Abstract
Background
Xanthomonas oryzae pv. oryzae (Xoo) is the causal agent of Bacterial Leaf Blight (BB), an emerging disease in rice in West-Africa which can induce up to 50 % of yield losses. So far, no specific resistance gene or QTL to African Xoo were mapped. The objectives of this study were to identify and map novels and specific resistance QTLs to African Xoo strains.
Results
The reference recombinant inbred lines (RIL) mapping population derived from the cross between IR64 and Azucena was used to investigate Xoo resistance. Resistance to African and Philippine Xoo strains representing different races was assessed on the RIL population under greenhouse conditions. Five major quantitative trait loci (QTL) for resistance against African Xoo were located on different chromosomes. Loci on chromosomes 1, 7, 9, 10 and 11 explained as much as 13 %, 37 %, 13 %, 11 % and 15 % of resistance variation, respectively. A major novel QTL located on chromosome 7 explained 37 % of the phenotypic variance to the African Xoo corresponding to race A3 whereas that on chromosome 11 is effective to all African races tested. Together with genes and QTLs for resistance to bacterial blight previously described, the QTLs described here were mapped onto the reference O. sativa subs japonica (var. Nipponbare) physical map.
Conclusion
We characterized new resistance QTLs. While some co-localize with known resistance genes/QTLs to Asian strains, others are specific to African strains. We result with new information on genes and QTLs for resistance to bacterial blight that will be useful for controlling the disease.
Keywords: Molecular mapping, QTL, disease resistance, Xanthomonas oryzae pv. oryzae, Oryza sativa
Background
Rice is a staple food for much of the world’s population, including that of sub-Saharan Africa. Population growth and migration have recently lead to a rapid growth in rice consumption in dozens of African countries. With the recent expansion and intensification of rice cultivation in Africa, rice diseases have concomitantly increased in most African rice growing areas (Sere et al. 2005; Traoré et al. 2009). Bacterial Blight (BB), caused by Xanthomonas oryzae pv. oryzae (hereafter, Xoo), is one of the most serious rice bacterial disease in Africa. The disease is prevalent in irrigated and rain-fed lowland rice growing areas. BB was first reported in Mali in 1979 and later in Senegal, Niger, Nigeria, Gabon, Mauritania, Benin, Burkina and Cameroon. Over the last decade a significant recurrence of this disease was observed in several regions in Africa (Reckhaus et al. 1983; Gonzalez et al. 2007; Basso et al. 2011; Verdier et al. 2011).
Resistance (R) genes are largely used in rice breeding programs in Asia to control BB disease. More than 30 R genes, which are given the prefix Xa for Xanthomonas, have been identified so far (for a review see Verdier et al. 2011) among which a few are deployed in breeding programs. Important prerequisites to the deployment of R genes are as follows: 1) to have an intensive knowledge of Xoo population structure, race distribution and frequency 2) to determine the durability of resistance of R genes to be deployed.
Most of the BB R genes provide complete race-specific resistance to Xoo strains. Different combinations of Xa4, xa5, Xa7, xa13 and Xa21 have been incorporated in popular rice commercial varieties in different countries in Asia (Century et al. 1999; Singh et al. 2006; Swamy et al. 2006; Perez et al. 2008; Sundaram et al. 2009, Shanti 2010; Suh et al. 2013; Ruengphayak et al. 2015). Few examples indicate that some R genes used for controlling BB disease are overcome by virulent strains as shown in Korea with the resistant gene Xa21 (Lee et al. 1999; Zhang et al. 2006). Xa4 is a gene used for more than 30 years and has introgressed in high yielding varieties in Asia but has lost efficacy in many cultivated areas (Mew et al. 1992). Although durability of BB R genes is, in part, because mutation of Xoo to overcome R genes (Vera Cruz et al. 2000), recent field and laboratory studies have also shown the influence of temperature on the interactions of rice R gene with Xoo. High temperatures are conducive to BB disease, and most BB R genes, including Xa4, are less effective at controlling BB disease at high temperatures (Vera Cruz et al. 2000; Webb et al. 2010).
Xanthomonas oryzae (Xo) is a diverse species, with distinct phylogenetic lineages comprising US Xo, Asian Xoo, African Xoo, and Xanthomonas oryzae pv. oryzicola (Xoc) (Triplett et al.2011; Hajri et al. 2012). Another lineage improperly named Xanthomonas campestris pv. leersiae (Xcl) comprises strains isolated on weeds (Wonni et al. 2014). Previous work highlighted differences in the race structure between Asian and African Xoo strains (Gonzalez et al. 2007). Virulence assays revealed three races (A1, A2 and A3) present in Mali, Burkina-Faso, Niger and Cameroon that do not represent any of the known Xoo races characterized in Asia so far (Gonzalez et al. 2007; Triplett et al. 2011). According to experiments conducted on BB isogenic lines (IRBB), BB resistance genes Xa4, xa5 and Xa7 provide resistance to some African Xoo strains (Gonzalez et al. 2007). Although in absence of a complete overview of Xoo race prevalence in Africa, we anticipated that Xa4, xa5 and Xa7 could provide resistance against strains of Xoo in Burkina-Faso, Cameroun and Niger. Despite the increasing importance of BB in Africa, little is known on the genetic determinism of resistance. O. glaberrima and O. sativa accessions were screened for resistance to African Xoo strains. The tropical japonica landrace Azucena is susceptible to all African Xoo strains. Few accessions, among them the indica cultivar IR64, are highly resistant to African Xoo strains. None of these accessions had the xa5 or Xa21 resistance alleles (Djedatin et al. 2011) suggesting that these accessions carry new resistance genes that could be good targets for R gene discovery and further deployment.
With the completion of genome sequences for japonica and indica rice (Kawahara et al. 2013) and for O. glaberrima (Wang et al. 2014a, b), it is essential to have a better picture of the different Xa resistance genes and QTLs characterized so far and their positions in the rice genome.
The objectives of this study are to:
Identify and analyse the genetic basis of rice resistance to African Xanthomonas oryzae pv. oryzae strains by developing a QTL approach using the reference mapping population made of recombinant inbred lines (RIL) derived from the cross between IR64 and Azucena.
Map novel and known bacterial blight resistance genes and QTLs to Xoo strains and analyze their colocalization on the reference Nipponbare physical map.
For the first time in history, we report on specific resistance QTLs to African Xoo strains. These QTLs will be used in breeding program to enhance rice genetic resistance to BB in Africa.
