each case, we found increasing agreement at more stringent bQTL p-values (Figure 3 and Figure S3B). For previously published H3K4me3 QTLs (Grubert et al., 2015), this reached 93.1–96.1% agreement for 4/5 TFs, and 94.9% agreement with our H3K4me3 QTLs. Since false positive bQTLs would agree only ~50% of the time, this suggests false discovery rates of between 7.8–13.8% for these bQTLs (though these estimates, which conservatively assume that the previously reported H3K4me3 QTLs have no false positives and that the true overlaps always agree in direction, cannot be extrapolated to all bQTLs) (see Supplemental Experimental Procedures). The one TF with lower (85.9%) agreement, Pou2f1, has been shown to sometimes act as a repressor (Lin et al., 2013), which may explain this difference. We observed almost identical concordance among bQTLs occurring outside of their TF’s consensus motifs (Figure S3C), suggesting that these bQTLs are not enriched for false positives.
Finally, we also compared our bQTLs with SNPs that themselves affect transcription (in contrast to the QTL comparisons above, where linkage disequilibrium generally prevents identification of causal variants). These SNPs, identified in a massively parallel reporter assay performed in LCLs (R. Tewhey and P. Sabeti, pers. comm.), showed 79.2–100% agreement in directionality with our bQTLs for all TFs (Figure 3D). Altogether, the concordance of bQTLs with four other types of molecular QTLs (Figure 3) further establishes the accuracy of our approach.
Inferring cooperative binding and pioneer factors
Our data allow us to test whether a bQTL for one TF is likely to act as a bQTL for other TFs as well. For example, rs2886870—our second strongest bQTL for NF-κB (p = 10−75)—was also a bQTL for JunD (p = 10−20) and PU.1 (p = 10−16). It disrupts an NF-κB motif, suggesting that its direct effect is on NF-κB binding, which then affects recruitment of additional TFs. Interestingly, this SNP has previously been associated with local chromatin accessibility, histone modifications, RNA polymerase II binding, and mRNA levels in LCLs (McVicker et al., 2013). It is also the strongest QTL for DNA replication timing in LCLs (p = 10−13) (Koren et al., 2014), affecting a ~422 kb region, suggesting that disrupting a single NF-κB binding site that recruits multiple TFs can have a major effect on the temporal program of DNA replication, in addition to local chromatin and transcription.
Examining the overlap between bQTLs for all of our factors, we found a 2.7-fold to 21-fold enrichment, with Stat1 showing the strongest overall enrichment (Figure 4A upper right). This suggests that single genetic variants may often affect binding of multiple TFs, since the alternative—multiple causal variants for different TFs in strong LD—is unlikely to occur so consistently. To further test this idea, we measured the concordance in directionality for bQTLs associated with binding of multiple TFs, which would be random (~50%) if one bQTL was tagging multiple causal variants. We found that the same allele had higher binding for both TFs in 96.1–100% of all TF pairs (Figure 4A lower left), strongly supporting the single-variant model.
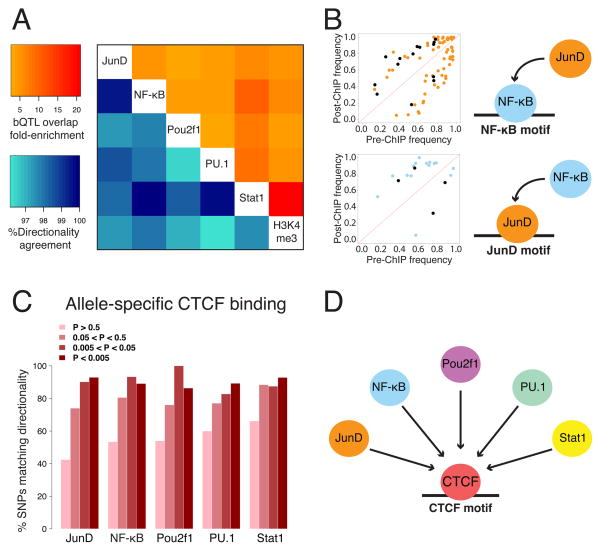
Figure 4. Causal relationships between TFs.
