Abstract
The physicochemical design and synthesis of effective cancer-directed and particle-based nanotherapeutic imaging agents remains a challenging task. Of critical importance is the ability to demonstrate maximum delivery, retention, and treatment efficacy for platforms designed to deposit their cargo at sites of disease without attendant dose-limiting toxicity. In this work, we describe dual-modality nanoparticle drug conjugates (NDCs) which utilize protease sensitive linkers to attached drug compounds and imaging labels to a clinically translated class of ultrasmall silica nanoparticle (C′ dots). We describe the synthesis and characterization of these linker-drug constructs. Linkers incorporating dipeptide enzyme substrates are attached to analogs of a prototypical epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI), through a cleavable amide bond or para-aminobenzyloxycarbonyl (PABC) group. These constructs are conjugated onto C′ dots leading to the desired NDCs. These NDCs exhibit fast and predictable release kinetics in the presence of model proteases, and are stable in various biological media. Finally, in vitro assays show NDCs to be highly active in reducing phosphorylated EGFR levels in H1650 cells, a human tumor-derived cell line. The data suggests that NDCs exhibit desirable properties that warrant further development towards oncological therapy.
Keywords: nanoparticle, drug delivery, drug conjugate, kinase inhibitor, cancer
Graphical Abstract
1. Introduction
Nanotherapeutic delivery vehicles tailored for drug selection and delivery to stratified subpopulations of patients have become a prominent feature in the current and emerging repertoire of cancer therapies.1–4 These are typically macro- or supra-molecular multicomponent systems, ranging in size from 1–1,000 nm, that are either inherently therapeutic (i.e., no active pharmaceutical ingredient) or function as carriers of therapeutics which have been directly coupled.5 To date, liposomal nanoparticles and biologics comprise a large proportion of the number of products used to treat a variety of cancer types, and these agents have either been approved or are still in clinical development;2,6 a number of polymer-based particle formulations are also currently being evaluated in early phase trials.1,4 It is rather surprising, therefore, that of these available nanotherapeutic products, there is a paucity of products possessing the molecular imaging capabilities needed to predict biological properties (i.e., clearance, biodistribution) in individual patients for optimizing cancer therapy. We propose and demonstrate an alternative class of nanotherapeutic, known as a nanoparticle drug conjugate™ (NDC),7 in which drug compounds and contrast-producing labels are covalently attached through protease sensitive linkers to ultrasmall (sub-10 nm diameter) nanoparticles to produce a controlled release, dual-modality platform for positron emission tomography (PET) and optically-driven applications.8–11 This combination of size, molecular composition and chemistry (i.e. mode of drug release), and endowed imaging capabilities may leverage the beneficial properties seen in other nanotherapeutic products with the aim of overcoming key obstacles hampering traditional formulations: narrow therapeutic indices, dose-limiting toxicities, and limited clinical utility.12
Desirable candidates for nanotherapeutic delivery systems share a common feature of incorporating and releasing a drug compound in a controlled manner, which can favorably alter drug bioavailability and pharmacokinetics, while minimizing off-target toxicities. Ideally, an imaging label has been incorporated to assess their precise localization and retention at disease sites. However the mechanisms by which these systems function differ significantly. For example, antibody drug conjugates (ADCs) achieve lower drug toxicity primarily through active targeting of tumor cells and conditional release of drug molecules.2,4 Upon binding a cell surface antigen, active drug release occurs after cellular internalization and endosomal uptake. On the other hand, liposomes and polymer-based drug delivery systems, which are typically much larger assembled complexes (~20–150 nm diameters) passively loaded with a greater payload (~10,000 drug molecules for Doxil)13 have generally lacked targeting capabilities (BIND-014 is an exception),4 relying primarily on the well-known enhanced permeability and retention (EPR) effect for the successful delivery of nano-formulated drugs.14 While interstitial permeation of liposomes may be poor due to their size, the free drug is released through various mechanisms that are not entirely understood. Abraxane (~140 nm) relies on a different approach to enhance the bioavailability of a hydrophobic compound. A specific formulation of albumin and drug (paclitaxel) forms the initial complex, which is then estimated to disperse into smaller protein-drug aggregates upon injection.2,4
In this work, we focus on the design, synthesis, characterization, and preliminary biological evaluation of NDCs as a first-in-kind ultrasmall organic-inorganic hybrid drug delivery vehicle with dual-modality (PET-optical) imaging capabilities. In vivo imaging will be presented in a subsequent paper, similar to that seen in prior studies.11,15 NDCs are comprised of an ultrasmall (~6 nm diameter) poly(ethylene glycol) coated (PEGylated) fluorescent core-shell silica nanoparticle, referred to as C′ dot, with drugs and radiolabels covalently attached to the surface. In a sense, these particles represent a ‘hybrid’ drug delivery system that draw upon particular features of the aforementioned nanomedicines; NDCs are macromolecular in size, similar to that of proteins (i.e., biologics), yet incorporate design features of other particle-based materials, such as a PEGylated outer layer that renders the surface neutrally charged. These properties, in turn, facilitate bulk renal clearance while enabling evasion of the reticuloendothelial system. The drug component is covalently attached to the PEGylated surface for stability, and drug release is achieved through enzymatic hydrolysis. We anticipate that the combination of these attributes may lead to distinctly favorable properties for this therapeutic platform, including improved control of drug release rates, quantitative pharmacokinetic profiles, and lack of toxicity, as well as enhanced target tissue delivery, retention, and distribution.
2. Results and Discussion
2.1. Design strategy
Previously, we have reported on the design and synthesis of ultrasmall nanoparticles, C dots16,17 with a range of modular functionalities. These are fluorescent, organo-silica core - silica shell particles that have diameters controllable down to the sub-10 nm range. The silica shell surface of the dye-containing core is covalently modified with silyl-polyethylene glycol (PEG) groups to enhance stability in aqueous and biologically relevant conditions. These particles have been evaluated in vivo and exhibit excellent clearance properties owing largely to their sub-10 nm size and inert surface.17,18 Among the additional functionalities incorporated into C dots are chemical sensing,19,20 non-optical (PET) image contrast and in vitro/in vivo targeting capabilities (enabling their use in visualizing lymph nodes for surgical applications, and melanoma detection in cancer11,15). In fact, these particles are the first organic-inorganic hybrid particles to have reached clinical trials in patients for molecular cancer imaging using either PET (or optical) methods.21 Cost-effective synthetic approaches in aqueous media have recently led to the latest generation dots, now referred to as C′ dots, that exhibit improved size control, enhanced covalent near infrared (NIR) dye incorporation efficiencies, and higher fluorescence brightness.22 The present work was conducted exclusively with this latest particle generation.
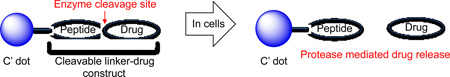
We hypothesize that therapeutic C′ dots, also adapted for molecular cancer imaging, may provide a unique platform for drug delivery owing to their physical properties, as well as demonstrated human in vivo characteristics.21 In conceptualizing C′ dot-based NDCs for drug delivery, we sought to develop a general strategy that would provide good biostability (to minimize premature drug release) and exhibit controlled release of the bioactive compound at the desired site (Figure 1) – properties that could potentially maximize target tissue bioavailability while abrogating dose-limiting toxicity. The addition of a radiolabel for PET imaging facilitates tumor visualization, quantifies tumor accumulation, and permits tracking of either the drug or particle in vivo. Studies with antibody and polymer drug conjugates suggest peptide-based linkers to be ideal candidates for NDC applications.23–25 These linkers, in the context of antibodies and polymers, have been shown to be stable both in vitro and in vivo, with highly predictable release kinetics that rely on enzyme-catalyzed hydrolysis, presumably by lysosomal proteases. For example, cathepsin B, a highly expressed protease in lysosomes, has been utilized to facilitate drug release from macromolecules.26
Figure 1.
Schematic of enzyme-mediated drug release from nanoparticle drug conjugate (NDC). Blue dots represent fluorescent PEGylated core-shell silica nanoparticles.
The model drug selected for this work is gefitinib – a receptor tyrosine kinase inhibitor (TKI) to the epidermal growth factor receptor (EGFR).27 EGFR has emerged as an attractive molecular candidate for targeted therapy.28 EGFR mutations resulting in constitutive activation have been found in 25% of metastatic non-small cell lung cancer (NSCLC) and 40–50% of primary glioblastoma multiforme (GBM) – two prevalent forms of brain cancer.29 While EGFR-TKIs have shown promise in preclinical studies, they have been shown to be largely ineffective in malignant brain cancer patients. This is likely due to their poor pharmacokinetic profiles and tissue or central nervous system (CNS) penetration, as well as dose-limiting toxicity,12,30,31 i.e. all factors that may be potentially improved via use of an appropriate NDC.
2.2. Synthesis of gefitinib analogues
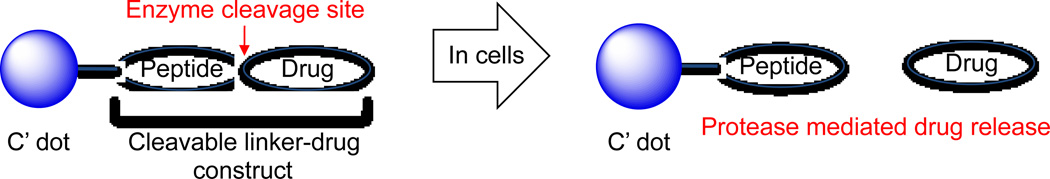
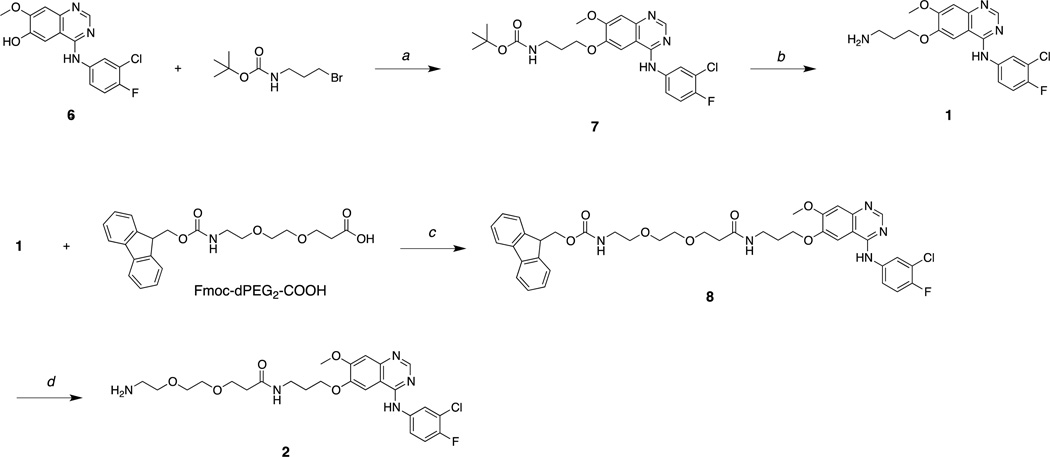
Gefitinib is designed to bind and inhibit the kinase domain active site of EGFR.32 To utilize the anilinoquinazoline component of gefitinib in the context of NDCs, it was necessary to incorporate a chemically reactive group that would not significantly perturb drug binding to the kinase domain. From reported X-ray crystallographic and structure-active relationship (SAR) studies,27,32 we hypothesized that replacing the terminal heterocycle (morpholino) group with an amine, as shown in Figure 2a, would not significantly alter drug activity but would provide the needed chemical functionality (amine) for modification and eventual covalent attachment to the C′ dot. With the commercial availability of 6, des-morpholino-gefitinib (dMG), we readily obtained the desired APdMG (1) through a one-step nucleophilic substitution of Boc protected amino propyl bromide, followed by acid deprotection (Scheme 1).