Results
Study of inheritance of BB resistance
The average lesion length induced by Xoo African strains on IR64 and Azucena are respectively: 0.25 ± 0.1 and 16.1 ± 2 cm with Xoo MAI1; 0.76 ± 0.2 and 22.36 ± 2.7 cm with Xoo BAI4; 4.07 ± 1 and 22.29 ± 3 cm with Xoo BAI3; 2.07 ± 0.5 and 26.29 ± 3.3 cm with Xoo NAI8. IR64 is observed to be highly resistant and Azucena highly susceptible to all virulent strains of African Xoo tested so far. The disease scores of the RILs range widely, from as low as 0.12 cm to as high as 30.05 cm, 0.14 to 32.2 cm, 0.2 to 32.6 cm and 0.44 to 37.9 cm with Xoo strains MAI1, BAI4, BAI3 and NAI8, respectively (Table 1). This continuous variation of lesion lengths indicates the existence of QTLs underlying the segregation of resistance. Both parents, IR64 and Azucena, are susceptible to Asian Xoo strain PXO86 with an average lesion length of 16.46 ± 1.5 and 26 ± 3 cm, respectively. Conversely, IR64 is resistant to PXO61; the Philippines race 1, with an average lesion length of 1.92 ± 0.4 cm, whereas Azucena is susceptible with an average lesion length of 28.32 ± 3.1 cm. The lesion length of the 178 RILs lines shows a continuous variation with an average lesion length of 6.95 to 34.5 cm and 0.5 to 30.8 cm with PXO86 and PXO6, respectively (Table 1), indicating the resistance to Asian strains is controlled by QTLs.
Table 1.
Lesion length induced by African and Asian Xoo on recombinant inbred lines and their parents
| Xoo strains | Lesion length (cm) induced on | ||
|---|---|---|---|
| IR64 | Azucena | RIL | |
| MAI1 | O.25 ± 0.1 | 16.1 ± 2 | 0.12 to 30.05 |
| BAI4 | 0.76 ± 0.2 | 22.36 ± 2.7 | 0.14 to 32.2 |
| BAI3 | 4.07 ± 1 | 22.29 ± 3 | 0.2 to 32.6 |
| NAI8 | 2.07 ± 0.5 | 26.29 ± 3.3 | 0.44 to 37.9 |
| PXO86 | 16.46 ± 1.5 | 26.0 ± 3 | 6.95 to 34.5 |
| PXO61 | 1.92 ± 0.4 | 28.32 ± 3.1 | 0.5 to 30.8 |
Xoo Xanthomonas oryzae pv. oryzae, RIL Recombinant Inbred Lines
Mapping QTLs using SSR markers
The IR64 x Azucena genetic map used in this study were comprised of 226 SSR markers and covered 1652.06 cM of the genome with an average inter-marker interval of 7.31 cM. QTL mapping, based on ANOVA, evidences twelve putative QTLs induced by African Xoo strains (MAI1, BAI4, BAI3 and NAI8). Two of them (qABB-7 and qABB-11) have a large effect on chromosomes 7 and 11, respectively. The others induce small effect on chromosomes 1, 3, 4, 8, 9, 10. The composite interval mapping using WinQTLCartographer 2.5 reveals five specific QTLs underlying resistance to African Xoo strains including those detected by ANOVA on chromosomes 1, 7, 9, 10 and 11. The estimated additive effect indicates that these loci derive from the resistant parent IR64. qABB-1, specific to MAI1, is linked to SSR markers RM129 and RM493, with a LOD score of 4.72 and a percentage of variance explained (R2) of 8 %. qABB-7 is linked to RM125 and RM214 with a LOD score of 16.20 and R2 of 30 % for MAI1, and a LOD score of 13.98 and R2 of 30 % for BAI4. qABB-11 is close to RM224 and RM144 with a LOD score of 5.32, 5.87 and 4.18 with Xoo MAI1, BAI3 and BAI4, respectively. qABB-11 controls 7, 9, and 14 % of the phenotypic variation explained by Xoo BAI4, MAI1, and BAI3, respectively. The Inclusive Composite Interval Mapping using Qgene-4.3.0, the more accurate QTL analysis method, confirms five specific QTLs mapped on chromosomes 1, 7, 9, 10 and 11 underlying resistance to African Xoo strains. The estimated additive effects confirm that these loci derive from the resistant parent IR64. Additive effect, linked markers, LOD score value and PVE are summarized in Table 2. qABB-11 on chromosome 11 was involved in the resistance to all African Xoo strains tested so far. QTLs on chromosomes 9 (qABB-9) and 10 (qABB-10) are specific to Xoo strain NAI8 (race A1). Asian Xoo strains induce five resistance QTLs different from those induced by African’s strains except that on chromosome 11. This is also induced by Xoo strain PXO61 (Philippines race 1). Three resistance QTLs to Xoo strain PXO86 (Phil race 2) are mapped on chromosomes 5 (qBB-5), 8 (qBB-8) and 12 (qBB-12) (Table 2). As indicated by the estimated additive effects, the QTLs on chromosomes 5 and 8 are controlled by Azucena allele while that on chromosome 12 is underlined by IR64 allele. A resistance QTL (qBB-4) is detected on chromosome 4 for the Asian Xoo strain PXO61.
Table 2.
Novel QTLs mapped in IR64 x Azucena population using African and Asian Xanthomonas oryzae pv.oryzae
| Xoo a strains | Country of origin | QTL localization: chromosome | QTL name | LODb score | Closely Linked marker | Marker position (cM) | Additive effect | Donor Allele | PVEc (%) |
|---|---|---|---|---|---|---|---|---|---|
| MAI1 | Mali | 1 | qABB-1 | 5.068 | RM129 | 58.45 | 1.7 | IR64 | 13.4 |
| 7 | qABB-7 | 16.006 | RM125 | 5.75 | 3.2 | IR64 | 36.6 | ||
| 11 | qABB-11 | 4.666 | RM144 | 100.21 | 1.5 | IR64 | 12.4 | ||
| BAI4 | Burkina Faso | 7 | qABB-7 | 13.943 | RM125 | 5.75 | 3.5 | IR64 | 33.4 |
| 11 | qABB-11 | 4.68 | RM144 | 100.21 | 1.8 | IR64 | 12.8 | ||
| BAI3 | Burkina Faso | 11 | qABB11 | 5.728 | RM144 | 100.21 | 2.5 | IR64 | 15.3 |
| NAI8 | Niger | 9 | qABB-9 | 4.359 | RM242 | 69.28 | 2.3 | IR64 | 12.9 |
| 10 | qABB-10 | 3.606 | RM294A | 74.36 | -2.23 | Azucena | 10.8 | ||
| 11 | qABB-11 | 6.03 | RM144 | 100.21 | 2.7 | IR64 | 17.4 | ||
| PXO86 | Philippines | 5 | qBB-5 | 4.209 | RM440 | 70.61 | -1.68 | Azucena | 11.5 |
| 8 | qBB-8-2 | 4.957 | RM281 | 129.96 | -2.16 | Azucena | 13.4 | ||
| 12 | qBB-12 | 4.869 | RM512 | 38.32 | 1.9 | IR64 | 13.2 | ||
| PXO61 | Philippines | 4 | qBB-4 | 3.403 | RM252 | 84.70 | -1.7 | Azucena | 10 |
| 11 | qABB-11 | 36.78 | RM144 | 100.21 | 7.3 | IR64 | 67.9 |
Xoo Xanthomonas oryzae pv. oryzae, LOD logarithm of odds, PVE Percentage of variance explained
Heredity studies
The screening of the recombinant inbred lines using Asian Xoo strain PXO61 evidenced the same QTL previously induced on chromosome 11 (qABB-11) by all African Xoo strains (with LOD score = 36.78 and R2 = 67) (Table 2).
