Abstract
A Xenopus borealis somatic 5S RNA gene was assembled with either the complete octamer of histones, (H2A/H2B/H3/H4)2, or the (H3/H4)2 tetramer of histones that comprises the central protein kernel of the nucleosome. Gel-mobility shifts, DNase I protection, and immunoblotting assays demonstrate that the class III transcription factor IIIA (TFIIIA) readily interacts with 5S DNA associated with the tetramer but that little or no binding is detected when 5S DNA is associated with the full octamer of histones. Thus, the presence of histones H2A and H2B in the 5S nucleosome significantly inhibits the interaction of TFIIIA with its cognate binding site within the 5S RNA gene. We propose that either the depletion of histones H2A and H2B from preexisting nucleosomes or the staged assembly of chromatin after replication in which a tetramer of histones H3/H4 associates with DNA before histones H2A/H2B will facilitate the binding of transcription factors to their cognate DNA sequences.
Full text
PDF

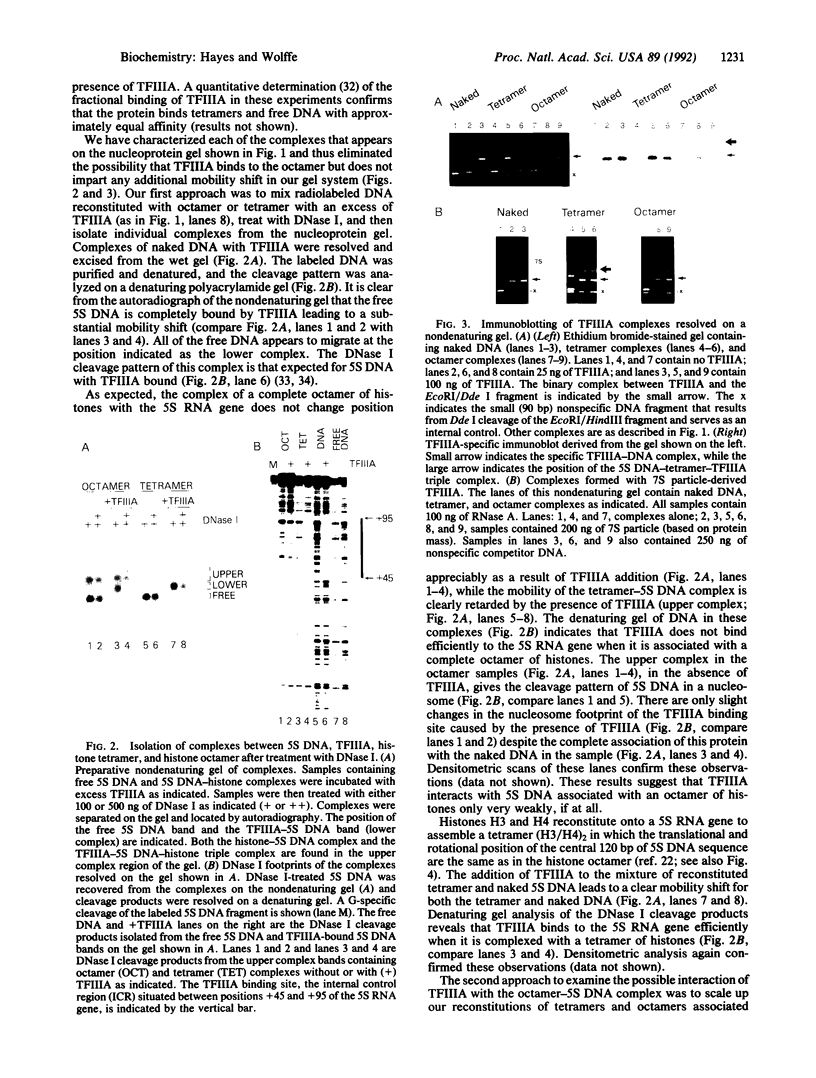
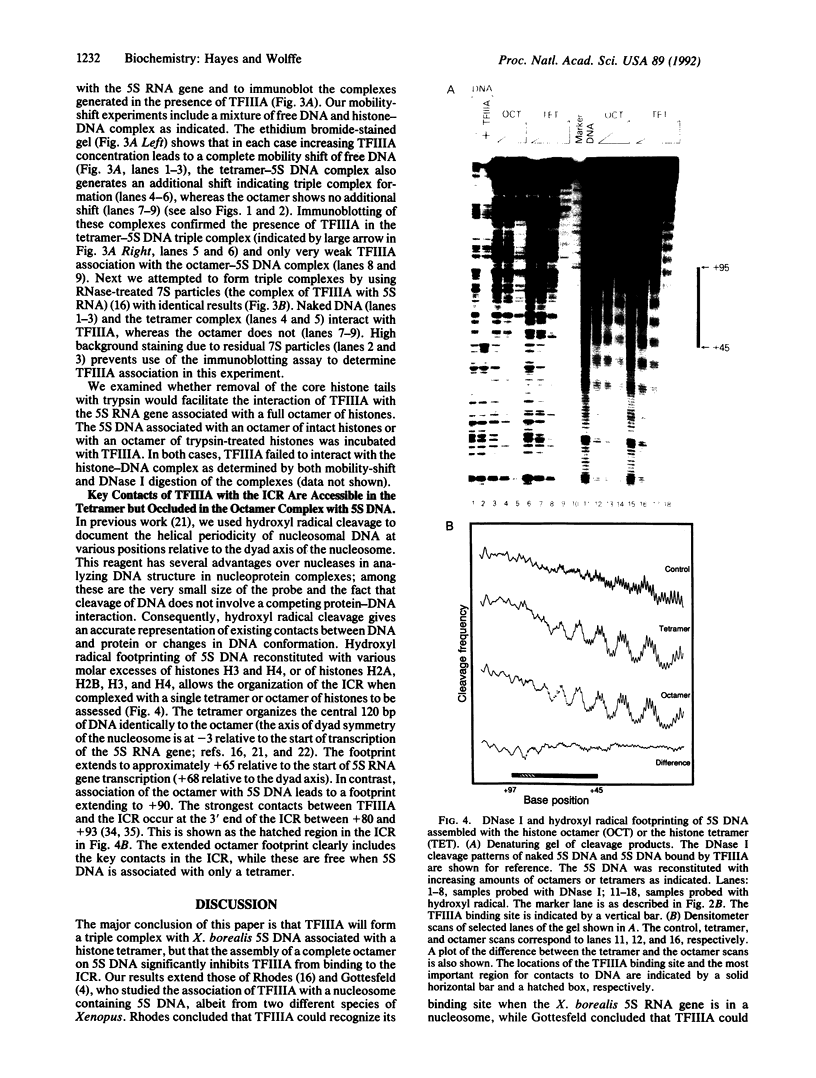

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almouzni G., Méchali M., Wolffe A. P. Competition between transcription complex assembly and chromatin assembly on replicating DNA. EMBO J. 1990 Feb;9(2):573–582. doi: 10.1002/j.1460-2075.1990.tb08145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G., Méchali M., Wolffe A. P. Transcription complex disruption caused by a transition in chromatin structure. Mol Cell Biol. 1991 Feb;11(2):655–665. doi: 10.1128/mcb.11.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. T., Brown D. D. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell. 1987 Nov 6;51(3):445–453. doi: 10.1016/0092-8674(87)90640-4. [DOI] [PubMed] [Google Scholar]
- Archer T. K., Cordingley M. G., Wolford R. G., Hager G. L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991 Feb;11(2):688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J., Dong F., van Holde K. E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J Mol Biol. 1989 Apr 5;206(3):451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gottesfeld J. M. Chromosomal footprinting of transcriptionally active and inactive oocyte-type 5S RNA genes of Xenopus laevis. Nucleic Acids Res. 1990 Oct 25;18(20):6031–6037. doi: 10.1093/nar/18.20.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Felts S. J., Weil P. A., Chalkley R. Transcription factor requirements for in vitro formation of transcriptionally competent 5S rRNA gene chromatin. Mol Cell Biol. 1990 May;10(5):2390–2401. doi: 10.1128/mcb.10.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R., Roberts J. M. Multistep pathway for replication-dependent nucleosome assembly. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6459–6463. doi: 10.1073/pnas.86.17.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González P. J., Palacián E. Interaction of RNA polymerase II with structurally altered nucleosomal particles. Transcription is facilitated by loss of one H2A.H2B dimer. J Biol Chem. 1989 Nov 5;264(31):18457–18462. [PubMed] [Google Scholar]