Figure 2.
Chemical structure of (a) gefitinib and gefitinib analogues (APdMG and dPEG2APdMG); (b) linker-drug directly connected through amide bond (3); (c) linker-drug connected through dPEG2 spacer (4); (d) linker-drug connected through degradable PABC spacer (5).
Scheme 1.
a) K2CO3, DMF, 60°C; b) TFA/H20 (9:1); c) HATU, DIEA, DMF; b) piperidine, DMF.
Additionally, the gefitinib analogue dPEG2APdMG (2), which contains an extended 10 atom PEG spacer, was prepared. This analogue was needed to address potential steric problems a protease, for example, might encounter due to the close proximity of the peptide linker and drug. Compound 2 was readily obtained by reacting 1 with Fmoc-dPEG2-COOH using standard coupling methods. The isolated product was then deprotected with piperidine/DMF (Scheme 1). H1650 cells were treated with these compounds and analyzed by western blot to assess phospho-Tyr168 levels in EGFR to ensure that in particular 1 retained activity against EGFR. The H1650 cells are a model human tumor-derived non-small-cell lung cancer (NSCLC) line (bronchioalveolar carcinoma), which contain a mutated EGFR (L858R and ΔE746-A750) resulting in constitutive activity of the receptor. Indeed, compound 1 showed effects similar to gefitinib, with inhibition of phospho-Tyr168 at 1 and 10 µM concentrations, and compound 2 showed reduced activity (data not shown).
2.3. Synthesis of linker-drug constructs
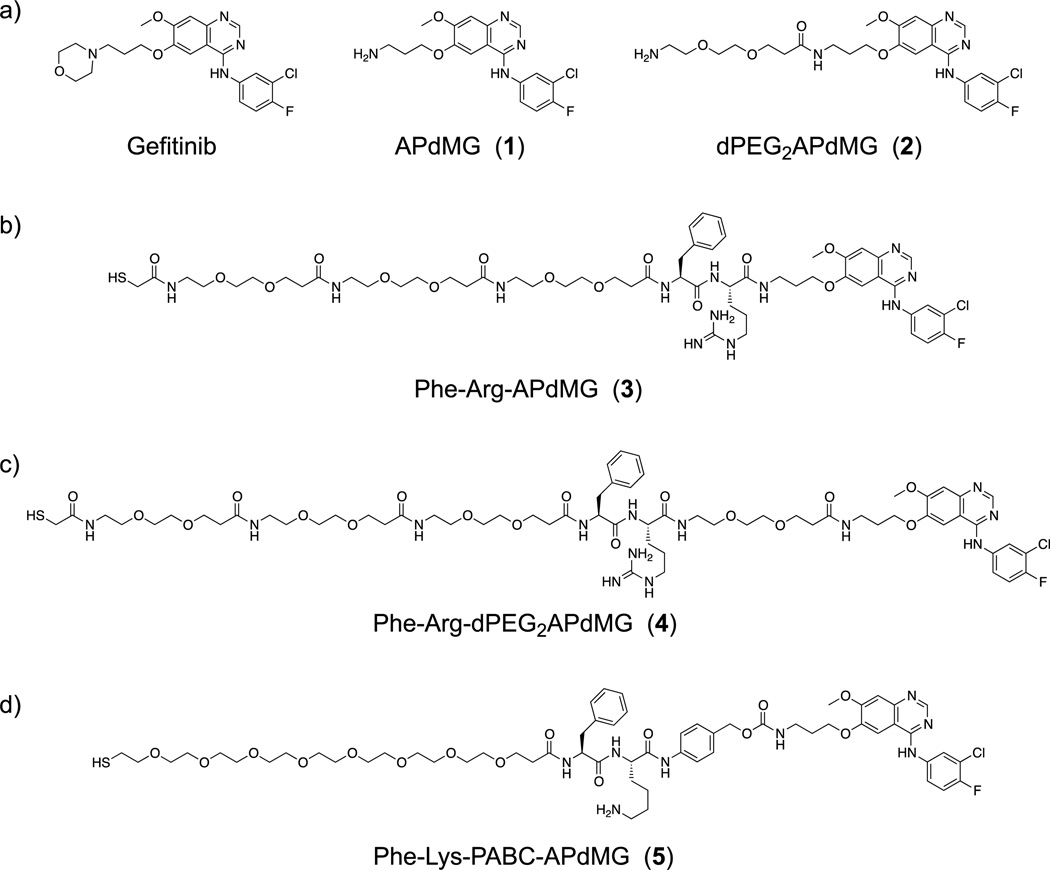
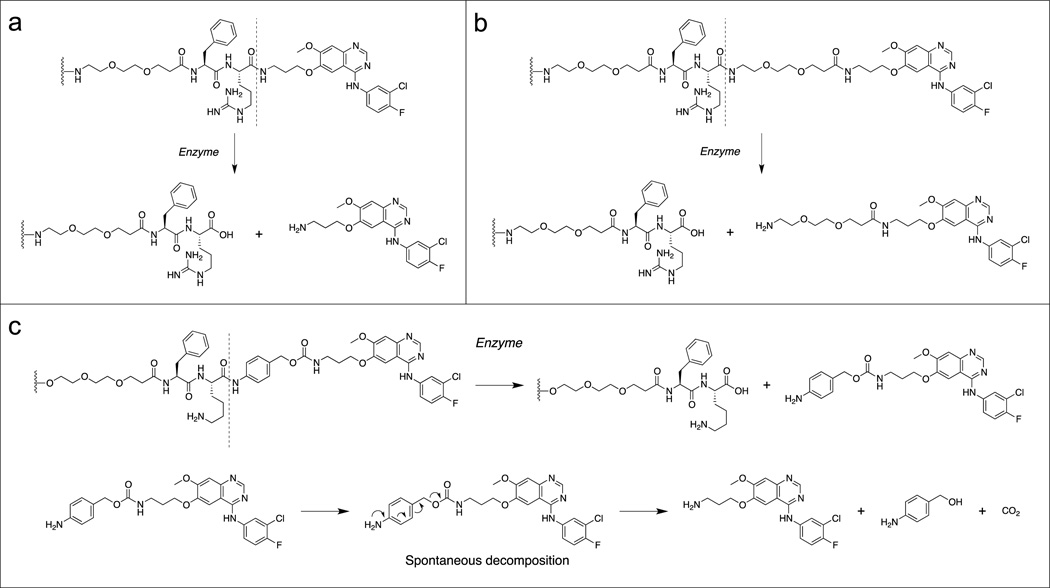
Three linker-drug designs were investigated for C′ dot-based drug delivery in this study (Figure 2b–d). Inspired by previously reported work with antibody drug conjugates,26 both linker types revolve around dipeptide sequences which utilize proteases for drug release. Proteases recognize and bind the dipeptide leading to hydrolysis at the C-terminal end, releasing the drug component from the linker. Two model proteases were used to evaluate the linker-drug constructs described here, trypsin and cathepsin B. Trypsin was selected as a representative serine protease. It is highly active against peptides containing basic amino acids such as arginine and lysine, and cleaves C-terminals to these residues. Cathepsin B is a cysteine protease with more stringent substrate specificity. The minimal substrate consensus sequence described to date is a dipeptide motif containing a hydrophobic and basic residue.26,33 Similar to trypsin, cathepsin B cleaves C-terminals to the basic amino acids. Thus the dipeptides phenylalanine-arginine (Phe-Arg) and phenylalanine-lysine (Phe-Lys), both recognition motifs for trypsin and cathepsin B, have been incorporated into the linker-drug constructs (Figure 3).
Figure 3.
Three linker-drug constructs, (a) Phe-Arg-APdMG utilizes an amide bond for drug attachment. Enzymes recognize and bind the dipeptide sequence (Phe-Arg) then hydrolyze the amide bond C-terminal to the dipeptide and release APdMG, 1; (b) to enhance drug release, Phe-Arg-dPEG2APdMG uses 2, which incorporates a longer 10 atom PEG spacer between the drug and dipeptide; (c) Phe-Lys-PABC-APdMG utilizes a para-aminobenzoxy-carbamate (or PABC) spacer group between the dipeptide (Phe-Lys) and aminopropyl-dMG. After enzyme catalyzed release of the spacer-drug, the spacer spontaneously decomposes from the drug.