The segregation ratio obtained by screening F2: IR24 x IRBB4 population with the Asian Xoo strain PXO61 is 3 resistant for 1susceptible. This is the segregation ratio of a dominant gene in a F2 population. It was the Xa4 dominant gene which was specific to PXO61. On the contrary, the segregation studies in the same population using the African Xoo strain BAI3 revealed a ratio of 3 susceptible for 1 resistant, that is the segregation ratio of a recessive gene in a F2 population. Then, the African Xoo strain BAI3 induces a recessive gene at the locus of qABB-11.
The study of bacterial growth and of the xylem colonization speed by the bacteria shows that both strains PXO61 and BAI3 grown in the same way in IR24 rice variety which is susceptible to bacterial blight. This fact is expressed by a more or less equal number of PXO61 and BAI3 colonies detected at each time. It’s the case on the twelfth day after inoculation when the number of PXO61 and BAI3 colonies is about Log10 (8) in the fraction A and Log10 (9) in the fraction B. This number increases to Log10 (10) and Log10 (10.5), respectively, in the fractions C and D (Fig. 1). On the other hand, in the resistant IRBB4 isogenic line, the colonies number of these two strains is far inferior to those got in IR24 variety (Fig. 2). In this case, both strains grown unequally. On the 12th day after inoculation, we count Log10 (8) of BAI3 and Log10 (6) of PXO61 in the fraction A, Log10 (8) of BAI3 against Log10 (4) of PXO61 in the fraction B. The Asian Xoo strain PXO61 is then stopped at the level of fraction B, which is expressed by the absence of colonies in the fractions C and D. On the other hand, Log10 (6) colonies of the African Xoo strains BAI3 have been shown in the fraction C and D (Fig. 2).
Fig. 1.
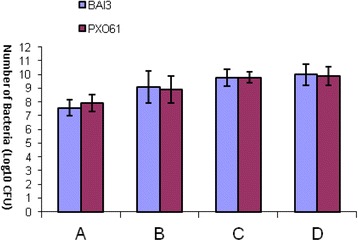
Bacteria growth during twelve days after inoculation of IR24 rice variety. A = 1st fragment of 5 cm (from the inoculation point) of infected leaves. B = 2nd fragment of 5 cm (from 5 to 10 cm to the inoculation point) of infected leaves. C = 3rd fragment of 5 cm (from 10 to 15 cm to the inoculation point) of infected leaves. D = 4th fragment of 5 cm (from 15 to 20 cm to the inoculation point) of infected leaves
Fig. 2.
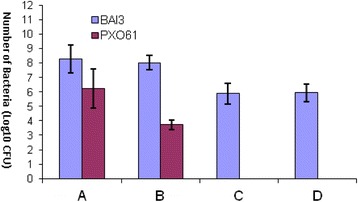
Bacteria growth during twelve days after inoculation of rice isogenic line IRBB4. A = 1st fragment of 5 cm (from the inoculation point) of infected leaves. B = 2nd fragment of 5 cm (from 5 to 10 cm to the inoculation point) of infected leaves. C = 3rd fragment of 5 cm (from 10 to 15 cm to the inoculation point) of infected leaves. D = 4th fragment of 5 cm (from 15 to 20 cm to the inoculation point) of infected leaves
The results of the quantification and of bacterial kinetic show clearly that the African Xoo strain BAI3 is quite different from the one of the Asian Xoo strain PXO61 that lights a dominant gene which was the Xa4 gene. In fact, in the IRBB4 isogenic line, resistant to both strains, the lesion caused by BAI3 was three times bigger than the one caused by PXO61. Xa4 prevent the multiplication and the progression of PXO61 in the vascular tissues which is expressed on the one hand by the reduced number of PXO61 in these tissues and, on the other hand by the total absence of this bacteria 10 cm to the inoculation point. This is in accordance with the functioning of dominant resistance gene. On the other hand, BAI3 colonies have been observed even at 20 cm to the inoculation point. This strain managed to multiply and to colonize the vascular tissues thus leading to a less resistant phenotype (Fig. 2 ). The African Xoo strain BAI3 would thus induced a recessive gene.
Discussion
In this study, novel resistance QTLs to BB are identified and mapped. Moreover, the information on known bacterial blight genes/QTLs characterized so far was updated (Additional file 1) and their genetic and physical localization positioned on the reference rice physical map (var Nipponbare).
IR64 has been confirmed as highly resistant to African Xoo strains. The IR64 x Azucena derived mapping populations segregated for BB resistance. Novel QTLs were discovered, mapped to chromosomes 1, 4, 5, 7, 8, 9, 10, 11, 12. Most were of relatively small effect except on chromosomes 7 and 11. qBB-4, qBB-5, qABB-9 and qBB-12 mapped to genomic regions where BB resistance QTL and or Xa genes were previously characterized in other crosses, indicating that BB-resistance genes to Asian and African Xoo strains may be shared by several rice accessions. Out of five, four QTLs induced by African Xoo strains are different from those induced by the Philippines Xoo strain PXO61 indicating these strains induced different resistant genes. QTLs induced by African Xoo strains are underlined by the allele of the indica rice IR64 while Azucena (japonica) is the donor parent of resistance against Asian Xoo strains except with strain PXO61.
The genetic characterization of Xoo strains together with the recent advances in X. oryzae genomic studies indicate that African X. oryzae pv. oryzae strains form a separate group within the X. oryzae (Gonzalez et al. 2007; Triplett et al. 2011; Hajri et al. 2012; Wonni et al. 2014). Also their repertoire of transcription activator-like (TAL) effectors is reduced compared to the Asian Xoo one (Gonzalez et al. 2007).
Two resistance QTLs with main effects were identified. The first, qABB-7, induced by the African Xoo strains MAI1 and BAI4 on chromosome 7 and the second, qABB-11, induced on chromosome 11 by all African strains tested so far are particularly interesting. The first one, qABB-7, controls 37 % of the phenotypic variance with a high LOD score of 16 (Table 2) compared to 2.5 to 3 fixed LOD score in other QTL studies (Wang et al. 2006a, b; Sakraborty and Zeng 2011). Our preliminary data indicate that the single resistance genes Xa4, xa5 and Xa7 provide strong levels of resistance to African Xoo strains collected in the 1980’s and in 2003 (Gonzalez et al. 2007). Xa4 is located in the qABB-11 region. The fine mapping of qABB-7 and qABB-11 is in progress. Our study also reveals the difference between African and Asian Xoo strains in terms of virulence. The resistant and susceptible phenotypes of the parents (IR64 and Azucena) induced by African Xoo strains greatly contrast with those induced by Asian Xoo strains (Table 1). According to the previous results, resistance to BB depends on the rice genetic background. O. sativa subspecies indica appeared to be the best source of resistance to bacterial blight in rice conversely to the African cultivated rice O. glaberrima that showed a relative narrow resistance basis to BB (Djedatin et al. 2011).
