Abstract
Purpose
Racial genetic admixture (RGA), a measure to account for ancestral genetic background that correlates with individual's racial classification, could provide insights on causation of racial disparity in endometrial cancer (EC). Our objective is to evaluate the association of RGA with EC outcomes.
Methods
EC patients enrolled onto the GOG-210 protocol were eligible. A randomized subcohort stratified by stage and self-reported race/ethnicity of black or white was used. Genotyping was performed using custom-selected Ancestry Informative Markers to calculate individual admixture estimates of African and European ancestral background.
Results
A total of 149 patients were evaluated (self-reported race: 70 black & 79 white). Mean RGA for African ancestry for self-reported black patients was 0.65 (range 0.04–0.86); while mean RGA for European ancestry for self-reported white patients was 0.77 (range 0.12–0.88). Progression-free survival (PFS) analysis using proportional hazards models stratified by stage and race revealed that each 0.10 increase in African ancestry was associated with worse PFS with hazard ratio (HR) of 1.11 (95% CI 0.90–1.37). Each 0.10 increase in European RGA was associated with improved PFS with HR of 0.86 (95% CI 0.69–1.07). Using tertiles of African RGA showed increasing risk of progression of death with increasing African RGA (with 0–5% as reference), HR (95% CIs) for top two tertiles are: 6%–66%: 1.38 (0.64, 2.97), and 67%–86%: 2.27 (0.74, 6.95).
Conclusion
RGA demonstrated a trend with PFS in self-reported black and white patients with EC. Patients with increased levels of African ancestry showed a trend towards worse survival after stratifying by stage/race.
Keywords: Racial disparity, Genetic admixture, Endometrial cancer, Ancestry informative markers
1. Introduction
Endometrial cancer (EC) is the most common gynecological cancer and the fourth most common cancer in women in the United States [1]. Racial and ethnic differences in incidence, mortality and survival of EC have been reported, particularly between women of African-American (AA) and European-American ancestry [2]. The specific etiologies underlying racial/ethnic disparities in EC are not clear. Although the overall incidence of EC is lower in black women when compared to whites, the survival rate is significantly lower in black females. In a review of advanced/recurrent endometrial cancer, black women were 60–80% more likely to die from EC when controlling for other variables such as performance status, disease stage, tumor histology, tumor grade and treatment rendered [3]. However, it is not clear if this disparity is related to biological differences or consequences of social, economic or cultural environments [4]. The National Cancer Institute's Strategic Plan for Leading the Nation calls to overcome cancer health disparities, including attempts to “understand the factors that cause cancer health disparities.” Thus, the need for further investigations to clarify the etiology of EC racial health disparity is imperative.
One critical question remains: how is race best defined? Although self-reported categorical race facilitates the understanding of “at risk” population differences, genetic similarity cannot be inferred solely on self-reported racial categories. As such, the greatest limitation in studying racial disparity lies within the use of race as a categorical variable. In order to understand and reduce racial-related health disparities in EC, we must investigate the factors that historically relate and statistically correlate with racial classification. Genetic admixture serves as a tool to estimate the genetic ancestral contributions to racial/ethnic classification and therefore has the potential to provide insights into the etiology of racial disparities in EC.
The genetic admixture approach reflects the historical experience of long-separated European, African and Amerindian populations that intermixed during colonization of the New World, passing autochthonous genetic information to newly created admixed populations. Individuals of admixed background carry unique patterns of ancestral information from each of the parental populations that contribute to their ancestral background. Geneticvariants informative of ancestry are known as Ancestry Informative Markers (AIMs), which are used to calculate estimates that reflect the proportion of ancestry in individuals. Correlation of genetic admixture estimates with clinicopathologic factors provide insight into the racial disparity that exists in EC. This approach has previously been used in the field of obesity and diabetes to more accurately explain racial/ethnic differences in health related outcomes [5–8].
The present study evaluates the extent to which genetic admixture, as a measure to represent the ancestral genetic component that underlies racial/ethnic classification, is related to prognosis in individuals with EC.
2. Methods
The Gynecologic Oncology Group Protocol #210 (GOG-210) involved prospective specimen collection from patients with EC for molecular staging. The goal was to use these specimens to “identify the molecular characteristics of EC in order to develop accurate models of risk and identify candidate targets for therapeutic intervention.” Patients who consented to GOG-210, thereby allowing their surgical specimens and clinical data to be used for future research, were eligible for this study. All patients selected for this study had histologically proven stage I–IV endometrioid endometrial cancer and were self-reported “Black/African American” or “White/Caucasian” race, with sufficient tumor-free samples for extracted DNA.