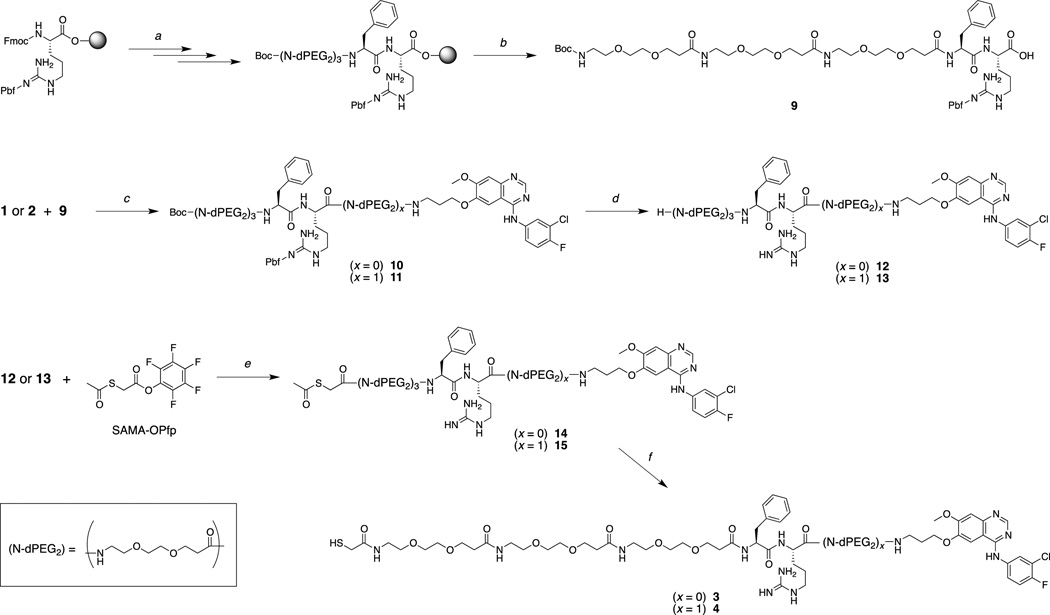
The Phe-Arg-APdMG (3) represents the most straightforward approach to obtaining a protease sensitive linker-drug construct (Figure 2b, 3a). In this design, the gefitinib analog 1 is directly attached to the C-terminus of the dipeptide sequence. Compound 3 was obtained using solid phase peptide synthesis methods leading to 9; followed by coupling of the C-terminus with 2 leading to 10 and a final acid deprotection step to yield 12 (Scheme 2). The N-terminal amine was then reacted with SAMA-OPfp; 14 is then treated with base resulting in the desired product 3. Compound 4 was obtained in a similar manner (Scheme 2), except the gefitinib analog 2 was coupled with 9 resulting in the precursor 11. Deprotection of 11 yielded 13, where the N-terminus was then converted into the S-acetyl acetamide (15). The final treatment with NaOH afforded Phe-Arg-dPEG2APdMG (4) – a linker-drug construct containing a 10 atoms PEG spacer between the Phe-Arg and the gefitinib analog (Figure 2c, 3b).
Scheme 2.
a) SPPS; b) HFIP in DCM; c) HATU, DIEA, DMF; d) TFA; e) DIEA, DMF; f) NaOH
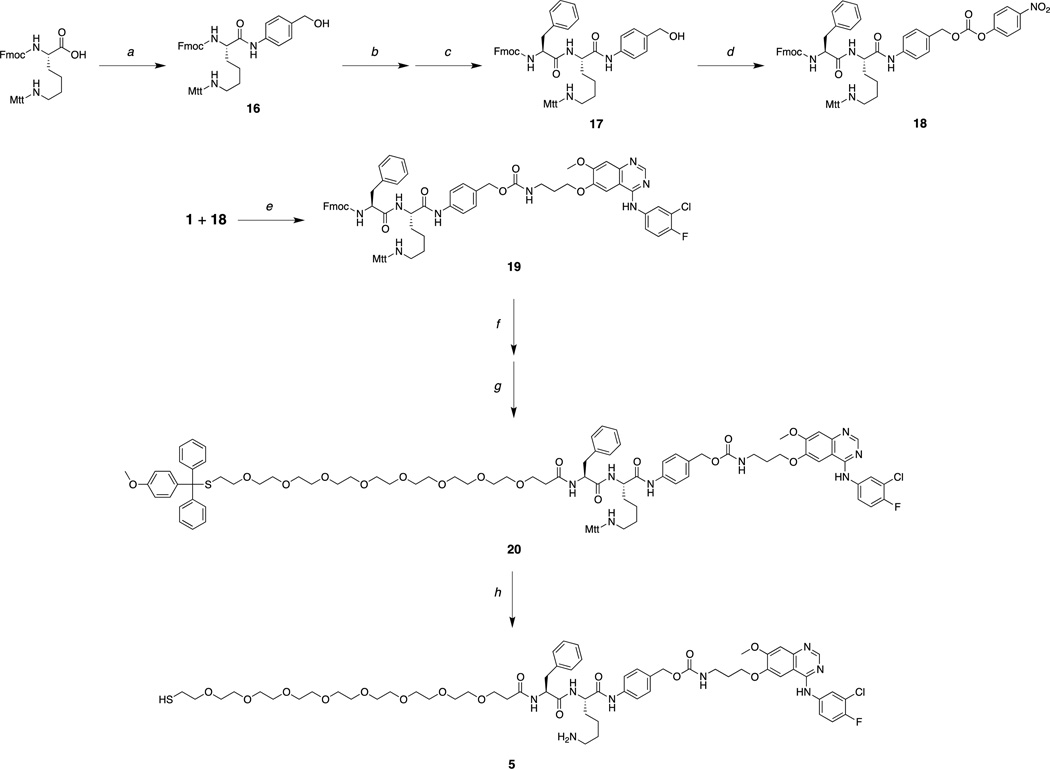
The increased spacing between the dipeptide and drug components without introducing structural changes to 2 were retained by designing and synthesizing a third construct, Phe-Lys-PABC-APdMG (5). This linker incorporates a so-called self-immolative34 para-aminobenzyloxy carbonyl (PABC) group between the peptide and drug (Figure 2d, 3c): Upon enzymatic hydrolysis, this group further decomposes into Para-aminobenzyl alcohol (PABA) and CO2, thereby releasing the drug compound APdMG. Synthesis of compound 5 begins with Fmoc-Lys(Mtt)-OH (Scheme 3).
Scheme 3.
a) para-Aminobenyl alcohol, HATU, DIEA, DMF; b) 30% piperidine in DMF; c) Fmoc-Phe, HATU, DIEA, DMF; d) para-nitrophenyl chloroformate, pyridine, DCM; e) DIEA, DMF; f) 30% piperidine in DMF; g) Mtt-S-dPEG8-COOH, HATU, DIEA, DMF; h) 0.5% TFA, 5% TIS in DCM.
The C-terminus of the protected amino acid is modified with para-aminobenzyl alcohol giving Fmoc-Lys(Mtt)-PABA (16). Upon removal of the Fmoc group with piperidine, and coupling with Fmoc-Phe-OH, the protected dipeptide Fmoc-Phe-Lys(Mtt)-PABA (17) is obtained. The free hydroxyl group of the terminal PABA is then reacted with para-nitrophenol carbonate chloride leading to an activated carbonate (18), which is further reacted with 1 resulting in compound 19. The Fmoc group is removed, and the free N-terminal amine is coupled with Mtt--S’-dPEG8-COOH, yielding 20. The final deprotection step required acidic conditions; however, the para-aminobenzyloxy carbamate group itself is susceptible to decomposition under such conditions. A sufficiently mild condition (0.5% TFA) was found so as to remove the Mtt group from the lysine and Mmt group from the terminal thiol while preserving the linker to afford the desired product 5. It should be noted that the Mtt group masking the lysine side chain proved to be ideally suited for this overall synthetic approach as it was found to be stable in the presence of para-nitrophenol carbonate chloride, but sufficiently labile for removal under mild acidic conditions. This is in contrast to the more commonly used hyper-labile Mmt group for lysine side chain protection,35 that can be readily removed in the presence of para-nitrophenol carbonate chloride.
2.4. Enzymatic evaluation of linker-drug constructs
In vitro assays were conducted for compounds 3 to 5 to evaluate drug release profiles; enzyme mediated hydrolysis of the constructs was monitored over different time points (Table 1, Figure S1 and S2 in Supplementary Material). For both trypsin and cathepsin B, the rate of drug release for the three linker-drug constructs follows the same general trend: 5 > 4 > 3 (fastest to slowest).
Table 1.
Drug Release Assay for Linker-Drug Constructs
| Substrate | Trypsin t1/2 a(min) |
Cathepsin B t1/2 a(min) |
|---|---|---|
| Phe-Arg-APdMG (3) | 9 | NH |
| Phe-Arg-dPEG2APdMG (4) | 2 | 110 |
| Phe-Lys-PABC-APdMG (5) | 1 | 1 |
NH – no hydrolysis
Time when 50% of the drug is released from linker or particle as determined by HPLC at 348 nm.
The results for 3 and 4 suggest that the proximity of the drug to the dipeptide unit affects enzyme activity and drug release. Hydrolysis (drug release) is enhanced when the spacing (distance) between the drug and dipeptide is increased through the incorporation of a 10 atom PEG linker. This effect is most prominently observed with cathepsin B, which is unable to hydrolyze construct 3. However, by incorporating the 10 atom PEG spacer between the drug and dipeptide (4), hydrolysis and drug release are observed.
2.5. Synthesis and characterization of NDCs
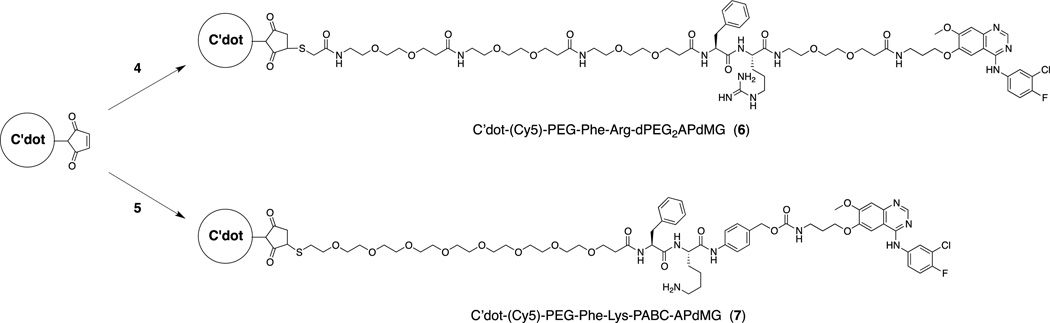
In order to obtain NDCs, maleimide-functionalized C′ dots (mal-PEG-Cy5-C′ dots) were first synthesized, as reported in detail elsewhere.22 The NDCs, C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG (6) and C′* dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG (7) were prepared by adding Phe-Arg-dPEG2APdMG (4) and Phe-Lys-PABC-APdMG (5) to the mal-PEG-C′* dots to allow the terminal thiols on the constructs to react with maleimide groups on the particle surface (Scheme 4).
Scheme 4.