A. For every pairwise combination of bQTLs, we calculated the enrichment of their overlaps (upper right, relative to bound SNPs that are not bQTLs), and the degree of allelic concordance among the overlaps (lower left). B. We tested whether SNPs in the binding motifs for one TF were predictive of binding of another TF. Scatter plots show allele frequencies before and after ChIP (as in Figure 1B), with those SNPs whose effect is in the direction predicted by the motif’s PWM colored to match the ChIP-ed TF, and those not matching colored black. C. Allelic bias of CTCF binding at heterozygous sites (Ding et al., 2014) generally agrees in direction with our bQTLs. See also Figure S4. D. Our model for CTCF as a major pioneer factor.
A single variant might affect multiple TFs by initially altering the binding of one TF—a “pioneer factor” —that in turn recruits others. To test this idea, we investigated bQTLs for one TF that are located within a binding motif for a different TF, since these suggest that one TF recruits another to its binding site (Kasowski et al., 2010)—as in the case of rs2886870 and NF-κB described above (This causality inference is only possible when a bQTL is within a TF binding motif, since otherwise we cannot discriminate between direct vs. indirect effects of a bQTL on TF binding). For example, NF-κB and JunD are known to act cooperatively (Rahmani et al., 2001); consistent with this, we found that SNPs in either of their motifs often associate with binding of the other, implying that either factor can help recruit the other (Figure 4B). In further support of this causal inference approach, the directionality of these shared bQTLs is overwhelmingly in the direction predicted by the motif’s PWM: for example, a SNP increasing the predicted affinity of a site for NF-κB usually increases the binding of JunD (Figure 4B top, orange points) rather than decreasing it (Figure 4B top, black points).
Extending this test to a collection of 2995 motifs (Weirauch et al., 2014), we found one TF whose motif disruption was associated with binding of all five TFs (Fisher’s exact p < 0.05 for each). This motif was for CTCF, a protein involved in chromosomal looping and chromatin structure (Rao et al., 2014). To further explore this relationship, we tested the directionality agreement between our bQTLs and QTLs for CTCF binding in LCLs (Ding et al., 2014). These showed 86.4–92.9% agreement for all five TFs (Figure 4C and Figure S4), supporting the motif analysis. Together, these results suggest that CTCF may be a major pioneer factor that influences the local landscape of binding for many other TFs (Figure 4D), though our results do not reveal the mechanism by which this occurs. It is possible that there are direct physical interactions between CTCF and the other TFs, but a more likely model is that CTCF binding alters nucleosome positioning and chromosomal looping to make local chromatin more accessible to other TFs.
bQTLs can mediate long-range effects on transcription
Cis-regulatory variants typically affect chromatin and transcription in a highly local manner—for example, most dsQTLs are located within 100 bp of the region whose accessibility they modulate (Degner et al., 2012). However long-range effects of cis-regulatory variants are also possible, particularly when distant chromosomal regions are in close physical proximity within the nucleus (Grubert et al., 2015).
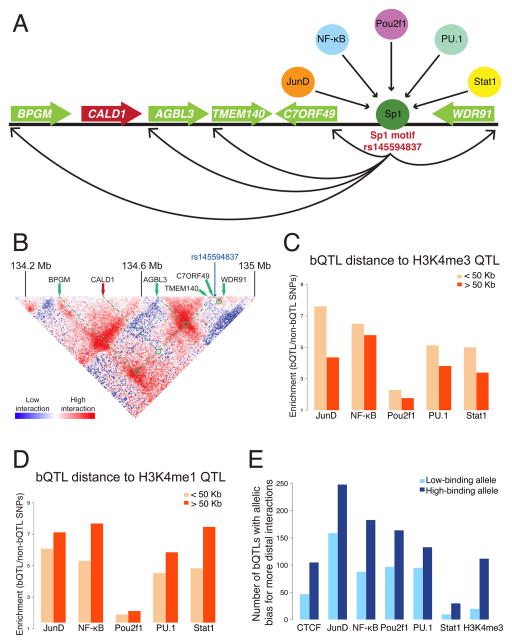
To investigate whether bQTLs can affect distant loci, we first compared them with long-range eQTLs. One example of a bQTL with distant effects is rs145594837, a 10-bp deletion that creates a strong Sp1 motif within an enhancer/promoter that is active in dozens of cell types (ENCODE Project Consortium, 2012; Roadmap Epigenomics Consortium 2015). The deletion allele has higher binding of all five TFs, suggesting that Sp1 may recruit them to this locus when it is bound. This deletion is an eQTL for five genes (Lappalainen et al., 2013), including BPGM, a gene involved in congenital erythrocytosis that is located 525 kb away (Figure 5A). Interestingly, the deletion is not an eQTL for a more proximal gene, CALD1, that is adjacent to BPGM. To determine if chromosomal contacts might mediate these selective effects on transcription, we analyzed Hi-C data from an LCL (Rao et al., 2014), which indicate which loci are in close proximity within the nucleus. Examining interactions between the deletion locus and each gene’s promoter, we found that all five genes affected by the deletion had high rates of contact, while CALD1 had the lowest level of interaction with the deletion locus (Figure 5B). This pattern suggests that this bQTL likely affects distant genes via long-range chromosomal contacts.