Tissue samples were obtained from the GOG Tissue Bank and reviewed by pathology to confirm histology and determine sufficient high-quality tissue yield devoid of tumor cells. DNA extraction was performed utilizing Qiagen DNA Midi kit for frozen tissue and EZ1 Extractor for formalin-fixed, paraffin-embedded tissue. After PCR amplification of genomic DNA, the GoldenGate assay on the BeadXpress system (Illumina, Inc.) was used for genotyping. The GoldenGate assay involves biotin-labeling of genomic DNA followed by capture of the labeled DNA onto streptavidin-coated sepharose beads. An artificial nucleotide-based molecule that contains universal priming sequences on either end and is complementary to the target DNA sequence of interest is then created, amplified and hybridized to holographically-labeled silica bars that form arrays with up to 30-fold redundancy of each target to be interrogated. Once the array has been visualized with the BeadXpress reader, wavelength and intensity values of the fluorescence are used to determine genotype. A custom LIMS is used to track both samples and laboratory throughput. Allele detection and genotype calling are performed using the GenomeStudio software v3 (Illumina, Inc.) [9]. Genotyping was performed utilizing 140 custom selected AIM panels previously used to estimate with precision the proportion of African, Amerindian and European admixture in individuals that account for the biodiversity of the samples and that reduce potential confounding from population stratification [10]. Maximum likelihood estimation is used to translate the information from the AIMs into estimates of West African, Amerindian and European ancestral estimates for each participant [11]. This method estimates the logarithms of the individual locus probabilities at all loci, computes the probability of the observed genotype for every possible admixture proportion from 0 to 100, and determines the maximum likelihood estimate of ancestry for each parental population for every individual. The range of West African, Amerindian and European ancestral estimates is from0.00 to 1.00, but the sum of the three estimates equals 1.00.
A subcohort of patients using case-cohort design was randomly sampled by random number generation from GOG-0210 and stratified by race and stage of disease in order to get representation across all race and stage combinations [12,13]. The selection was limited to patients who were self-reported black or white with endometrioid tumors. The sample size was driven by budget constraints and respective utilization of DNA material for this pilot project; however, the observed number of 53 PFS events yields approximately 80% power to detect a hazard ratio (HR) of 2.2 for a binary exposure with a 50/50 split. Proportional hazards models were used to compare PFS by RGA. Continuous levels of RGA were analyzed by 10 percentage point differences, and tertiles of African RGA were also compared. Models were analyzed four ways: stratified by stage, and stratified by stage and race; and each with or without adjustment for body mass index (BMI). Analyses of individual single-nucleotide polymorphism (SNP) were done using 0/1/2 values for the genotype and were fully adjusted for admixture proportion with significance levels adjusted using Bonferroni adjustment to account for multiple testing. Results were considered statistically significant if two-sided p < 0.05 or if two-tailed 95% confidence intervals (Cis) excluded the reference value; no adjustment for multiple testing was made except for the case of the individual SNP data. [14,15]. Analysis of variance was used to compare age and BMI between blacks and whites, and k − 1 degree of freedom chi-square tests (where k is the number of categories of the characteristic of interest) were used to compare performance status, stage, and grade between blacks and whites.
3. Results
3.1. Demographics
As of August 2010, a total of 3107 patients with endometrioid endometrial cancer with self-reported black or white race and completed data entry were available from the GOG-210 protocol. Of these, 188 patients were randomly selected stratified by race and stage. A total of 39 were ineligible, leaving 149 patients available for analysis in this pilot study. (Table 1).
Table 1.
Patient selection for analysis.
| Self-reported race stage |
In the original GOG-0210 cohorta (m/n) |
Selected for subcohort (m/n) |
Eligible for subcohort with admixture doneb (m/n) |
|---|---|---|---|
| African-American | |||
| 1 | 28/185 | 3/20 | 1/16 |
| 2 | 10/32 | 10/32 | 10/24 |
| 3 | 15/32 | 15/31 | 10/23 |
| 4 | 6/10 | 6/10 | 4/7 |
| White | |||
| 1 | 268/2230 | 4/19 | 4/17 |
| 2 | 39/203 | 4/19 | 3/14 |
| 3 | 111/351 | 6/23 | 6/21 |
| 4 | 41/64 | 19/34 | 15/27 |
| Total | 518/3107 | 67/188 | 53/149 |
m = number with PFS event in strata.
n = number in specified strata.
Black and white patients with endometrioid tumors.
Thirty-nine patients chosen for the subcohort were not included. The non-mutually exclusive reasons were: 2 withdrew consent, 5 were not deemed to be endometrioid cell type based on central pathology review, and 34 were not assayed for RGA.