C′ dots-(Cy5)-PEG-mal are reacted with linker-drug constructs Phe-Arg-dPEG2APdMG (4) and Phe-Lys-PABC-APdMG (5) resulting in the NDCs 6and 7.
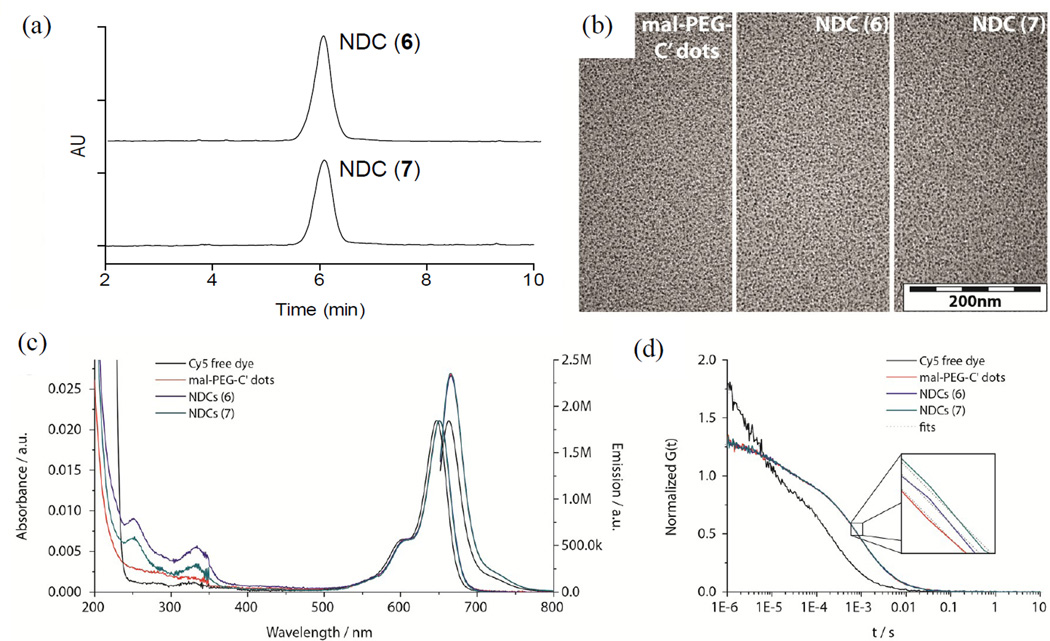
The naming convention used for the NDCs was adopted to reflect the chemical directionality and connectivity of the linker-drug constructs. Consequently, the name of the linker-drug is placed after the C′ dot, as opposed to before, the latter convention adopted in our previous nomenclature (e.g. cRGD-Cy5-C′ dot). The desired NDC products 6 and 7 were isolated following purification by gel filtration (Figure S3). All final particle products (C′ dots and NDCs) were studied by analytical reversed phase HPLC, transmission electron microscopy (TEM), optical absorption/emission spectroscopy and fluorescence correlation spectroscopy (FCS) in order to characterize overall purity, morphology, size, fluorescence properties. The NDCs 6 and 7 were prepared with good overall purity and uniformity as determined by HPLC at 348 nm and TEM (Figure 4a and 4b).
Figure 4.
Characterization of mal-PEG-C′ dots and NDC 6 and 7. (a) Analytical C18 reversed phase HPLC at 348 nm; (b) TEM; (c) absorbance and emission spectra; (d) FCS correlation curves.
Absorption and emission spectra of optical density matched (around 650 nm) samples of Cy5 free dye, mal-PEG-C′ dots, and NDCs 6 and 7 were obtained (Figure 4c). The absorption spectra reveal a peak at around 340 nm, in addition to the Cy5 peak at around 650 nm, for the NDCs, which matches well with the absorbance of gefitinib and suggests successful attachment of the drug-linker constructs onto the PEGylated C′ dot surface. According to the emission spectra, as expected maleimide functionalized C′ dots and NDCs show the same brightness enhancement relative to free Cy5 dye of around 1.4 confirming that outer surface functionalization does not change the particle fluorescence properties. Finally, FCS measurements suggest that particle size only slightly increases after conjugation with drug-linker constructs. In Figure 4d FCS autocorrelation curves shift to larger times, t, implying slower diffusive motion as a result of the larger particle size. From fits of the auto-correlation curves also shown in Figure 4d (inset), the hydrodynamic size of the particles (Table 2) only slightly increases from originally 6.3 nm (C′ dot) to 6.4 nm (NDC 6) and 6.5 nm (NDC 7). Considering that the fully extended drug-linker constructs are at least 2 nm in length, this slight increase of particle size further implies that the actual configuration of the drug-linker constructs on the particle surface most likely is not extended, but rather folded.
Table 2.
Nanoparticle Characterization Summary
| Particle | Diameter a (nm) |
DPR Average b |
|---|---|---|
| C′dot-(Cy5)-PEG-Mal | 6.3 | - |
| C′dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG (6) | 6.4 | 5 |
| C′dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG (7) | 6.5 | 2 |
DPR – drug to particle ratio
Determined by FCS
Determined by HPLC
Analytical HPLC was also used to assess the number of drug molecules per particle or drug to particle ratio (DPR) (Table 2). The concentration of gefitinib analogs can be readily measured at 348 nm, while particle concentrations can be obtained at 650 nm, due to the Cy5 embedded within the C′ dots. For the NDCs used in these studies, the average DPRs were modest, 5 and 2 for NDC 6 and 7, respectively. More recently DPRs in the range of 10 have been obtained and will be utilized in upcoming studies. It is worth noting that despite the poor solubility of the gefitinib analogues precipitation of NDCs was not observed, most likely due to the high dispersibility of C′ dots.
2.6. In vitro evaluation of NDCs
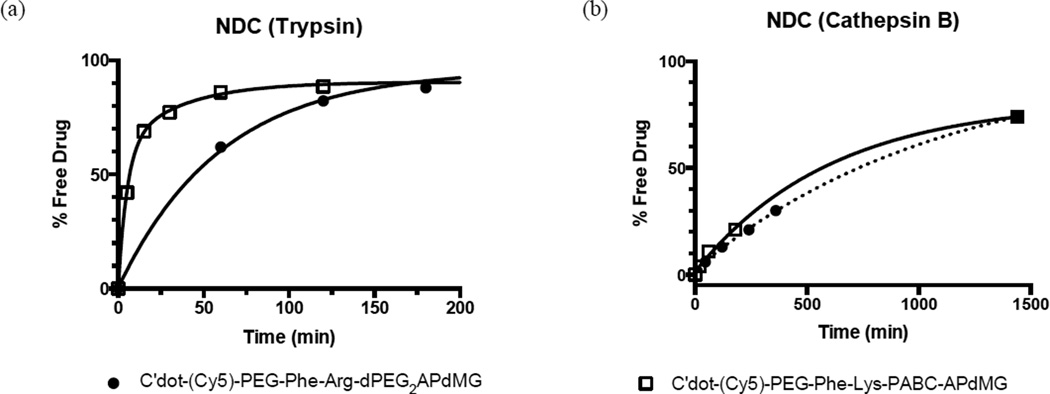
Enzyme dependent drug release over time was measured for C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG 6 and C′ dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG 7 to obtain in vitro drug release profiles (Figure 5). Representative HPLC data demonstrating drug release with trypsin is shown in Figure 6. NDCs 6 and 7 proved to be excellent substrates for trypsin, requiring 44 min and 6 min, respectively, to achieve 50% drug release (Figure 5a, Table 3). Surprisingly, in the presence of cathepsin B, release kinetics were markedly slower for both NDCs: 50% of drug release was achieved in 560 min for 6 and 510 min for 7 (Figure 5b, Table 3). Taken together, the data demonstrates accessibility of the linker-drug constructs on the particle surface to enzymes, leading to controlled release of the drug components.
Figure 5.
In vitro drug release from NDCs 6 and 7 in the presence of enzymes. C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG 6 and C′ dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG 7 were treated with either trypsin (a) or cathepsin B (b). Enzymatic reactions were monitored over time by HPLC at 348 nm. Trypsin assays were conducted in 10 mM phosphate buffer (pH 7.2) at 37°C; cathepsin B assays were conducted in 25 mM sodium acetate buffer (pH 5.0) at 37°C.
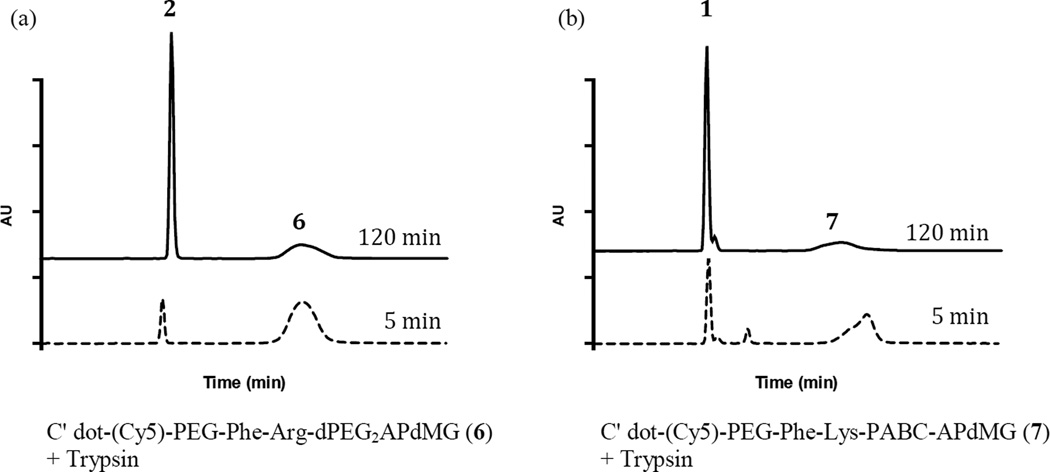
Figure 6.
Representative HPLC profiles of in vitro drug release from NDCs 6 and 7 in the presence of trypsin. NDCs were treated with trypsin then analyzed by HPLC after 5 and 120 min. (a) NDC 6; (b) NDC 7. Data indicate that compounds 2 or 1 are released from the C′dots. Trypsin assays were conducted in 10 mM phosphate buffer (pH 7.2) at 37°C. HPLC analysis was performed at 348 nm.
Table 3.