Preliminary segregation and bacteria growth studies revealed that the two most important QTLs induced by African Xoo on rice chromosomes 11 are recessive while most of the resistance genes characterized so far are dominant with the exception of xa5, xa8, xa13, xa19, xa20, xa24, xa33(t), xa34(t) and xa35(t) (Verdier et al. 2011). These results are compatible with the hypothesis that African and Asian strains have different effector genes that induced different resistance genes (Yu et al. 2011a, b; Hajri et al. 2011).
So far, 40 R genes and 17 QTLs conferring host resistance against various strains of Xoo have been identified (Li et al. 1999; Chen at al. 2002; Gu et al. 2004; Blair et al. 2003; Ramalingam et al. 2003; Wu et al. 2008a, b; Cheema et al. 2008; Ruan et al. 2008; Korinsak et al. 2009; Wang et al. 2009; Sundaram et al. 2009; Chen et al. 2011; Bhasin et al. 2012; Han et al. 2014; Kim et al. 2015). Approximately one third of naturally occurring R genes against Xoo (xa5, xa8, xa13, xa15, xa19, xa20, xa24, xa26, xa28, xa31(t), xa33(t) and xa34) are recessive (Sanchez et al. 1999; Wu et al. 2008a, b; Ruan et al. 2008; Korinsak et al. 2009). More than 20 R genes were mapped onto rice chromosomes, and some of them have been well characterized. Nine resistance genes have been molecularly cloned including six dominant genes, Xa21 (Song et al., b), Xa1 (Yoshimura et al. 1998), Xa3/Xa26 (Sun et al. 2004; Xiang et al. 2006), Xa27 (Gu et al. 2004; Bimolata et al. 2013), Xa10 (Tian et al.2014), Xa23 (Wang et al. 2014a, b) and three recessive xa5 (Iyer and McCouch 2004) and xa13 (Chu et al. 2006a, b), xa25 (Liu et al. 2011) have been cloned. All known resistance genes/QTLs and their flanking markers mapped on the reference Nipponbare physical map show that they are unequally distributed on rice chromosomes. Chromosomes 4 and 11 appeared to carry most of the known BB resistance genes. Indeed, these chromosomes are known to carry clusters of resistance genes analogs (RGA) (Mago et al. 1999; Ghazi et al. 2009). Resistance QTLs to rice sheath blight resistance (Zou et al. 2000) and resistance genes to blast (Wang et al. 1994) also clustered on chromosome 11. Many QTLs mapped closely to single dominant or recessive Xa genes. It is the case of Xa1 and Xa2 which bracketed AQBT008 on chromosome 4. Some of them are considered as a single gene like AQBT023 designated as Xa4 on the Lemont x Teqing map (Li et al. 1999). After interpolation on the Nipponbare physical map, AQBT023 was shown to be distinct to Xa4 located on chromosome 11.
This interpolation on the reference Nipponbare physical map highlights common, specific and novel QTL/genes for resistance to African and Asian Xoo strains. Indeed, xa34(t) resistance gene to Chinese Xoo races V co-localize with qABB-1, the resistance QTL induced by the African Xoo strain MAI1 on rice chromosome 1. In the same way, AQBT021 effective on Asian Xoo strains, co-localize with qABB-10, a resistance QTL induce by the African Xoo strain NAI8 on chromosome 10. xa8 (Ogawa and Yamamoto 1987), qBB7 (QTL identified on chromosome 7, Ramalingam et al. 2003) and qABB-7 overlap on chromosome 7 as well as xa24 (Wu et al. 2008a, b) and AQBT001 on chromosome 2. Lemont and Azucena, the donors’ parents of AQBT021 and qABB-10 respectively, belong to the japonica subspecies. These resistance QTLs may be the same. On chromosome 8, xa13, qBB8 and qBB-8-2 co-localize perfectly. qBB8 and qBB-8-2 may be underlined by the recessive xa13 gene, but the heredity of qBB8 and qBB-8-2 has not been studied yet. On chromosome 11, the QTL induced by African Xoo strains co-localized with Xa4, Xa3, Xa32 and xa35(t), the known single resistance genes to Asian Xoo strains. These Xa genes indicated that some major R genes also contribute to quantitative resistance as reported in common bean in which RGAs co-localized with anthracnose-specific QTL (Geffroy et al. 2000). This was also the case of QTL associated with resistance to stripe rust and Barley yellow dwarf virus in barley (Toojinda et al. 2001), and partial resistance to Cucumber mosaic virus in pepper (Pflieger et al. 1999). Specifics and novels QTLs inducing resistance to African and/or Asian Xoo strains are identified as qABB-9 induced by the African Xoo strain NAI8 on chromosome 9, qBB-4 and qBB-5 induced on chromosomes 4 and 5 by Asian Xoo strains PXO61 and PXO86 respectively, (Fig. 3).
Fig. 3.
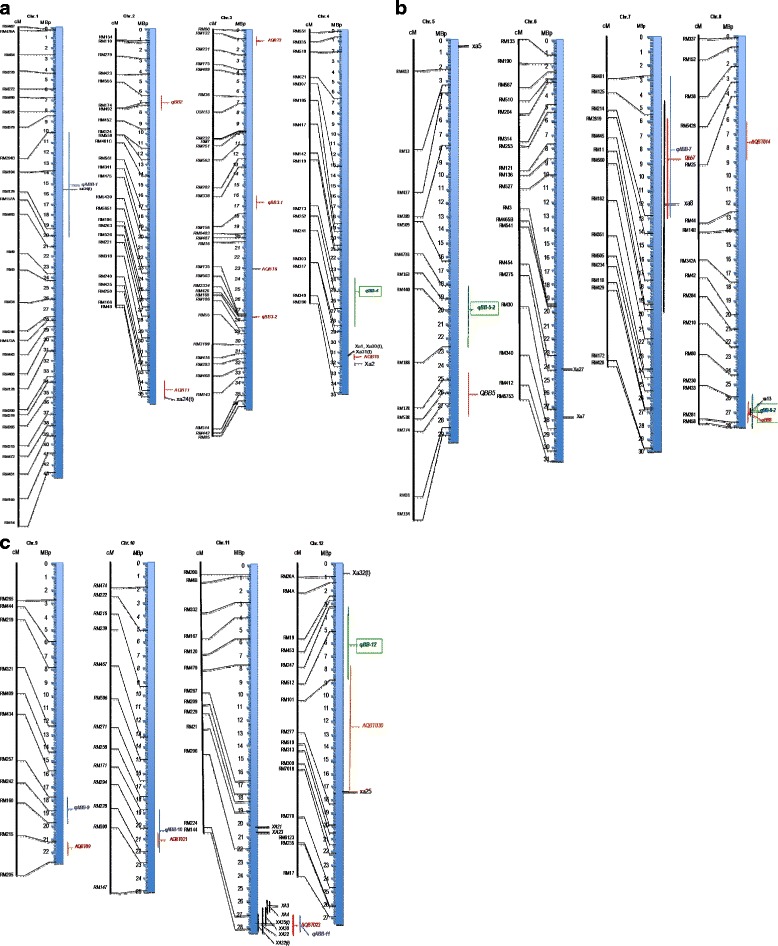
a, b and c: Integrative map showing all known resistance QTLs/genes to African and Asian Xoo. The vertical graduated blue thick lines represent the physical map of each chromosome and is linked to the genetic map on the left showing the microsatellites markers (RM). On the right size of each physical map the genes and QTLs are indicated in red, black and green. In black are the R genes that were previously identified with Asian Xoo strains. R genes that were cloned are indicated by a horizontal line to a specific location on the physical map. The genes that are not cloned yet are represented by a vertical line indicating the confidence interval. In red we indicated QTLs that were detected in others rice breeding populations. Novel QTLs identified using the African Xoo strains are represented in blue those induced by the Philippines strains PXO61 and PXO86 are shown in green
In addition to specific resistance QTLs to African Xoo trains, the known resistance genes/QTLs to Asian Xoo strains which co-localized with QTL induced by African Xoo will be used in rice breeding programs to develop bacterial blight resistant cultivars for Africa.