Figure 5. Long-range effects of bQTLs.
A. An example of a bQTL for all five TFs that affects the expression of five genes, including one (BPGM) 525 kb away. B. The pattern of chromosomal contacts around the bQTL shown above indicates high levels of interactions with the promoter regions of the distal genes that it affects. No enrichment is observed with CALD1, a gene within this region not affected by the bQTL. Color bar indicates the ratio of observed/expected contact, given the distance between loci (even though the deletion is not enriched more than expected for contacts with the two closest genes, C7ORF49 and TMEM140, there is still a high level of contact with these and other proximal loci). Hi-C data were normalized with the “balanced” method (Rao et al., 2014, see also Figure S5). C. Enrichment of bQTLs for local and distal hQTLs affecting H3K4me3 show a consistent bias for local effects. D. Enrichment of bQTLs for local and distal hQTLs affecting H3K4me1 show a consistent bias for distal effects. E. bQTLs affect the extent of long-range contacts. For each factor, the number of bQTLs where the higher-bound allele has more distal (>30 kb) interactions is plotted in dark blue, and the number where the lower-bound allele has more is plotted in light blue. The difference is significant in every case.
To more systematically explore the long-range effects of bQTLs, we compared them to QTLs previously found to affect histone modifications (hQTLs); these hQTLs were previously classified as either local (<50 kb from the site they affect) or distal (>50 kb away) (Grubert et al., 2015). For hQTLs affecting the promoter-associated mark H3K4me3, we found a greater enrichment for bQTLs acting as local as opposed to distal hQTLs (Figure 5C). Although the magnitude of the difference was small, it was consistent across all five TFs (Fisher’s exact p = 3.9×10−7), indicating that short-range effects on H3K4me3 are more likely to be explained by bQTLs. In contrast, for hQTLs affecting the enhancer-associated mark H3K4me1, we found the opposite trend, with a slightly higher enrichment of bQTLs with long-range effects for all five TFs (Figure 5D; Fisher’s exact p = 1.1×10−8). These opposing patterns may be due to enhancers generally participating in more long-range chromosomal contacts than promoters, giving more opportunity for bQTLs to act on enhancers from a distance. Consistent with this model, we found no distance bias for hQTLs affecting H3K27ac (Fisher’s exact p = 0.27), a mark found at both enhancers and promoters. Across all hQTLs, the most distant effect we observed was a PU.1 bQTL (rs10161851) associated with H3K4me1 levels 1.98 mb away; however since these hQTLs were only mapped within 2 mb (Grubert et al., 2015), it is likely there are bQTLs affecting chromatin across even greater distances.