Drug Release Assay for NDCs
| Substrate | Trypsin t1/2 a (min) |
Cathepsin B t1/2 a (min) |
|---|---|---|
| C′dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG (6) | 44 | 560 |
| C′dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG (7) | 6 | 510 |
Time when 50% of the drug is released from linker or particle. Determined by HPLC.
The stabilities of NDCs were evaluated in aqueous solutions under acidic and neutral pH conditions (5.0 and 7.2) at 37°C. Both NDCs exhibited minimal degradation or drug release after 48 hours, as assessed by HPLC (Table 4). Recently, thiol-maleimide based conjugations have drawn scrutiny due to the observed loss of linker drug constructs from antibody drug conjugates due to possible reverse Michael or exchange reactions that can occur with free thiols. To assess in vitro stability of the NDCs in the presence of excess thiols, 7 was incubated with 30 mM gluthathione at 37°C for 48 hours at pH 7.2. Less than 5% of the linker-drug was separated from the C′ dot after 48 hours (Table 4).
Table 4.
NDC Stability Data
| Particle | pH 5.2 a 48 hr |
pH 7.2 b 48 hr |
Glutathionec 48 hr |
Mediad 18 hr |
|---|---|---|---|---|
| NDC 6 | ~ 1% | ~ 2% | ~ 5% | < 5% |
| NDC 7 | ~ 6% | ~ 2% | ~ 4% | < 1% |
25 mM sodium acetate buffer
50 mM phosphate buffer
10 mM glutathione (reduced) in 50 mM phosphate buffer
DEM, serum free, 18 hrs after cell treatment
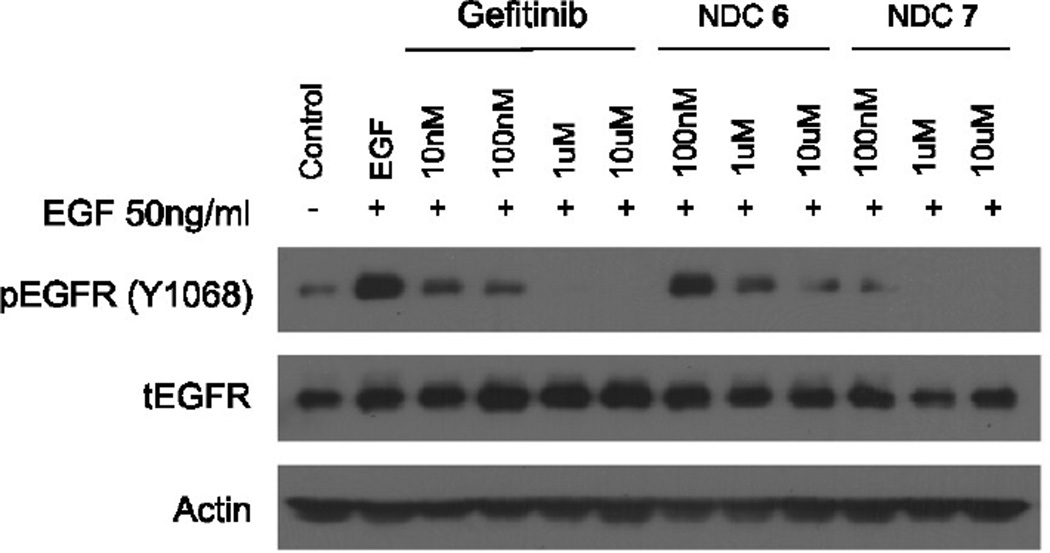
The biological activities of C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG 6 and C′ dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG 7 were assessed by treatment of H1650 cells followed by western blot detection of phospho-Tyr168 in EGFR and compared with gefitinib. Serum starved cells were incubated with each NDC over a period of 18 hours, and then subjected to EGF stimulation. As expected, the gefitinib control exhibited a dose-dependent decrease in Tyr168 phosphorylation of EGFR, with complete inhibition at 1 µM (Figure 7).
Figure 7.
Western blot analysis of H1650 cells treated with gefitinib, C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2APdMG (NDC 6), and C′ dot-(Cy5)-PEG-Phe-Lys-PABC-APdMG (NDC 7). Cells treated with gefitinib or specified NDC at indicated concentrations for 18 hrs followed by EGF (50 ng/mL) for 5 minutes. (pEGFR – phosphorylated EGFR; tEGFR – total EGFR)
NDCs 6 and 7 also showed dose-dependent inhibition (Figure 7). However, cells treated with 6 showed detectable levels of phospho-Tyr168 at 10 µM. By contrast, compound 7, which had a lower DPR (Table 2), exhibited increased potency relative to 6 and the gefitinib control; a marked loss of phospho-Tyr168 expression was observed at 100 nM of compound 7, with complete inhibition at NDC concentrations of 1 µM. Thus, even though NDC 7 had a lower DPR, it incorporated a more active gefitinib analogue 3, as against analogue 2 used in NDC 6. Given the possibility that premature drug release can occur and lead to the observed decreases in phospho-Tyr168 EGFR, we monitored the stability of the NDCs used in these assays. An aliquot of the media with NDC 6 or 7 (10 µM) used for treating the H1650 cells for 18 hours was analyzed by HPLC. Both particles proved to be stable under these conditions as no free drug was detected in the media and NDCs were intact (Table 4).
2.7. Radiolabeled NDCs
In addition to the NDCs 6 and 7, we investigated the potential for incorporating a secondary imaging modality for future in vivo studies. To this end, we synthesized a linker-drug construct, Phe-Arg-dPEG2-d-Tyr-aminopropyl-dMG that incorporates a d-tyrosine residue with the drug component for attaching a radiolabel (compounds 21 and 23 in Supplementary Material). The NDC C′ dot-(Cy5)-PEG-Phe-Arg-dPEG2-d-Tyr-APdMG was prepared, and successfully radioiodinated with 131I at >90% radiochemical purity (Figure S4).
3. Conclusion
In this report we introduce nanoparticle drug conjugates composed of a clinically translated silica-based nanoparticle platform with covalently attached drug molecules and contrast-producing radiolabels. The NDCs are ultrasmall (6–7 nm) and utilize enzyme sensitive linkers, where drug release is catalyzed by proteases. Analogs of gefitinib, an important EGFR-TKI cancer drug, was modified with these linkers and conjugated onto the PEGylated particle surface. The resulting NDCs exhibited excellent in vitro stability, solubility, and proved to be active in NSCLC cells, demonstrating improved potency at 100 nM concentrations over the native drug itself. The attachment of a radiolabel will enable accurate quantification of NDC biological properties for optimizing cancer therapies and improving patient selection on the basis of observing successful particle localization at the tumor site. Investigations into the effects of drug loading, the use of additional targeting surface ligands, and in vivo studies (stability and pharmacokinetics) using relevant brain tumor mouse models are underway.
4. Experimental section
4.1. General information
Gel permeation chromatography (GPC) characterization was performed using a BioLogic LP system equipped with a 275nm UV detector and with resin Superdex 200 from GE healthcare. Transmission electron microscopy (TEM) images were taken using a FEI Tecnai T12 Spirit microscope operated at an acceleration voltage of 120kV. Absorbance and fluorescence spectra were measured by a Varian Cary 5000 spectrophotometer or Photon Technologies International Quantamaster spectrofluorometer. FCS measurements were conducted using a home built FCS setup with a 633nm solid-state laser as the laser source for Cy5. Silica gel purifications were conducted on a Teledyne ISCO CombiFlash Rf. Analytical HPLC were run on a Waters Alliance HPLC System or Autopure LCMS System (2767 Sample Manager, 2996 Photodiode Array Detector, 2420 ELS Detector, Micromass ZQ, 2525 Binary Gradient Module, Column Fluidics Organizer, 515 HPLC Pump, Pump Control Module II). HPLC purifications were performed on a Waters Preparative System (2996 Photodiode Array Detector, 2545 Binary Gradient Module) or Autopure LCMS System. 1H-NMR and 13C-NMR data were obtained on a Bruker Ultrashield 500 Plus. Further instrument details are included in Supplemental Materials.
4.2. Materials
Solvents and reagents purchased from Sigma Aldrich and Fisher Scientific unless otherwise noted. Maleimide functionalized Cy5 fluorophores were purchased from GE healthcare. Silanes were obtained from Gelest. PEGylated reagents were purchased from Quanta BioDesign. O-Des-morpholinopropyl gefitinib was obtained from Toronto Research Chemicals (TRC). Amino acids and peptide synthesis reagents were purchased from EMD Chemicals. DMSO-d and CDCl3 were purchased from Cambridge Isotopes. Silica TLC plates, 4 g, 12 g, 24 g, and 40 g RediSep Rf normal phase cartridges were obtained from Teledyne ISCO.
4.3 Synthesis
4.3.1. O-des-Morpholino-gefitinib, dMG (6)
Compound was obtained commercially. 1H-NMR (500 MHz, DMSO-d6): δ 9.69 (s, 1H), 9.47 (s, 1H), 8.48 (s, 1H), 8.22 (dd, J= 6.9, 2.7 Hz, 1H), 7.84 (ddd, J= 9.1, 4.4, 2.7 Hz, 1H), 7.78 (s, 1H), 7.41 (t, J= 9.1 Hz, 1H), 7.22 (s, 1H), 3.98 (s, 3H). 13C-NMR (125 MHz, DMSO-d6): δ 155.83, 153.91, 151.89, 146.73, 146.20, 122.74, 121.69, 116.50, 116.33, 109.51, 107.18, 105.25, 55.92. ESI-MS (m/z) for C15H11ClFN3O2 (exact mass 319.1): [M+H]+ calc. 320.1, obs. 320.2.
4.3.2. N-(tert-Butyloxycarbonyl)-aminopropyl-dMG (7)
500 mg of 6 was dissolved in 100 mL of anhydrous DMF. K2CO3 (650 mg, 4.7 mmol, 3 eq) was added to the solution. Stir bar. The reaction proceeded at 60°C for 16 hrs and checked by TLC and/or HPLC. The solvent was removed in vacuo leaving a brown oil, which was flash purified on a 24 g RediSep Rf normal phase cartridge using a linear gradient of DCM to 20% MeOH in DCM. The final product was isolated as a white solid (312 mg, 62% yield). 1H-NMR (500 MHz, DMSO-d6): δ 9.57 (s, 1H), 8.49 (s, 1H), 8.12 (dd, J= 6.8, 2.7 Hz, 1H), 7.81 (m, 1H), 7.44 (t, J= 9.1 Hz, 1H), 7.20 (s, 1H), 6.92 (t, J= 5.7 Hz, 1H), 4.17 (t, J= 6.0 Hz, 2H), 3.95 (s, 3H), 3.15 (q, J= 6.5 Hz, 2H), 1.95 (p, J = 6.5 Hz, 2H), 1.38 (s, 9H). 13C-NMR (125 MHz, DMSO-d6): δ 156.01, 155.59, 154.39, 152.61, 146.92, 123.40, 122.29, 118.62, 116.54, 116.37, 107.23, 102.59, 77.51, 66.69, 55.84, 37.22, 35.75, 28.95, 28.22. ESI-MS (m/z) for C23H26ClFN4O4 (exact mass 476.2): [M+H]+ calc. 477.2, obs. 4773.