Conclusion
We mapped several resistance QTLs to bacterial blight in rice using a reference recombinant inbred lines derived from the cross between Azucena and IR64 rice varieties. Some of them are specific and novel to African or Asian strain of Xanthomonas oryzae pv. oryzae such as qABB-9 induced by the African Xoo strain NAI8 on chromosome 9, qBB-4 and qBB-5 induced on chromosomes 4 and 5 by Asian Xoo strains PXO61 and PXO86, respectively. The others co-localize with known Xoo resistance genes/QTLs. It is the case of qABB-1, the resistance QTL induced by the African Xoo strain MAI1 on rice chromosome 1 which co-localize with xa34(t) resistance gene to Chinese Xoo races V. In the same way, AQBT021 effective on Asian Xoo strains, co-localize with qABB-10, a resistance QTL induced by the African Xoo strain NAI8 on chromosome 10.
So far, and for the first time, specific resistance QTLs to African Xoo are identified and mapped. The fine mapping of the QTL induced by African Xoo MAI1 and BAI4 on chromosome 7 which controlled 37 % of phenotypic variance as well as the one induced on chromosome 11 by all African Xoo tested is ongoing and will provide news markers for breeding program.
Methods
Plant materials
The reference mapping population consisted in 172 recombinant inbred lines (RIL) obtained by single seed descent (SSD) from the cross between the tropical japonica landrace Azucena (susceptible parent) and the indica cultivar IR64 (resistant parent) was used to identify and map the bacterial blight (BB) resistance QTLs. IR64 is an elite improved indica rice grown in tropical Asia. It carries the BB resistance gene Xa4 and has a large spectrum of resistance to Xoo. The Azucena landrace is susceptible to BB disease, and tolerant to drought and other abiotic stresses. The IR64 x Azucena mapping population was used to develop genetic map for characterizing gene/QTLs associated to drought, iron toxicity tolerance and resistance to the Rice yellow Mottle Virus (Boisnard A et al. 2007).
Bacterial strains and plant inoculations
Six Xanthomonas oryzae pv. oryzae (Xoo) strains were used to evaluate resistance to BB in the mapping population. These strains included four African Xoo strains: BAI3 and NAI8 (race A1) from Burkina-Faso and Niger respectively, BAI4 (race A2) from Burkina-Faso, MAI1 (race A3) from Mali and two Asian strains PXO61 (Philippines race 1) and PXO86 (Philippines race 2). The bacterial strains were cultured on PSA medium (per liter of H2O, 10 g of peptone, 10 g of sucrose, 1 g of glutamic acid, and 16 g of bacto agar at pH 7.0) overnight. Inoculum was prepared by re-suspending bacterial culture in sterile, distilled water at an optical density of 0.2 (DO600), bearing approximately 108 cfu per ml. The progenies and their parents were grown under controlled conditions (28 °C; 80 % humidity and 12 h day length) in greenhouse at IRD Montpellier/France. They were inoculated at the booting stage (6 weeks after germination) using the leaf-clipping method (Kauffman et al. 1973) in which the fully-opened leaves were cut with scissors dipped in the bacterial suspension. Ten plants per genotype and two uppermost fully expanded and randomly chosen leaves were inoculated with each strain. Phenotypic evaluations, consisting of the disease scoring, were done 3 weeks after inoculation by measuring the leaf lesion length. Disease reactions were classified according to the mean lesion length (LL) as following: resistant (R) if the lesion length was < 5 cm, moderately resistant (MR) with LL of 5 to 10 cm; moderately susceptible (MS) with LL of 10 to 15 cm, susceptible (S) with LL > 15 cm.
Molecular mapping analysis using SSR markers
Leaves of rice plants were harvested and ground in liquid nitrogen. Genomic DNA was extracted as previously described (Edwards et al. 1991). PCR were performed in 15 μL reactions in an automated thermal cycler and the program consisted of the following cycles: initial denaturation at 94 °C for 5 min; 30 cycles of denaturation at 94 °C for 30 s, annealing temperature for 30 s and extension at 72 °C for 45 s; and a final extension step at 72 °C for 5 min. Amplified products were analyzed by electrophoresis on 6.5 % polyacrylamide gels, using electrophoresis system LICOR; or by electrophoresis in a 2 % agarose gel.
Statistical analysis and Xoo resistance QTLs mapping
A linkage map comprising 226 SSR markers and constructed from the RIL population was used for mapping resistance QTL to Xoo. An analysis of variance, using marker genotypes as the groups, was carried out using MapDisto (Lorieux 2007). Data files were prepared using the Export map and data function of MapDisto. Analyses of distribution of the phenotypic traits as well as QTL detection were mainly performed using the Qgene v. 4.3.0 program (Nelson 2005, http://www.qgene.org) and Windows QTL cartographer 2.5 (Wang et al. 2006b). Different methods were compared such as Single-marker regression (SMR), Simple interval mapping (SIM), and Composite interval mapping (CIM). The Forward cofactor selection option was used in CIM. The LOD score statistic was used for all methods in order to make the results comparable. Empirical thresholds to declare presence of a QTL were obtained using the resampling by permutation method, performing 1,000 iterations for each trait/chromosome combination (log-likelihood of odds (LOD) score of 3).
Heredity studies
QTL mapping using Asian Xoo strain PXO61
At the locus of qABB-11, the QTL on chromosome 11 that was involved in the resistance on all African Xoo tested, were localized a cluster of Xa genes including Xa3, Xa4 and Xa21. Xa3 was not effective against Xoo race 1 (Gonzalez et al. 2007). Xa21 was identified in Oryza longistaminata, a wild rice race. Therefore, Xa4 would be the only one Xa candidate gene at the above locus. In order to validate the presence of Xa4 gene at this locus, the Asian Xoo strain PXO61 belonging to Philippines race 2 was used to screen the RIL population according to Kauffman et al. (1973). The resistance of rice to PXO61 strain was specifically under Xa4 control.
Development and screening of F2: IR24 x IRBB4 population
The Asian rice variety IR24 belonging to indica sub-species was crossed with isogenic line IRBB4 carrying Xa4 gene. IRBB4 was used as donor while IR24 represent a recurrent parent. The Hybrid F1 obtained was used to generate a F2 population. Two sets of 100 F2 individuals were screened with the African Xoo strain BAI3 and the Asian one PXO61, respectively.