Another question is whether these long-range chromosomal contacts act simply as passive conduits for bQTLs affecting distal loci, or if instead bQTLs can help shape the three-dimensional architecture of chromosomes and thus affect which other loci they contact. CTCF has been found at the base of many chromosomal loops (Rao et al., 2014), and altering these sites can disrupt looping (Guo et al., 2015; Sanborn et al., 2015), but the role of natural genetic variants in chromosomal architecture is unknown. To investigate this, we tested whether bQTLs for CTCF or other TFs may affect the extent of long-range contacts at these loci. Utilizing allele-specific Hi-C data from an LCL (Rao et al., 2014), for each bQTL that was heterozygous in this sample we counted the number of Hi-C reads connecting each allele of the bQTL with any distal locus (>30 kb away). For each TF, we then counted the number of bQTLs where the high-binding allele had significantly more contacts (based on a binomial test of allele-specific read counts), and likewise counted the cases where the low-binding allele was more interactive. For previously published CTCF bQTLs (Ding et al., 2014), we found that high-binding alleles were 2.2-fold more likely to have more contacts than the low-binding alleles (Figure 5E; binomial p = 1.4×10−6), consistent with CTCF’s well-established role in shaping chromosomal contacts. Surprisingly, we observed a similar pattern for all five of our TFs (Figure 5E; 1.4-fold to 3.0-fold differences; binomial p ≤ 0.001 for each). At sites where multiple heterozygous bQTLs were present and their high-binding alleles were encoded on the same chromosome, the trends were stronger still (3.0-fold to 5.0-fold differences for the 5 TFs, and 6.0-fold for CTCF; binomial p ≤ 0.001 for each). We found the strongest effects of all for H3K4me3 bQTLs, with a 5.6-fold interaction difference between alleles with more vs. less of this modification (binomial p = 5.2×10−17), and 9.5-fold when these were accompanied by a nearby TF bQTL (binomial p = 8.2×10−12). Although our bQTLs are also often associated with CTCF binding (Fig 4C), CTCF could not explain these patterns at our bQTLs, since excluding sites with detectable CTCF binding did not weaken the overall trend (1.6-fold to 2.7-fold differences for 4/5 TFs, and 12-fold for H3K4me3). Altogether, these results suggest that natural genetic variation can affect chromosomal architecture, and that the alleles with greater TF binding or more H3K4me3 tend to have more distal contacts, even at loci lacking CTCF.
bQTLs provide candidate molecular mechanisms for thousands of GWAS-implicated variants
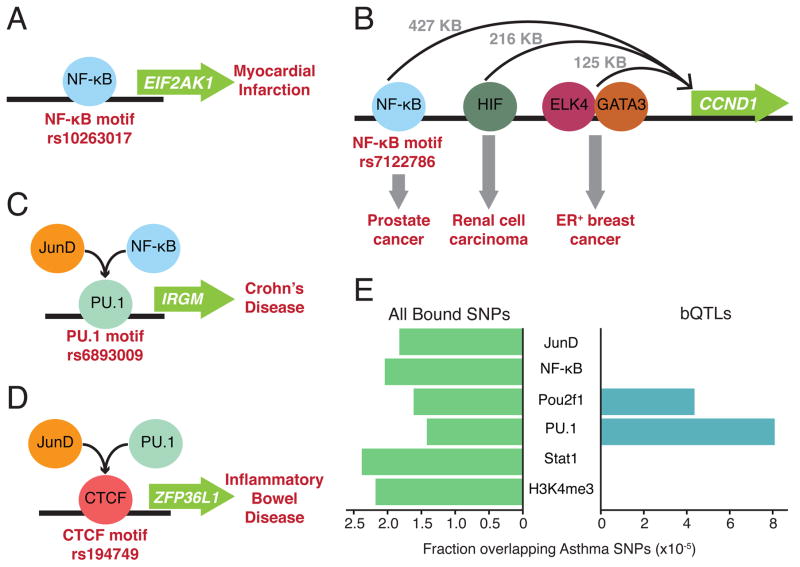
Integrating our results with GWAS can help elucidate molecular mechanisms of complex disease. For example, rs10263017—a bQTL for NF-κB whose derived allele disrupts an NF-κB binding motif (circled in Figure 1B)—is also associated with mRNA levels of EIF2AK1 (Lappalainen et al., 2013), as well as risk of myocardial infarction (Yamada et al., 2011). Thus our results imply that its likely molecular mechanism is alteration of NF-κB binding (Figure 6A). This variant is located in an enhancer that is active in dozens of tissues, including the heart left ventricle (Roadmap Epigenomics Consortium 2015), illustrating how bQTLs from LCLs can reveal molecular mechanisms that may be most relevant in other cell types.