4.3.3. Aminopropyl-O-des-morpholino-gefitinib, APdMG (1)
7 (100 mg, 0.21 mmol) was treated with 1 mL TFA:water (9:1) for 30 min. The TFA:water was removed in vacuo, leaving a light yellow oil. The oil was washed with diethyl ether, then dissolved in a solution of water:acetonitrile (1:1), frozen and lyophilized. A tan solid was obtained (TFA salt, 98 mg, 95% yield). 1H-NMR (500 MHz, DMSO-d6): δ 10.73 (s, 1H), 8.80 (s, 1H), 8.03 (s, 1H), 8.05 – 8.00 (m, 1H), 7.88 (s, 3H), 7.72 (ddd, J= 9.0, 4.3, 2.6 Hz, 1H), 7.54 (t, J= 9.0 Hz, 1H), 7.35 (s, 1H), 4.27 (t, J= 5.9 Hz, 2H), 4.00 (s, 3H), 3.04 (p, J= 6.7, 6.3 Hz, 2H), 2.13 (dt, J= 12.2, 6.0 Hz, 2H). 13C-NMR (125 MHz, DMSO-d6): δ 158.02, 148.80, 116.96, 116.79, 107.60, 103.66, 66.17, 56.42, 36.39, 26.59. ESI-MS (m/z) for C18H18ClFN4O2 (exact mass 376.1): [M+H]+ calc. 377.1, obs. 377.2.
4.3.4. 9-Fluorenylmethoxycarbonyl-N-amido-dPEG2-aminopropyl-dMG, Fmoc-dPEG2APdMG (8)
A solution containing 1 (25 mg, 0.05 mmol, TFA salt) and Fmoc-N-amido-dPEG2-COOH (20 mg, 0.05 mmol) in DMF (500 µL) was prepared. DIEA (19 mg, 0.15 mmol, 26 µL) was added followed by a solution of HATU (19 mg, 0.05 mmol) in DMF (100 µL). The reaction proceeded for 30 min at room temp, and determined complete by LCMS. The volume was reduced in vacuo, and purified by silica gel chromatography using a gradient of ethyl acetate and 10% methanol in ethyl acetate. Fractions were collected, pooled and solvent removed in vacuo. The isolated product was a white solid (84% yield). 1H-NMR (500 MHz, DMSO-d6): δ 9.55 (d, J= 10.7 Hz, 1H), 8.51 (d, J= 2.4 Hz, 1H), 8.18 – 8.07 (m, 1H), 7.96 (t, J= 5.5 Hz, 1H), 7.87 (d, J= 7.6 Hz, 2H), 7.83 – 7.76 (m, 2H), 7.67 (d, J= 7.5 Hz, 2H), 7.49 – 7.36 (m, 3H), 7.31 (td, J= 7.5, 1.2 Hz, 3H), 7.21 (s, 1H), 4.28 (d, J= 6.9 Hz, 2H), 4.24 – 4.10 (m, 3H), 3.94 (s, 3H), 3.60 (t, J= 6.4 Hz, 2H), 3.46 (s, 4H), 3.36 (t, J= 6.0 Hz, 2H), 3.27 (q, J= 6.6 Hz, 2H), 3.10 (q, J= 5.9 Hz, 2H), 2.32 (t, J= 6.5 Hz, 2H), 1.97 (p, J= 6.5 Hz, 2H). 13C-NMR (125 MHz, CDCl3): δ 170.08, 143.85, 127.54, 126.98, 125.10, 120.05, 102.60, 69.42, 69.04, 66.78, 66.58, 55.87, 46.69, 40.01, 39.94, 39.85, 39.77, 39.68, 39.60, 39.51, 39.43, 39.35, 39.25, 39.18, 39.07, 39.01, 36.16, 35.69, 28.67, 0.08. ESI-MS (m/z) for C40H41ClFN5O7 (exact mass 757.27): [M+H]+ calc. 758.3, obs. 758.4.
4.3.5. Amino-dPEG2-aminopropyl-dMG, dPEG2APdMG (2)
8 (10 mg, 0.013 mmol) was dissolved in 30% piperidine in DMF (1 mL) and allowed to react for 15 min at room temp. The solvent was removed in vacuo, dissolved in water/acetonitrile and purified by reversed phase (C18) HPLC. The product was recovered as a white powder (7 mg, 80% yield). 1H-NMR (500 MHz, DMSO-d6): δ 10.66 (s, 1H), 8.78 (s, 1H), 8.05 – 7.97 (m, 3H), 7.80 (s, 3H), 7.72 (ddd, J= 9.0, 4.3, 2.6 Hz, 1H), 7.54 (t, J= 9.1 Hz, 1H), 7.32 (s, 1H), 4.20 (t, J = 6.1 Hz, 2H), 4.00 (s, 3H), 3.65 – 3.48 (m, 7H), 3.27 (q, J= 6.6 Hz, 2H), 2.96 (q, J= 5.5 Hz, 2H), 2.34 (t, J= 6.5 Hz, 2H), 1.98 (t, J = 6.5 Hz, 2H). 13C-NMR (125 MHz, DMSO-d6): δ 116.96, 69.55, 69.29, 66.73, 66.61, 56.38, 38.57, 36.03, 35.63, 28.58. ESI-MS (m/z) for C25H31ClFN5O5 (exact mass 535.20): [M+H]+ calc. 536.2, obs. 536.3.
4.3.6. Boc-N-amido-(dPEG2)3-Phe-Arg(Pbf)-OH (9)
Chlorotrityl resin (100 mg, 0.1 mmol, 1 mmol/g) was transferred into a fritted syringe reaction vessel and suspended in 2 mL anhydrous DCM for 10 min. The solvent was dispensed, and a solution of DIEA in anhydrous DCM followed by a solution of Fmoc-Arg(Pbf)-OH (97.5 mg, 1.5 eq) in anhydrous DCM were drawn into the syringe; and agitated for 40 min. The solution was dispensed and the resin washed for 2 min, 2× with DCM, then 2× with DMF. Standard solid phase peptide synthesis procedures were the carried out to obtain the final peptide. In short Fmoc deprotections were accomplished by washing the resin 2× using 30% piperidine/DMF (1 mL) for 10 min. This was followed by DMF (1 mL) washes, 4× for 2 min each. Coupling reactions were carried out at room temp using an excess of protected amino acid (3 eq in 2 mL DMF), DIEA (120 mg, 0.9 mmol, 160 µL, 9 eq in 1 mL DMF), HATU (114 mg, 0.3 mmol, 3 eq in 2 mL DMF), added to the syringe in that order, and shaken for 1 hr. This was followed by DMF (1 mL) washes, 4× for 2 min each. Fmoc-Phe (116 mg, 0.3 mmol, 3 eq) was added, followed by three residues of Fmoc-N-dPEG2-OH (120 mg, 0.3 mmol, 3 eq). After completion of the sequence, final Fmoc deprotection and washes, BOC anhydride (87 mg, 0.4 mmol, 4 eq) and DIEA (120 mg, 0.9 mmol, 160 µL, 9 eq) in 2 mL of DMF were used to cap the N-terminal amine. The peptide-resin was washed with DMF (1 mL, 2 min, 2×), then DCM (1 mL, 2 min, 4×). The protected peptide product was then cleaved off of the resin by adding 50% HFIP in DCM (2 mL) to the syringe and shaking for 1 hr at room temp. The crude peptide was then purified by reversed phase (C18) HPLC, lyophilized, resulting in a white solid (62 mg). Analytical HPLC: Figure S5. ESI-MS (m/z) for C54H86N8O17S (exact mass 1150.56): [M+H]+ calc. 1151.6, obs. 1151.7.
4.3.7. Boc-N-amido-(dPEG2)3-Phe-Arg(Pbf)-APdMG (10)
A solution containing 9 (23 mg, 0.02 mmol, 1 eq) and 1 (9 mg, 0.024 mmol, 1.2 eq) in DMF (1 mL) was prepared. To this DIEA (10 mg, 0.08 mmol, 14 µL, 4 eq) was added followed by HATU (9 mg, 0.024 mmol, 1.2 eq). The reaction was monitored by HPLC, and completed within 30 minutes. The solvent was removed in vacuo, then resuspended in DCM. The DCM solution was washed with water 4×, then evaporated leaving a tan oil. The crude material was purified by reversed phase (C18) HPLC. The final product was isolated as a white solid (21 mg, 70% yield). Analytical HPLC: Figure S6. ESI-MS (m/z) for C72H102ClFN12O18S (exact mass 1508.68): [M+H]+ calc. 1509.7, obs. 1509.7; [M+2H]2+ calc. 755.4, obs. 755.0.
4.3.8. Boc-N-amido-(dPEG2)3-Phe-Arg(Pbf)-dPEG2APdMG (11)
A solution containing 9 (30 mg, 0.026 mmol, 1 eq) and 2 (18 mg, 0.034 mmol, 1.3 eq) in DMF (1 mL) was prepared. To this DIEA (14 mg, 0.1 mmol, 18 µL, 4 eq) was added followed by HATU (13 mg, 0.034 mmol, 1.3 eq). The reaction was monitored by HPLC, and completed within 30 minutes. The solvent was removed in vacuo, then resuspended in DCM. The DCM solution was washed with water 4×, then evaporated leaving a tan oil. The crude material was purified by reversed phase (C18) HPLC. The final product was isolated as a white solid (28 mg, 65% yield). Analytical HPLC: Figure S7. ESI-MS (m/z) for C79H115ClFN13O21S (exact mass 1667.77): [M+2H]2+ calc. 834.9, obs. 834.7.