In planta growth experiments
Rice variety IR24 with its isogenic line IRBB4 were screened using African Xoo strain BAI3 and Asian Xoo strain PXO61. Two, three and four pieces of 5 cm from the apex to the base of infected leaf were harvested 4, 8 and 12 days after inoculation, respectively. On each day, infected leaves fragments were harvested on three different plants. Infected leaves collected were briefly rinsed in 70 % of ethanol for 10 s followed by submersion in sterilized water. Leaves were put into 2 ml eppendorf tubes containing 2 metallic beads (ϕ = 3 mm), frozen by submersion into liquid nitrogen and ground into fine powder using the Qiagen Tissue Lyser system (30 rounds/s for 2 min). Ground material was resuspended in 1 ml of sterilized water and 10 μl drops of a dilution series were spotted onto PSA medium plate in triplicates. The plates were incubated at 28 °C until colonies could be counted. This experiment was performed three times.
Mapping of known resistance gene/QTLs on the reference Nipponbare physical map
In a first step, information on all known BB resistance genes and QTLs was reviewed. This review included gramene accessions, number of genes/QTLs, their names, synonyms and symbols, the genetic populations in which they were mapped. Their donor’s parents as well as their genetic position and their co-localized markers in various mapping populations were also reported here. In the same way, physical positions were recorded if available (Supplementary data). The different genetic maps used were SSR Cornell 2000, RIL IR64 x Azucena, DH IR4 x Azucena, RIL Lemont x Tequin, RIL Zhenshan 97 x Minghui 63, JRGP RFLP 2000 Nipponbare x Kasalath, Cornell RFLP 2001 O. sativa x O. longistaminata, and the reference MSU7. Physical positions of the cloned genes such as Xa1, Xa7, Xa10, Xa23, Xa26, Xa27, xa5, xa13 and xa25 were directly reported on the integrative map. For the non cloned genes and QTLs, we chose the closest ones with known genetic positions among the co-localized maker and interpolated them on the reference Nipponbare MSU7 physical map. Other genes and QTLs with conflicting positions, as well as those that have been not mapped such as Xa11, Xa12, Xa15, xa32(t), AQBT026 and AQBT030, were not positioned on the integrative map.
Acknowledgments
We thank USAID which partly supported this work through the project “Marker-Assisted Selection for improvement of rice varieties resistance to RYMV for West Africa” conducted by the second author (AfricaRice) and the Generation Challenge Program (subprogram SP5), which partly funded this study. We are grateful to Africa Rice and IRD for their support and Thierry Mathieu (IRD) for his help during greenhouse experiments and screening. We thank also Daniel Fonséca and Nour Ahmadi for providing information about the SSR markers used in this study.
Abbreviations
- ANOVA
analysis of variance
- BB
bacterial blight
- CFU
colony-forming units
- CIM
composite interval mapping
- DO
optical density
- LL
lesion length
- LOD
log-likelihood of odds
- MR
moderately resistant
- MS
moderately susceptible
- PSA
peptone sucrose agar
- QTL
quantitative trait loci
- R
resistant
- RIL
recombinant inbred lines
- S
susceptible
- SIM
simple interval mapping
- SMR
single marker regression
- SSD
single seed descent
- SSR
simple sequence repeats
- TAL
transcription activator-like
- Xoo
Xanthomonas oryzae pv. oryzae
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GD carried out the phenotyping assays, participated in QTL analysis and physical maps drawing and drafted the manuscript. MN, AS, VV and AG participated in the design of the study and its coordination. ML participated in QTL analysis. AG drowns the physical maps. VV conceived and participated in phenotyping assays. All authors contributed to the manuscript draft, read and approved the final manuscript.
Authors’ information
GD: Assistant Professor at Université Polytechnique d’Abomey/Benin. MN: head of biotechnology lab of AfricaRice. AS: Professor and head of Applied biochemistry and molecular biology lab of University of Abomey-Calavi/Benin ML: Currently works overseas at CIAT, Cali, Colombia, where he is leading the Rice Genetics and Genomics group. VV: Director of research at IRD Montpellier, head of IPME unit. AG: Director of research at IRD Montpellier, head of DIADE unit.
References
- Basso A, Onasany A, Issaka S, Sido AY, Haougui A, Adam T, Séré Y, Saadou M. Le flétrissement bactérien du riz au Niger: diversité pathologique d’isolats collectés sur les périmètres irrigués. J Appl Biosci. 2011;38:2551–2563. [Google Scholar]
- Bhasin H, Bhatia D, Raghuvanshi S, Lore J, Sahi G, Kaur B, Vikal Y, Singh K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol Breed. 2012;30:607–611. doi: 10.1007/s11032-011-9646-y. [DOI] [Google Scholar]
- Bimolata W, Kumar A, Sundaram RM, Laha GS, Qureshi IA, Reddy GA, Ghazi IA. Analysis of nucleotide diversity among alleles of the major bacterial blight resistance gene Xa27 in cultivars of rice (Oryza sativa) and its wild relatives. Planta. 2013;238(2):293–305. doi: 10.1007/s00425-013-1891-3. [DOI] [PubMed] [Google Scholar]
- Blair MW, Garris AJ, Iyer AS, Chapman B, Kresovich S, McCouch SR. High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.) Theor Appl Genet. 2003;107:62–73. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Boisnard A, Albar L, Thiéméle D, Tondeau M, Ghesquiere A. Evaluation of genes from eIF4E and eIF4G multigenic families as potential candidates for partial resistance QTLs to Rice yellow mottle virus in rice. Theoretical and Applied Genetics. 2007;116:53–62. doi: 10.1007/s00122-007-0646-6. [DOI] [PubMed] [Google Scholar]
- Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC. Developmental control of Xa21-mediated disease resistance in rice. Plant J. 1999;20:231–236. doi: 10.1046/j.1365-313x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- Cheema KK, Grewal NK, Vikal Y, Sharma R, Lore JS, Das A, Bhatia D, Mahajan R, Gupta V, Bharaj T, Singh K. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4 L and transferred to Oryza sativa L. Genet Res. 2008;90:397–407. doi: 10.1017/S0016672308009786. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. A new gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, and elite restorer line. Phytopathology. 2002;92:750–754. doi: 10.1094/PHYTO.2002.92.7.750. [DOI] [PubMed] [Google Scholar]
- Chen S, Liu X, Zeng L, Ouyang D, Yang J, Zhu X (2011) Genetic analysis and molecular mapping of a novel recessive gene xa34(t) for resistance against Xanthomonas oryzae pv. oryzae. Theor Appl Genet. DOI 10.1007/s00122-011-1534-7 [DOI] [PubMed]