Figure 6. Effects of bQTLs on disease risk.
Each example involves a SNP in a TF binding motif that is also a bQTL, and is associated with disease risk. A. bQTL for NF-κB (highlighted in Figure 1B) in an NF-κB binding motif that is also an eQTL for EIF2AK1 and associated with myocardial infarction. B. bQTL for NF-κB in an NF-κB binding motif that is also an eQTL for CCND1 and is associated with prostate cancer. See also Figure S5. C. bQTL for PU.1, NF-κB, and JunD in a PU.1 binding motif that is also associated with Crohn’s disease. D. bQTL for JunD and PU.1 in a CTCF binding motif that is also associated with inflammatory bowel disease. E. Comparing enrichments for asthma-associated SNPs: all bound SNPs (left), and bQTLs (right). See also Figure S6 and Table S2.
In other cases, bQTLs affecting disease risk may act over long genomic distances. One example of this is rs7122786, a SNP located in an LD block that contains multiple independent variants associated with risk of prostate cancer (Amin Al Olama et al., 2015). The derived allele disrupts an NF-κB binding motif within an LCL-specific DNase hypersensitive region (ENCODE Project Consortium, 2012), and is a bQTL for NF-κB. Although it is located near the MYEOV gene, it is actually an eQTL for another gene 427 kb away: CCND1 (Cyclin D1), a cell-cycle regulator that plays a major role in prostate cancer and other malignancies. This variant was the strongest eQTL for CCND1 in LCLs (p = 3×10−10) (Dixon et al., 2007), and was replicated in another study (Lappalainen et al., 2013), further supporting the hypothesis that this is a functional variant. Interestingly, two other enhancers located 125 kb and 216 kb from CCND1 each contain variants associated with risk of specific cancer types (ER+ breast cancer and renal cell carcinoma, respectively) (Schödel et al., 2012; French et al., 2013); our results suggest the existence of a third enhancer that specifically affects risk of prostate cancer (Figure 6B). Hi-C data (Rao et al., 2014) show that all three enhancers and CCND1 are part of a large region with a high density of chromosomal interactions (Figure S5), which likely facilitates this long-range effect.
We can also infer more complex molecular mechanisms involving TFs recruited to the binding sites of other factors. For example, variants in two LD blocks flanking IRGM, a regulator of autophagy, are independently associated with Crohn’s disease (Parkes et al., 2007; McCarroll et al., 2008). For one of these blocks, two candidate causal variants have been proposed (McCarroll et al., 2008; Brest et al., 2011); however in the second block, the strong LD and lack of obvious candidates has prevented any progress in determining its causal variant. We found that rs6893009, which is in perfect LD with the reported variant (rs4958847), is one of our strongest bQTLs for PU.1 (p = 2×10−31), as well as a moderate bQTL for NF-κB and JunD (p < 2×10−5). The derived allele creates a high-affinity PU.1 binding site, suggesting that PU.1 helps recruit NF-κB and JunD (Figure 6C). In addition to B cells and LCLs, this region has an active chromatin state in the small intestine (Roadmap Epigenomics Consortium 2015), a key tissue for Crohn’s disease. A second example involves rs194749, a bQTL for JunD and PU.1 that is also associated with inflammatory bowel disease (Jostins et al., 2012). This SNP falls within a CTCF binding motif, and is also associated with local chromatin accessibility (Degner et al., 2012) and DNA methylation (Gutierrez-Arcelus et al., 2013); the nearest gene is ZFP36L1, which has a well-established anti-inflammatory role (Sanduja et al., 2011). Therefore it is likely that CTCF is the pioneer factor, whose binding impacts the recruitment of JunD and PU.1, potentially altering the cis-regulation of ZFP36L1 (Figure 6D).