4.3.9. H2N-(dPEG2)3-Phe-Arg-APdMG (12)
TFA/water (9:1, 1 mL) was added 10 (21 mg, 0.014 mmol) and left at room temp for 1 hour. The reaction was evaporated then dissolved in ACN/water, frozen and lyophilized leaving a white solid. The material was used without further purification. The final product was left as a white solid. Analytical HPLC: Figure S8. ESI-MS (m/z) for C54H78ClFN12O13 (exact mass 1156.55): [M+H]+ calc. 1157.6, obs. 1157.8; [M+2H]2+ calc. 579.3, obs. 579.1.
4.3.10. H2N-(dPEG2)3-Phe-Arg-(dPEG2)-APdMG (13)
TFA/water (9:1, 1 mL) was added 11 (28 mg, 0.017 mmol) and left at room temp for 1 hour. The reaction was evaporated then dissolved in ACN/water, frozen and lyophilized leaving a white solid. Analytical HPLC: Figure S9. ESI-MS (m/z) for C61H91ClFN13O16 (exact mass 1315.64): [M+H]+ calc. 1316.7, obs. 1316.5; [M+2H]2+ calc. 658.6, obs. 658.5.
4.3.11. S-Acetyl-mercaptoacetamido-(dPEG2)3-Phe-Arg-APdMG (14)
A solution containing 12 (5 mg, 0.004 mmol, 1 eq) and DIEA (1.5 mg, 0.012 mmol, 2 µL, 3 eq) in DMF (200 µL) was prepared. SAMA-OPfp (2 mg, 0.006 mmol, 1.5 eq) in DMF (100 µL) was then added to the solution, and allowed to react for 1 hour. The solvent was removed in vacuo then purified by reversed phase HPLC. 3 mg of a white solid was recovered (60% yield). Analytical HPLC: Figure S10. ESI-MS (m/z) for C58H82ClFN12O15 (exact mass 1272.54): [M+H]+ calc. 1273.6, obs. 1273.3; [M+2H]2+ calc. 637.3, obs. 637.6.
4.3.12. S-Acetyl-mercaptoacetamido-(dPEG2)3-Phe-Arg-dPEG2APdMG (15)
A solution containing 13 (5 mg, 0.004 mmol, 1 eq) and DIEA (1.5 mg, 0.012 mmol, 2 µL, 3 eq) in DMF (200 µL) was prepared. SAMA-OPfp (2 mg, 0.006 mmol, 1.5 eq) in DMF (100 µL) was then added to the solution, and allowed to react for 1 hour. The solvent was removed in vacuo then purified by reversed phase HPLC. 3 mg of a white solid was recovered (53% yield). Analytical HPLC: Figure S11. ESI-MS (m/z) for C65H95ClFN13O18S (exact mass 1431.63): [M+2H]2+ calc. 716.8, obs. 716.7.
4.3.13. Mercaptoacetamido-(dPEG2)3-Phe-Arg-APdMG, Phe-Arg-APdMG (3)
2 mg of 14 was dissolved in 100 µL of water/MeOH (1:1), to which 2 uL of 1 N NaOH was added. After 15 minutes, 2 uL of 1 M HCl was added to neutralize. The solution was used directly for subsequent experiments or conjugations. Analytical HPLC: Figure S12. ESI-MS (m/z) for C56H80ClFN12O14S (exact mass 1230.53): [M+H]+ calc. 1231.5, obs. 1231.4; [M+2H]2+ calc. 616.3, obs. 616.3.
4.3.14. Mercaptoacetamido-(dPEG2)3-Phe-Arg-dPEG2APdMG, Phe-Arg-dPEG2APdMG (4)
2 mg of 15 was dissolved in 100 µL of water/MeOH (1:1), to which 2 uL of 1 N NaOH was added. After 15 minutes, 2 uL of 1 M HCl was added to neutralize. The solution was used directly for subsequent experiments or conjugations. Analytical HPLC: Figure S13. ESI-MS (m/z) for C63H93ClFN13O17S (exact mass 1389.62): [M+H]+ calc. 1390.6, obs. 1390.5; [M+2H]2+ calc. 695.8, obs. 695.7.
4.3.15. Fmoc-Lys(Mtt)-PABOH (16)
A solution of Fmoc-Lys(Mtt)-OH (748 mg, 1.2 mmol, 1 eq) and para-amino benzyl alcohol (300 mg, 2.4 mmol, 2 eq) in DMF (5 mL) was prepared. DIEA (465 mg, 3.6 mmol, 630 µL, 3 eq) was added followed by a solution of HATU (502 mg, 1.3 mmol, 1.1 eq) in DMF (2 mL). The reaction was complete within 30 minutes and as determined by HPLC/LCMS. The solvent was partially removed in vacuo, and extracted with ethyl acetate/water. The ethyl acetate layer was washed 4× with water, then evaporated leading to an orange solid. The crude material was purified on a 40 g RediSep Rf normal phase cartridge using a linear gradient of hexane and ethyl acetate. The final product was isolated as a white solid (850 mg, 97% yield). 1H-NMR (500 MHz, CDCl3): δ 7.96 (s, 1H), 7.73 (d, J = 7.9 Hz, 2H), 7.54 (d, J= 7.5 Hz, 2H), 7.47 – 7.41 (m, 6H), 7.39 – 7.20 (m, 12H), 7.18 – 7.11 (m, 2H), 7.05 (d, J= 7.9 Hz, 2H), 5.29 (s, 1H), 4.63 (s, 1H), 4.44 (d, J= 6.4 Hz, 2H), 2.28 (s, 3H), 2.11 (t, J= 6.9 Hz, 2H), 1.88 (s, 1H), 1.58 (s, 5H), 1.51 (s, 1H), 1.38 (s, 2H). 13C-NMR (500 MHz, CDCl3): δ 146.36, 143.22, 141.32, 135.70, 128.57, 128.52, 128.49, 127.79, 127.74, 127.12, 126.14, 124.92, 120.16, 120.02, 77.27, 77.02, 76.76, 70.62, 64.92, 60.41, 47.16, 43.30, 30.57, 23.43, 21.07, 20.93, 14.21, 0.01. ESI-MS (m/z) for C48H47N3O4 (exact mass 729.36): [M+H]+ calc. 730.4, obs. 730.2.
4.3.16. Fmoc-Phe-Lys(Mtt)-PABOH (17)
The Fmoc group was removed from 16 (425 mg, 0.6 mmol, 1 eq) using 9 mL of 30% piperidine in DMF for 10 minutes. The solvent was removed in vacuo, and the resulting oil was resuspended in 10 mL DMF A solution of Fmoc-Lys(Mtt)-OH (748 mg, 1.2 mmol, 1 eq) and para-amino benzyl alcohol (300 mg, 2.4 mmol, 2 eq) in DMF (5 mL) was prepared. DIEA (465 mg, 3.6 mmol, 630 µL, 3 eq) was added followed by a solution of HATU (502 mg, 1.3 mmol, 1.1 eq) in DMF (2 mL). The reaction was complete within 30 minutes and as determined by HPLC/LCMS. The solvent was partially removed in vacuo, and extracted with ethyl acetate/water. The ethyl acetate layer was washed 4× with water, then evaporated leading to an orange solid. The crude material was purified on a 40 g RediSep Rf normal phase cartridge using a linear gradient of hexane and ethyl acetate. The final product was isolated as a white solid (850 mg, 97% yield). 1H-NMR (500 MHz, CDCl3) δ 8.20 (s, 1H), 7.73 (d, J= 7.6 Hz, 2H), 7.50 (s, 2H), 7.48 – 7.34 (m, 8H), 7.34 – 7.20 (m, 11H), 7.20 – 7.07 (m, 7H), 7.04 (d, J= 8.1 Hz, 2H), 6.28 (s, 1H), 5.22 (s, 1H), 4.62 (s, 2H), 4.43 (dd, J= 10.7, 6.7 Hz, 1H), 4.32 (d, J= 11.5 Hz, 1H), 3.05 (s, 2H), 2.27 (s, 3H), 2.11 – 2.02 (m, 2H), 1.89 (s, 1H), 1.46 (d, J= 6.3 Hz, 1H), 1.26 (s, 2H). 13C-NMR (500 MHz, CDCl3) δ 146.35, 141.31, 136.97, 135.70, 129.10, 128.93, 128.57, 128.50, 127.82, 127.74, 127.71, 127.39, 127.12, 126.14, 124.91, 124.84, 120.10, 120.04, 77.28, 77.23, 77.02, 76.77, 70.60, 67.15, 64.96, 54.06, 47.08, 43.35, 31.30, 30.60, 23.52, 20.93. ESI-MS (m/z) for C57H56N4O5 (exact mass 876.43): [M+H]+ calc. 877.4, obs. 877.3.
4.3.17. Fmoc-Phe-Lys(Mtt)-PABC-APdMG (19)
Compound 17 (420 mg, 0.5 mmol, 1 eq) was dissolved in anhydrous DCM (20 mL). Pyridine (216 mg, 2.7 mmol, 5.4 eq) was added, followed by a solution of 4-nitrophenyl chloroformate (180 mg, 0.9 mmol, 1.8 eq) in anhydrous DCM. The reaction proceeded for 2 hours at room temperature, then checked by HPLC and TLC. The solvent was removed in vacuo then purified by on a 24 g RediSep Rf normal phase cartridge using a linear gradient of hexane and ethyl acetate. The product Fmoc-Phe-Lys(Mtt)-PABC-pNP (18), was isolated as a yellow solid (360 mg, 70% yield). 18 (50 mg, 0.05 mmol, 1 eq) was dissolved in anhydrous DCM (3 mL). A solution of 1 (25 mg, 0.05 mmol, TFA salt, 1 eq) with DIEA (65 mg, 0.5 mmol, 90 µL, 10 eq) in anhydrous DCM was then added to 18. The reaction proceeded at room temperature for 4 hours, then checked by HPLC and TLC. The solvent was removed in vacuo, and the crude material was purified on a 4 g RediSep Rf normal phase cartridge with a linear gradient of hexane and ethyl acetate. The final product was isolated as a yellow solid (38 mg, 60% yield). ESI-MS (m/z) for C76H72ClFN8O8 (exact mass 1278.51): [M+H]+ calc. 1279.5, obs. 1279.4; [M+2H]2+ calc. 640.3, obs. 640.3.