- Chu ZH, Fu BY, Yang H, Xu CG, Li ZK, Sanchez A, Park YJ, Bennetzen JL, Zhang QF, Wang SP. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet. 2006;112:455–461. doi: 10.1007/s00122-005-0145-6. [DOI] [PubMed] [Google Scholar]
- Chu ZH, Yuan M, Yao LL, Ge XJ, Yuan B, Xu CG, Li XH, Fu BY, Li ZK, Bennetzen JL, Zhang QF, Wang SP. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djedatin G, Ndjiondjop MN, Mathieu T, Vera Cruz CM, Sanni A, Ghesquière A, Verdier V. Evaluation of African cultivated rice Oryza glaberrima for resistance to bacterial blight. Plant Dis. 2011;95:441–447. doi: 10.1094/PDIS-08-10-0558. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapidmethod for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy V, Sevignac M, d’Oliveira J, Fouilloux G, Skroch P, Thoquet P, Gepts P, Langin T, Dron M. Inheritance of partial resistance against Colletotrichum lindemuthianum in Phaseolus vulgaris and co-localization of quantitative trait loci with genes involved in specific resistance. Mol Plant Microbe Interact. 2000;3:287–296. doi: 10.1094/MPMI.2000.13.3.287. [DOI] [PubMed] [Google Scholar]
- Ghazi IA, Srivastava PS, Dalal V, Gaikwad K, Singh AK, Sharma TR, Singh NK, Mohapatra T. Physical mapping, expression analysis and polymorphism survey of resistance gene analogues on chromosome 11 of rice. J Biosci. 2009;34:251–261. doi: 10.1007/s12038-009-0029-z. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Szurek B, Manceau C, Mathieu T, Sere Y, Verdier V. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol Plant Microbe Interact. 2007;20:534–546. doi: 10.1094/MPMI-20-5-0534. [DOI] [PubMed] [Google Scholar]
- Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang D, Wang GL, Yin Z. High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor Appl Genet. 2004;108:800–807. doi: 10.1007/s00122-003-1491-x. [DOI] [PubMed] [Google Scholar]
- Hajri A, Brin C, Zhao S, David P, Feng J, Koebnik R, Szurek B, Verdier V, Boureau T, Poussier S (2012) Multilocus sequence analysis and type III effector repertoire mining provide new insights into the evolutionary history and virulence of Xanthomonas oryzae. Molecular Plant Pathology 13:288–302 [DOI] [PMC free article] [PubMed]
- Han X, Yang Y, Wang X, Zhou J, Zhang W, Yu C, Cheng C, Cheng Y, Yan C, Chen J. Quantitative Trait Loci Mapping for Bacterial Blight Resistance in Rice Using Bulked Segregant Analysis. Int J Mol Sci. 2014;15(7):11847–11861. doi: 10.3390/ijms150711847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AS, McCouch SR. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol Plant Microbe Interact. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Suh J, Qin Y, Noh T, Reinke RF, Jena KK (2015) Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.) Theoretical and Applied Genetics 128:1933–1943. [DOI] [PubMed]
- Korinsak S, Sriprakhon S, Sirithanya P, Jairin J, Korinsak S, Vanavichit A, Toojinda T. Maejo Int J Sci Technol. 2009;3:235–247. [Google Scholar]
- Lee SW, Choi SH, Han SS, Lee DG, Lee BY. Distribution of Xanthomonas oryzae pv. oryzae strains virulent to Xa21 in Korea. Phytopathology. 1999;89:928–933. doi: 10.1094/PHYTO.1999.89.10.928. [DOI] [PubMed] [Google Scholar]
- Li ZK, Luo LJ, Mei HW, Paterson AH, Zhao XZ, Zhong DB, Wang YP, Yu XQ, Zhu L, Tabien R, Stansel JW, Ying CS. A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol Gen Genet. 1999;261:58–63. doi: 10.1007/s004380050941. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 2011;34:1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- Lorieux M. MapDisto, A Free User-Friendly Program For Computing Genetic Maps. Computer demonstration (P958) given at the Plant and Animal Genome XV conference, Jan 13-17 2007, San Diego, CA. 2007. [Google Scholar]
- Mago R, Nair S, Mohan M. Resistance gene analogues from rice: cloning, sequencing and mapping. Theor Apply Genet. 1999;99:50–57. doi: 10.1007/s001220051207. [DOI] [Google Scholar]
- Mew TW, Vera Cruz CM, Medalla ES. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to the planting of rice cultivars in the Philippines. Plant Dis. 1992;76:1029–1032. doi: 10.1094/PD-76-1029. [DOI] [Google Scholar]
- Nelson J (2005) Methods and Software for Genetic Mapping, pp. 53–74 in The Handbook of Plant Genome Mapping; Genetic and Physical Mapping, edited by Meksem K., Kahl G., editors. Wiley-VCH Verlag GmbH and Co, Weinheim, Germany
- Ogawa T, Yamamoto T. Selection of Recurrent Parents to Develop near-Isogenic Lines Resistant to Bacterial Leaf-Blight of Rice. Jarq Japan Agric Res Q. 1987;21:65–69. [Google Scholar]
- Perez LM, Redoña ED, Mendioro MS, Vera Cruz CM, Leung H. Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermosensitive genetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica. 2008;164:627–636. doi: 10.1007/s10681-008-9653-1. [DOI] [Google Scholar]
- Pflieger S, Lefebvre V, Caranta C, Blattes A, Goffinet B, Palloix A. Disease resistance gene analogs as candidates for QTLs involved in pepper–pathogen interactions. Genome. 1999;42:1100–1110. doi: 10.1139/g99-067. [DOI] [PubMed] [Google Scholar]
- Ramalingam J, Cruz CMV, Kukreja K, Chittoor JM, Wu JL, Lee SW, Baraoidan M, George ML, Cohen MB, Hulbert SH, Leach JE, Leung H. Candidate Defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact. 2003;16:14–24. doi: 10.1094/MPMI.2003.16.1.14. [DOI] [PubMed] [Google Scholar]
- Reckhaus PM. Occurrence of bacterial blight of rice in Niger, West Africa. Plant Dis. 1983;67:1039. doi: 10.1094/PD-67-1039. [DOI] [Google Scholar]
- Ruan H,Yan C, An D, Liu R, Chen J (2008) Identifying and Mapping New Gene xa32(t) for Resistance to Bacterial Blight(Xanthomonas oryzae pv. oryzae, Xoo) from Oryza meyeriana L. Acta Agriculturae Boreali-Occidentalis Sinica. DOI CNKI:SUN:XBNX.0.2008-06-035
- Ruengphayak S, Chaichumpoo E, Phromphan S, Kamolsukyunyong W, Sukhaket W, Phuvanartnarubal E, Korinsak S, Korinsak S, Vanavichit A. Pseudo-backcrossing design for rapidly pyramiding multiple traits into a preferential rice variety. Rice. 2015;8:7. doi: 10.1186/s12284-014-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakraborty S, Zeng Z. QTL Mapping for Days to Flowering under Drought Condition in Rice (Oryza sativa L.) Genome. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2011;39:58–63. [Google Scholar]
- Sanchez AC, Ilag LL, Yang D, Brar DS, Ausubel F, Khush GS, Yano M, Saskai T, Li Z, Huang N. Genetic and physical mapping of xa13, a recessive bacterial blight resistance gene in rice. Theor Appl Genet. 1999;98:1022–1028. doi: 10.1007/s001220051163. [DOI] [Google Scholar]