Examining disease-specific bQTL/GWAS enrichment also revealed informative trends. In these comparisons, we can go beyond comparing GWAS loci with all binding sites of a TF (Schaub et al., 2012; Farh et al., 2015; Guo et al., 2015), since most SNPs in binding sites do not detectably affect binding (Figure S1D). For example, asthma risk loci were enriched only for bQTLs of PU.1 and Pou2f1 (5.7-fold and 2.7-fold enriched, respectively; Figure 6E right). However across all binding sites (not just bQTLs), PU.1 and Pou2f1 actually had the fewest overlaps with asthma-associated SNPs (Figure 6E left). We found a similar lack of correspondence for many other diseases (median correlation of bQTL/GWAS overlap vs. bound SNP/GWAS overlap of r = −0.03 among 12 autoimmune diseases; Figure S6), suggesting that intersecting GWAS loci with all binding sites of a TF may yield misleading results because these overlaps are dominated by SNPs with no effect on TF binding.
Altogether, 3,601 of our bQTLs have been previously implicated by GWAS, either directly or indirectly via LD (r2 > 0.8 in YRI) (Table S2). These represent 2,282 different disease-associated variants, 995 (44%) of which are associated with bQTLs for multiple factors. Interestingly, this is 8.0-fold higher than the overall fraction of bQTLs associated with multiple factors, suggesting that variants affecting multiple TFs are more likely to impact phenotypes. Certain TF combinations may be especially likely to affect particular traits: for example, although PU.1 and Stat1 are known to act cooperatively (Aittomäki et al., 2004), they have few shared bQTLs. Nonetheless, two of their shared bQTLs are associated with atopic dermatitis (Sun et al., 2011; Hirota et al., 2012) —over 5000-fold more than expected by chance (p = 7×10−8). No other disease shows a significant enrichment for this TF pair, suggesting the possibility of a specific role for these TFs in atopic dermatitis, though additional examples will be required to confirm this trend.
Discussion
In this work, we have introduced an approach for mapping cis-acting QTLs for molecular traits. Compared to the standard QTL mapping approach, we achieved a greater than 25-fold reduction in cost and effort. We applied this method to map thousands of QTLs for TF binding and histone methylation, revealing a widespread role for common genetic variants to modulate TF binding in humans. We found that single genetic variants can often affect the binding of multiple TFs; that CTCF is a major pioneer factor capable of recruiting diverse TFs to its binding sites; that bQTLs can have long-range effects on cis-regulation, mediated by chromosomal contacts; and that bQTLs can also affect the extent of these long-range contacts.
Our collection of bQTLs is also a valuable resource for any researchers investigating the molecular mechanisms of disease risk loci. Indeed, our observation that disease-specific bQTL enrichments differ substantially from simple TF binding site enrichments (Figure 6E) suggests that bQTLs provide a critical level of information regarding disease etiologies. However since any given bQTL/GWAS variant overlap could be due to chance, these should be regarded as mechanistic hypotheses to guide further investigation.
Although it is clear that the vast majority of GWAS-implicated variants are noncoding, and that many fall into tissue-specific enhancers, their effects on transcription or other cellular processes are still mostly unknown. Considering the extensive GWAS overlap we found with bQTLs for only five TFs in one cell type, our work suggests that a substantial fraction of genetic variation affecting human phenotypes may act via TF binding. Indeed, the number of GWAS hits in our bQTL catalog is greater than has been reported in recent eQTL studies analyzing RNA-seq data from hundreds or thousands of samples (Lappalainen et al., 2013; GTEx Consortium 2015). How could these bQTLs affect phenotypes, if they are not also eQTLs? Although some bQTLs may act through processes other than transcription (e.g. replication timing), a more likely explanation is that their effects on transcription have not been detectable thus far. This could be due to small effect sizes, tissue-specificity, or environmental-specificity; or alternatively, some may only affect transcriptional variability, either between cells or between individuals (Ansel et al., 2008; Fraser and Schadt 2010; Hulse and Cai 2013). Of course, many bQTLs may have no effects on transcription or downstream phenotypes, though this will be difficult to distinguish from other scenarios listed above.
Our pooled ChIP-seq approach can be extended in many ways. Larger pools will allow inclusion of low-frequency variants, and deeper sequencing will allow detection of smaller effects. Our approach can be applied to any cell type from any species, with only small amounts of biological material needed from each individual. In addition, our pooling framework is not limited to ChIP—any method that isolates a subset of DNA or RNA could be used in its place. For example, our approach could map variants in DNA affecting chromatin accessibility or noncoding RNA localization, or variants in RNAs affecting RNA-protein interactions or translational pausing. Other pooling-based frameworks that do not involve isolating a subset of DNA/RNA are also possible, as we have shown for mapping QTLs affecting DNA methylation (Kaplow et al., 2015). Large collections of cis-regulatory variants, such as those we present here, may ultimately enable the development of predictive models of TF binding and transcription.