4.3.18. Mmt-S-dPEG8-Phe-Lys(Mtt)-pABC-dMG (20)
18 mg of 19 (0.014 mmol, 1 eq) was deprotected with 2 mL of 30% piperidine in DMF. After 5 minutes the reaction was confirmed complete by HPLC/LCMS, and the solvent was removed in vacuo. The resulting oil was dissolved in DMF (0.5 mL), to which Mmt-S-dPEG8-COOH (13 mg, 0.017 mmol, 1.2 eq) and DIEA (9 mg, 0.070 mmol, 13 µL, 5 eq) were added. A solution of HATU (6 mg, 0.014 mmol, 1.2 eq) in DMF (200 µL) was prepared and added to the reaction. After 1 hr, the reaction was deemed complete by HPLC/LCMS, and solvent was removed in vacuo. The remaining oil was flash purified on a 4 g RediSep Rf normal phase cartridge using a linear gradient of DCM to 10% MeOH in DCM. The final product was isolated as a white solid (23 mg, 92% yield). Analytical HPLC: Figure S14. ESI-MS (m/z) for C100H114ClFN8O16S (exact mass 1768.77): [M+2H]2+ calc. 885.9, obs. 886.0.
4.3.19. HS-dPEG8-Phe-Lys-PABC-aminopropyl-dMG (5)
10 mg of 20 (5.6 µmol) was treated with 2 mL of 1% TFA/10% TIS in DCM for 2 hrs, then checked by HPLC/LCMS to confirm complete deprotection. The solution was removed in vacuo, and then washed with cold ether, 3×. The white solid was dissolved in water/acetonitrile (1:1), frozen and lyophilized. The resulting white solid was used without further purification (6 mg, 86% yield). Analytical HPLC: Figure S15. ESI-MS (m/z) for C60H82ClFN8O15S (exact mass 1240.53): [M+H]+ calc. 1241.5, obs. 1241.6; [M+2H]2+ calc. 621.3, obs. 621.3.
4.3.20. Mal-PEG-C′ dots22
A maleimide and NHS ester functionalized polyethylene glycol (mal-dPEG12-NHS) was conjugated with aminosilane (APTES) in DMSO (molar ratio mal-PEG-NHS:APTES:DMSO 1:0.9:60). The reaction mixture was left under nitrogen at room temperature for 48 hours to generate silane functionalized mal-dPEG (mal-dPEG-APTES). A maleimide functionalized Cy5 (mal-Cy5) was reacted with a thiol-silane (MPTMS) in DMSO (molar ratio Cy5:MPTMS:DMOS 1:25:1150). The reaction was left under nitrogen at room temperature for 24 hours to generate a silane functionalized Cy5 (Cy5-MPTMS). TMOS and Cy5-MPTMS were then titrated into an ammonia hydroxide solution (~pH 8) (molar ratio TMOS:Cy5:NH3:H2O 1: 0.001:0.44:1215). The solution was stirred at 600 rpm at room temperature for 24 hours to form homogeneous Cy5 encapsulated silica nanoparticles. The mal-dPEG-APTES and silane functionalized polyethylene glycol (PEG-silane, MW around 500, Gelest) were then added into the synthesis solution to PEGylate and surface-functionalize the particles (PEG-silane:TMOS:mal-PEG-APTES 1:2.3:0.006). The solution was stirred at 600 rpm at room temperature for 24 hours followed by incubation at 80°C for another 24 hours without stirring. The solution was dialyzed in 2000 mL with deionized water for two days (10k MWCO), filtered with 200 nm syringe filters, and finally chromatographically purified (Superdex 200) resulting in the desired mal-C′ dots.
4.3.21. NDCs (6) and (7)
For a typical conjugation reaction, 100 µL of 0.2 M phosphate buffer (pH 7.2) was added to 1 mL of mal-PEG-C′ dots in water (15 µL), then incubated on ice. A 10 mM solution of the desired construct (4 or 5) was prepared in acetonitrile/water (1:1). 30 µL of this solution was added to the mal-PEG-C′ dots, and incubated at 4°C overnight. The reaction solution was then purified by GPC using Biogel P10 with water as the eluent.
Supplementary Material
Acknowledgments
We would like to thank Prof. R. Duncan for helpful technical input and S. Cheal for radiochemistry support. M.B. would like to gratefully acknowledge support by The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, and a Dana Foundation Award, Brain and Immuno-Imaging program. U.W., M.B., and B.Y. are inventors of patent applications held by Cornell University and Memorial Sloan Kettering Cancer Center on technology described in this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Figures S1-S4, HPLC data for 3–5, 9–15 (Figure S5-S15), synthesis of 22 and 23, further details on instruments, enzyme release assays and in vitro cell-based experiments.
References
- 1.Ediriwickrema A, Saltzman WM. ACS Biomater. Sci. Eng. 2015 doi: 10.1021/ab500084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawidczyk CM, Kim C, Park JH, Russell LM, Lee KH, Pomper MG, Searson PC. J. Control. Release. 2014;187:133. doi: 10.1016/j.jconrel.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venditto VJ, Szoka FC., Jr Adv. Drug Delivery Rev. 2013;65:80. doi: 10.1016/j.addr.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang AZ, Langer R, Farokhzad OC. Annu. Rev. Med. 2012;63:185. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 5.Bawa R. Handbook of Clinical Nanomedicine: Nanoparticles, Imaging, Therapy and Clinical Applications. Singapore: Pan Stanford Publishing; 2015. [Google Scholar]
- 6.Thakor AS, Gambhir SS. CA Cancer J Clin. 2013;63:395. doi: 10.3322/caac.21199. [DOI] [PubMed] [Google Scholar]
- 7.Yoo B, Bradbury M, Wiesner U. Provisional patent. 2014 [Google Scholar]
- 8.Louie A. Chemical Reviews. 2010;110:3146. doi: 10.1021/cr9003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jennings LE, Long NJ. Chemical Communications. 2009:3511. doi: 10.1039/b821903f. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y-H, Jeon J, Hong SH, Rhim W-K, Lee Y-S, Youn H, Chung J-K, Lee MC, Lee DS, Kang KW, Nam J-M. Small. 2011;7:2052. doi: 10.1002/smll.201100927. [DOI] [PubMed] [Google Scholar]
- 11.Benezra M, Penate-Medina O, Zanzonico PB, Schaer D, Ow H, Burns A, DeStanchina E, Longo V, Herz E, Iyer S, Wolchok J, Larson SM, Wiesner U, Bradbury MS. J. Clin. Invest. 2011;121:2768. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabizon A, Bradbury M, Prabhakar U, Zamboni W, Libutti S, Grodzinski P. Lancet. 2014;384:2175. doi: 10.1016/S0140-6736(14)61457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barenholz Y. J. Control. Release. 2012;160:117. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Fang J, Nakamura H, Maeda H. Adv. Drug Delivery Rev. 2011;63:136. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Bradbury MS, Phillips E, Montero PH, Cheal SM, Stambuk H, Durack JC, Sofocleous CT, Meester RJC, Wiesner U, Patel S. Integr. Biol. 2013;5:74. doi: 10.1039/c2ib20174g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Nano Lett. 2005;5:113. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 17.Burns AA, Vider J, Ow H, Herz E, Penate-Medina O, Baumgart M, Larson SM, Wiesner U, Bradbury M. Nano Lett. 2008;9:442. doi: 10.1021/nl803405h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J, Burns AA, Williams RM, Zhou Z, Flesken-Nikitin A, Zipfel WR, Wiesner U, Nikitin AY. J. Biomed. Opt. 2007;12:064007. doi: 10.1117/1.2823149. [DOI] [PubMed] [Google Scholar]
- 19.Burns A, Ow H, Wiesner U. Chem. Soc. Rev. 2006;35:1028. doi: 10.1039/b600562b. [DOI] [PubMed] [Google Scholar]
- 20.Burns A, Sengupta P, Zedayko T, Baird B, Wiesner U. Small. 2006;2:723. doi: 10.1002/smll.200600017. [DOI] [PubMed] [Google Scholar]
- 21.Phillips E, Penate-Medina O, Zanzonico PB, Carvajal RD, Mohan P, Ye Y, Humm J, Gönen M, Kalaigian H, Schöder H, Strauss HW, Larson SM, Wiesner U, Bradbury MS. Sci. Transl. Med. 2014;6:260ra149. doi: 10.1126/scitranslmed.3009524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma K, Mendoza C, Hanson M, Werner-Zwanziger U, Zwanziger J, Wiesner U. Chem. Mater. 2015;27:4119. [Google Scholar]
- 23.Duncan R. Nat. Rev. Cancer. 2006;6:688. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 24.Duncan R, Vicent MJ. Adv. Drug Delivery Rev. 2013;65:60. doi: 10.1016/j.addr.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Ducry L, Stump B. Bioconjugate Chem. 2010;21:5. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 26.Dubowchik GM, Firestone RA, Padilla L, Willner D, Hofstead SJ, Mosure K, Knipe JO, Lasch SJ, Trail PA. Bioconjugate Chem. 2002;13:855. doi: 10.1021/bc025536j. [DOI] [PubMed] [Google Scholar]
- 27.Schnur RC, Arnold LD. (Pfizer Inc.) Alkynyl and azido-substituted 4-anilinoquinazolines. 1998 US 5747498. [Google Scholar]
- 28.Mendelsohn J, Baselga J. Semin. Oncol. 2006;33:369. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Neoplasia. 2010;12:675. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omuro AM, Kris MG, Miller VA, Franceschi E, Shah N, Milton DT, Abrey LE. Cancer. 2005;103:2344. doi: 10.1002/cncr.21033. [DOI] [PubMed] [Google Scholar]
- 31.Masuda T, Hattori N, Hamada A, Iwamoto H, Ohshimo S, Kanehara M, Ishikawa N, Fujitaka K, Haruta Y, Murai H, Kohno N. Cancer Chemother. Pharmacol. 2011;67:1465. doi: 10.1007/s00280-011-1555-6. [DOI] [PubMed] [Google Scholar]
- 32.Yun C-H, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Cancer Cell. 2007;11:217. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubowchik GM, Firestone RA. Bioorg. Med. Chem. Lett. 1998;8:3341. doi: 10.1016/s0960-894x(98)00609-x. [DOI] [PubMed] [Google Scholar]
- 34.Carl PL, Chakravarty PK, Katzenellenbogen JA. J. Med. Chem. 1981;24:479. doi: 10.1021/jm00137a001. [DOI] [PubMed] [Google Scholar]
- 35.Aletras A, Barlos K, Gatos D, Koutsogianni S, Mamos P. Int. J. Pept. Protein. Res. 1995;45:488. doi: 10.1111/j.1399-3011.1995.tb01065.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.