- Gottesfeld J. M. DNA sequence-directed nucleosome reconstitution on 5S RNA genes of Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1612–1622. doi: 10.1128/mcb.7.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Clark D. J., Wolffe A. P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D., Wolffe A. P. The structure of DNA in a nucleosome. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J., Tullius T. D., Wolffe A. P. A protein-protein interaction is essential for stable complex formation on a 5 S RNA gene. J Biol Chem. 1989 Apr 15;264(11):6009–6012. [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989 Aug;8(8):2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Wiegand R., Brutlag D. Ribonucleic acid and other polyanions facilitate chromatin assembly in vitro. Biochemistry. 1981 Apr 28;20(9):2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988 Oct;7(10):3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Read C. M., Baldwin J. P., Crane-Robinson C. Structure of subnucleosomal particles. Tetrameric (H3/H4)2 146 base pair DNA and hexameric (H3/H4)2(H2A/H2B)1 146 base pair DNA complexes. Biochemistry. 1985 Jul 30;24(16):4435–4450. doi: 10.1021/bi00337a027. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Senshu T., Fukuda M., Ohashi M. Preferential association of newly synthesized H3 and H4 histones with newly replicated DNA. J Biochem. 1978 Oct;84(4):985–988. doi: 10.1093/oxfordjournals.jbchem.a132213. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Smith S., Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991 Apr;10(4):971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Reconstitution of chromatin core particles. Biochemistry. 1977 Nov 29;16(24):5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- Thomas G. A., Kubasek W. L., Peticolas W. L., Greene P., Grable J., Rosenberg J. M. Environmentally induced conformational changes in B-type DNA: comparison of the conformation of the oligonucleotide d(TCGCGAATTCGCG) in solution and in its crystalline complex with the restriction nuclease EcoRI. Biochemistry. 1989 Mar 7;28(5):2001–2009. doi: 10.1021/bi00431a007. [DOI] [PubMed] [Google Scholar]
- Tremethick D., Zucker K., Worcel A. The transcription complex of the 5 S RNA gene, but not transcription factor IIIA alone, prevents nucleosomal repression of transcription. J Biol Chem. 1990 Mar 25;265(9):5014–5023. [PubMed] [Google Scholar]
- Weisbrod S., Wickens M. P., Whytock S., Gurdon J. B. Active chromatin of oocytes injected with somatic cell nuclei or cloned DNA. Dev Biol. 1982 Nov;94(1):216–229. doi: 10.1016/0012-1606(82)90085-9. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell. 1986 Oct 24;47(2):217–227. doi: 10.1016/0092-8674(86)90444-7. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988 Sep 23;241(4873):1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Transcriptional activation of Xenopus class III genes in chromatin isolated from sperm and somatic nuclei. Nucleic Acids Res. 1989 Jan 25;17(2):767–780. doi: 10.1093/nar/17.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991 Feb 8;64(3):533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]