Experimental Procedures
Cell culture conditions
LCLs from unrelated Yoruban individuals were obtained from the Coriell Institute (Camden, NJ). The LCLs were grown in RPMI-GlutaMAX-HEPES media (Life Technologies) with 15% FBS, 100 I.U./mL penicillin, and 100 μg/mL streptomycin at 37°C, 5% CO2.
ChIP-seq
Each individual cell line was grown to a density of 6–8×105 cells/mL and 7×105 cells were collected. Samples were pooled such that each individual was approximately equally represented in the pool. TF ChIPs were done using a pool of 60 individuals. The cells used for NF-κB ChIP were first treated with 25 ng/mL human recombinant TNF-alpha (eBioscience #14-8329) for six hours at 37°C, 5% CO2. After stimulation, cells were cross-linked in 1% formaldehyde for 10 minutes at room temperature. Nuclear lysates were sonicated using a Branson 250 Sonifier (power setting 2, 100% duty cycle for 7 × 30-s intervals), yielding chromatin fragments of 50–2000 bp. Lysates were treated overnight at 4°C with 8 μg of antibody (Santa Cruz Biotechnology: JunD (sc-74), NF-κB (sc-372), Pou2f1/Oct-1 (sc-232), PU.1 (sc-22805), Stat1 (sc-345)). Protein-DNA complexes were captured on Dynabeads-Protein G (Life Technologies, cat #10003D) and eluted in 1% SDS TE buffer at 65°C. Purified ChIP-seq libraries were generated as described in Kasowski et al. (2010), with two biological replicates per TF, and sequenced on an Illumina HiSeq 2000 (101 bp, paired-end reads). H3K4me3 ChIP was performed as described in McVicker et al. (2013), using 4 μg of antibody for H3K4me3 (Abcam ab8580) in a pool of 71 LCLs. Libraries were sequenced on an Illumina HiSeq 2000 (50 bp, single-end reads). All data are available at NCBI SRA (SRP058204).
Calculating post-ChIP allele frequencies
Genotypes were imputed for all individuals for 18,931,934 SNPs, using a published imputation pipeline (Degner et al., 2012). Reads were mapped using the WASP pipeline (van de Geijn et al. 2015). Briefly, raw fastq reads were mapped to the human reference sequence (GRCh37) using bowtie2 (with parameters: -N 0 -L 20 --end-to-end --np 0 --n-ceil 0, 0.15) (Langmead et al., 2012). We used the “find_intersecting_snps.py” script from the WASP pipeline to identify reads that have mapping biases, remapped with the same bowtie2 parameters, and used “filter_remapped_reads.py” to retrieve reads that remapped correctly. These mapped BAM files were sorted using samtools sort and duplicate reads were filtered out for each replicate using samtools rmdup (Li et al., 2009). After duplicate removal, replicates were combined for all subsequent analysis. Samtools mpileup was used to retrieve all mapped reads aligning to each SNP (with parameters: -Q 25 -f <hg19_genome> -l <SNPs.bed>).
Reads were filtered out if they had a base quality score less than 25 at the SNP. SNPs were excluded if they had a minor allele frequency (MAF) < 0.02 among the 60 YRI samples, or fewer than 15 reads mapping to either allele, or if they were located in an ENCODE “blacklisted” region (ENCODE Consortium, 2012). The post-ChIP allele frequencies were calculated by counting the number of reads from each allele. Although post-ChIP frequencies could be inferred for unobserved SNPs that are in perfect LD with observed ones, this would not result in any additional independent bQTLs, so we restricted our analysis to directly observed SNPs.
Mapping and analyzing bQTLs
Additional methods are described in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We would like to thank B. van de Geijn, Y. Gilad, T. Kawli, G. McVicker, J. Pritchard, M. Snyder, D. Spacek, R. Tewhey, and members of the Fraser Lab for sharing data, protocols, cell lines, and advice. This work was supported by NIH grant 1DP2OD008456-01. AKT is an NSF predoctoral fellow. HBF is a Pew Scholar in the Biomedical Sciences.