- Sere Y, Onasanya A, Verdier V, Akator K, Ouedraogo LS, Segda Z, Mbare MM, Sido AY, Baso A. Rice Bacterial Leaf Blight in West Africa: Preliminary studies on disease in farmer’s field and screening. Asian J Plant Sci. 2005;4:577–579. doi: 10.3923/ajps.2005.577.579. [DOI] [Google Scholar]
- Shanti ML, Shenoy VV, Lalitha Devi G, Mohan Kumar V, Premalatha P, Naveen Kumar G, Shashidhar HE, Zehr UB, Freeman WH. Marker-assisted breeding for resistance to bacterial leaf blight in popular cultivar and parental lines of hybrid rice. J Plant Pathol. 2010;92:495–501. [Google Scholar]
- Singh S, Sidhu JS, Huang N, Vikal Y, Li Z, Brar DS, Dhaliwal HS, Khush GS. Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theoretical and Applied Genetics. 2006;102:1011–1015. doi: 10.1007/s001220000495. [DOI] [Google Scholar]
- Suh JP, Jeung JU, Noh TH, Cho YC, Park SH, Park HS, Shin MS, Kim CK, Jena KK. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 2013;6:5. doi: 10.1186/1939-8433-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL, Cao YL, Yang ZF, Xu CG, Li XH, Wang SP, Zhang QF. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313X.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Sundaram RM, Vishnupriya MR, Laha GS, Rani NS, Rao PS, Balachandran SM, Reddy GA, Sarma NP, Sonti RV. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol J. 2009;4:400–407. doi: 10.1002/biot.200800310. [DOI] [PubMed] [Google Scholar]
- Swamy P, Panchbhai AN, Dodiya P, Naik V, Panchbhai SD, Zehr UB, Azhakanandam K, Char BR. Evaluation of bacterial blight resistance in rice lines carrying multiple resistance genes and Xa21 transgenic lines. Curr Sci. 2006;90:818–824. [Google Scholar]
- Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, et al. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 2014;26:497–515. doi: 10.1105/tpc.113.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toojinda T, Broers LH, Chen XM, Hayes PM, Kleinhofs A, Korte J, Kudrna D, Leung H, Line R, Powell W, Ramsey L, Vivar H, Waugh R. Mapping quantitative and qualitative disease resistance genes in a doubled haploid population of barley (Hordeum vulgare) Theor Appl Genet. 2001;101:580–589. doi: 10.1007/s001220051519. [DOI] [Google Scholar]
- Traoré O, Pinel-Galzi A, Sorho F, Sarra S, Rakotomalala M, Sangu E, Kanyeka Z, Séré Y, Konaté G, Fargette D. A reassessment of the epidemiology of Rice yellow mottle virus following recent advances in field and molecular studies. Virus Res. 2009;141:258–67. doi: 10.1016/j.virusres.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Triplett LR, Hamilton JP, Buell CR, Tisserat NA, Verdier V, Zink F, Leach JE. Genomic Analysis of Xanthomonas oryzae from US Rice Reveals Substantial Divergence from Known X. oryzae Pathovars. Appl Environ Microbiol. 2011;77:3930–7. doi: 10.1128/AEM.00028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Cruz CM, Bai J, Oña I, Leung H, Nelson RJ, Mew T, Leach JE. Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. PNAS. 2000;97:13500–13505. doi: 10.1073/pnas.250271997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier V, Vera Cruz C, Leach JE (2012) Controlling rice bacterial blight in Africa: Needs and prospects. J Biotechnol 159(4):320-328 [DOI] [PubMed]
- Wang G, Mackill DJ, Bonman JM, McCouch SR, Nelson RJ. RFLP mapping of genes conferring complete and partial resistance to blast resistance in a durably resistant rice cultivar. Genetics. 1994;136:1421–1434. doi: 10.1093/genetics/136.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wan X, Crossa J, Crouch J, Weng J, Zhai H, Wan J. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res. 2006;88:93–104. doi: 10.1017/S0016672306008408. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer2.5. Raleigh: Department of Statistics, North Carolina Stat University; 2006. [Google Scholar]
- Wang CL, Xu AB, Gao Y, Fan YL, Liang YT, Zheng CK, Sun LQ, Wang WQ, Zhao KJ. Generation and characterisation of Tn5-tagged Xanthomonas oryzae pv. oryzae mutants that overcome Xa23-mediated resistance to bacterial blight of rice. Eur J Plant Pathol. 2009;123:343–351. doi: 10.1007/s10658-008-9371-5. [DOI] [Google Scholar]
- Wang C, Fan Y, Zheng C, Qin T, Zhang X, Zhao K. High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol Genet Genomics. 2014;289:745–753. doi: 10.1007/s00438-014-0848-y. [DOI] [PubMed] [Google Scholar]
- Wang M, Yu Y, et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat Genet. 2014;46(9):982–911. doi: 10.1038/ng.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb KM, et al. A benefit of high temperature: Increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010;185:568–576. doi: 10.1111/j.1469-8137.2009.03076.x. [DOI] [PubMed] [Google Scholar]
- Wonni I, Cottyn B, Detemmerman L, Dao S, Ouedraogo L, Sarra S, Tekete C, Poussier S, Corral R, Triplett L, Koita O, Koebnik R, Leach J, Szurek B, Maes M, Verdier V. Analysis of Xanthomonas oryzae pv. oryzicola population in Mali and Burkina Faso reveals a high level of genetic and pathogenic diversity. Phytopathology. 2014;104:520–531. doi: 10.1094/PHYTO-07-13-0213-R. [DOI] [PubMed] [Google Scholar]
- Wu L, Goh ML, Sreekala C, Yin Z. XA27 Depends on an Amino-Terminal Signal-Anchor-Like Sequence to Localize to the Apoplast for Resistance to Xanthomonas oryzae pv oryzae. Plant Physiol. 2008;148:1497–1509. doi: 10.1104/pp.108.123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li X, Xu C, Wang S. Fine genetic mapping of xa24, a recessive gene for resistance against Xanthomonas oryzae pv. oryzae in rice. Theor Appl Genet. 2008;118:185–191. doi: 10.1007/s00122-008-0888-y. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Cao YL, Xu CG, Li XH, Wang SP. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor Appl Genet. 2006;113:1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci U S A. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector which induces the rice nodulin-3 Os11N3 gene. Mol Plant Microbe Interact. 2011;24(9):1102–1113. doi: 10.1094/MPMI-11-10-0254. [DOI] [PubMed] [Google Scholar]
- Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B (2011) Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector which induces the rice nodulin-3 Os11N3 gene. Mol Plant Microbe Interact. Epub ahead of print [DOI] [PubMed]
- Zhang J, Li X, Jiang G, Xu Y, He Y. Pyramiding of Xa7 and Xa21 for the improvement of disease resistance to bacterial blight in hybrid rice. Plant Breed. 2006;125:600–605. doi: 10.1111/j.1439-0523.2006.01281.x. [DOI] [Google Scholar]
- Zou JH, Pan XB, Chen Z, Xu JY, Lu JF, Zhai WX, Zhu LH. Mapping quantitativre trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.) Theor Appl Genet. 2000;101:569–573. doi: 10.1007/s001220051517. [DOI] [Google Scholar]