Footnotes
Author contributions
HBF conceived the project. AKT performed the experiments and analysis. MM assisted with ChIP-seq of H3K4me3. TM, DG, BH, and HBF assisted with analysis. AKT and HBF wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aittomäki S, Yang J, Scott EW, Simon MC, Silvennoinen O. Molecular basis of Stat1 and PU.1 cooperation in cytokine-induced Fcgamma receptor I promoter activation. Int Immunol. 2004;16:265–74. doi: 10.1093/intimm/dxh037. [DOI] [PubMed] [Google Scholar]
- Amin Al Olama A, Dadaev T, Hazelett DJ, Li Q, Leongamornlert D, et al. Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among Europeans. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv203. Advance online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel J, Bottin H, Rodriguez-Beltran C, Damon C, Nagarajan M, Fehrmann S, Francois J, Yvert G. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 2008;4:e1000049. doi: 10.1371/journal.pgen.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banovich NE, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoure N, et al. Quantifying ChIP-seq data, a spiking method providing an internal reference for sample-to-sample normalization. Genome Res. 2014;24:1157–68. doi: 10.1101/gr.168260.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, et al. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 2014;10:e1004798. doi: 10.1371/journal.pgen.1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 2015;25:1268. doi: 10.1101/gr.184671.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Messer PW, Petrov DA. Genome-wide signals of positive selection in human evolution. Genome Res. 2014;24:885–95. doi: 10.1101/gr.164822.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Xie X. Common polymorphic transcript variation in human disease. Genome Res. 2009;19:567. doi: 10.1101/gr.083477.108. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Lam S, Neumann S, Kobor MS. Population-specificity of human DNA methylation. Genome Biology. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Schadt EE. The quantitative genetics of phenotypic robustness. PLoS One. 2010;5:e8635. doi: 10.1371/journal.pone.0008635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB. Gene expression drives local adaptation in humans. Genome Res. 2013;23:1089–96. doi: 10.1101/gr.152710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–65. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Conti DV, Wang K. Enlight: web-based integration of GWAS results with biological annotations. Bioinformatics. 2015;31:275–6. doi: 10.1093/bioinformatics/btu639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–10. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Amer J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- Hulse AM, Cai JJ. Genetic variants contribute to gene expression variability in humans. Genetics. 2013;193:95–108. doi: 10.1534/genetics.112.146779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplow IM, MacIsaac JL, Mah SM, McEwen LM, Kobor MS, Fraser HB. A pooling-based approach to mapping genetic variants associated with DNA methylation. Genome Res. 2015;25:907–17. doi: 10.1101/gr.183749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–2. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–5. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–7. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie R, O’Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–94. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Segal E. In pursuit of design principles of regulatory sequences. Nat Rev Genet. 2014;15:453–68. doi: 10.1038/nrg3684. [DOI] [PubMed] [Google Scholar]
- Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015;25:1018–29. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Yang H, Zhang H, Zhou L, Guo Z. A novel transcription mechanism activated by ethanol: induction of Slc7a11 gene expression via inhibition of the DNA-binding activity of transcriptional repressor octamer-binding transcription factor 1 (OCT-1) J Biol Chem. 2013;288:14815–23. doi: 10.1074/jbc.M113.466565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–9. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani M, et al. Functional cooperation between JunD and NF-kB in rat hepatocytes. Oncogene. 2001;20:5132. doi: 10.1038/sj.onc.1204678. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TE, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012;22:860–9. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. PNAS. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, Tomlinson IP, Ratcliffe PJ, Mole DR. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–5. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- van de Geijn B, McVicker G, Gilad Y, Pritchard JK. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods. 2015 doi: 10.1038/nmeth.3582. Advance Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–50. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–43. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. PNAS. 2013;110:11952–7. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y. Association of a polymorphism of BTN2A1 with myocardial infarction in East Asian populations. Atherosclerosis. 2011;215:145–52. doi: 10.1016/j.atherosclerosis.2010.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.