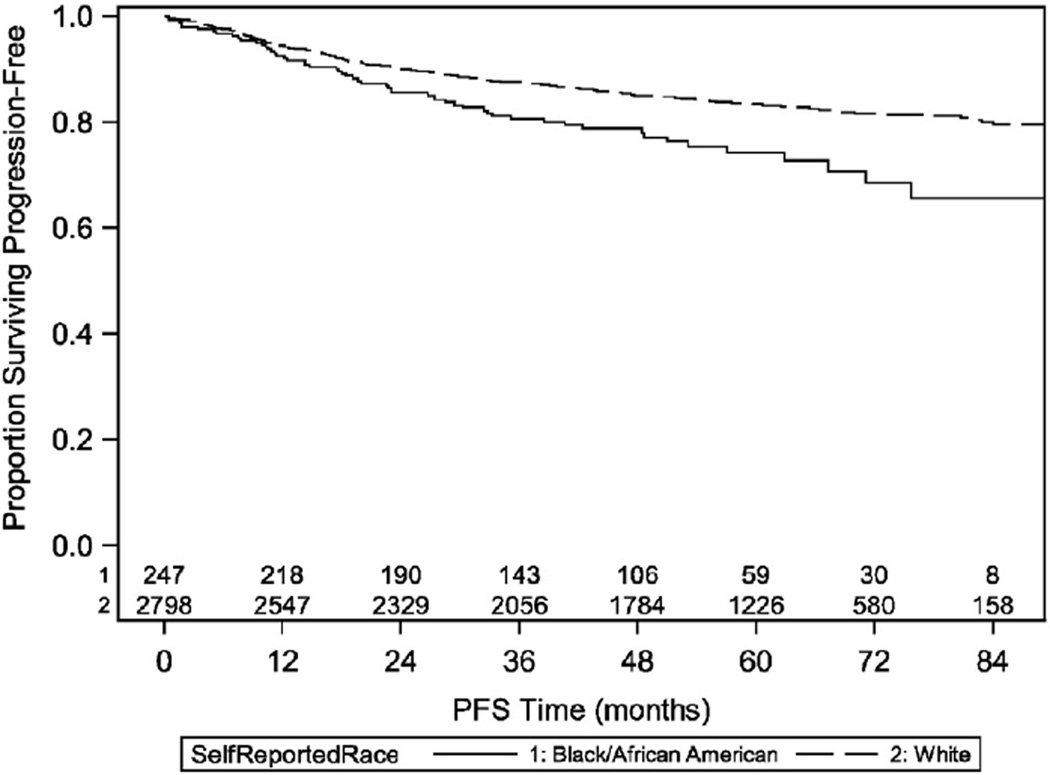
The self-reported racial breakdown was 70 black patients and 79 white patients. Mean age was 62.1 years, and 79% of patients had GOG performance status of zero. Groups were similar with regard to age (61.7 years black; 62.4 years white), while mean body mass index (BMI) was higher in black than in white patients (37.5 vs. 32.9 mg/m2). The distribution of grade was similar between black and white patients, and the distribution of stage was similar but was determined by the stratification. (Table 2) Importantly, using baseline analysis of self-reported race for the entire cohort (n = 3045), a racial disparity existed with five-year PFS of 83% for white patients and 74% for black patients (log-rank p < 0.001). (Table 3 & Fig. 1) The relationship of PFS with race and with BMI is shown for the full cohort and the subcohort in Table 3, and the results are consistent between the two cohorts.
Table 2.
Baseline characteristics by self-reported race for patients.
| Black (n = 70) |
White (n = 79) |
|
|---|---|---|
| Age, years | ||
| Mean (SE) | 61.7 (1.39) | 62.4 (1.33) |
| Median (25th, 75th) | 61.6 (53.5, 69.5) | 60.5 (55.5, 69.7) |
| BMI, kg/m2 | ||
| Mean (SE) | 37.5 (1.18) | 32.9 (0.93) |
| Median (25th, 75th) | 35.4 (31.0, 46.2) | 31.3 (27.5, 37.7) |
| Performance Status, n (%) | ||
| 0 | 51 (73%) | 66 (84%) |
| 1 | 17 (24%) | 13 (16%) |
| 2 | 2 (3%) | 0 (0%) |
| Stage, n (%) | ||
| I | 16 (23%) | 17 (22%) |
| II | 24 (34%) | 14 (18%) |
| III | 23 (33%) | 21 (27%) |
| IV | 7 (10%) | 27 (34%) |
| Grade, n (%) | ||
| 1 | 15 (22%) | 20 (25%) |
| 2 | 21 (30%) | 21 (27%) |
| 3 | 33 (48%) | 38 (48%) |
SE: standard error, 25th and 75th are the 25th and 75th percentiles, and BMI: body mass index.
Note: One black patient missing grade.
Table 3.
Comparison of race and BMI results for progression-free survival in full cohort (n = 3045)a and subcohort (n = 149).
| Variable/cohort | HR (95% CI) | |
|---|---|---|
| Stratified by stage | Stratified by stage and race | |
| Race (Black) | ||
| Full cohort | 1.49 (1.13, 1.98) | Not applicable |
| Subcohort | 1.36 (0.76, 2.42) | Not applicable |
| BMI (1 kg/m2) | ||
| Full cohort | 0.996 (0.986, 1.006) | 0.994 (0.984, 1.004) |
| Subcohort | 0.998 (0.969, 1.028) | 0.992 (0.959, 1.025) |
Of the 3107 patients in the original cohort (see Table 1), 62 patients were excluded: 4 withdrew consent, and 58 were not deemed to be endometrioid cell type based on central pathology review.
Fig. 1.
Progression free survival by self-reported race for all patients in the GOG 210 cohort.
3.2. Racial genetic admixture
The proportion of calculated genetic admixture varied between self-reported groups. Mean admixture for self-reported black patients was 65% African, 15% Amerindian and 20% European ancestry. Self-reported white patients demonstrated 6% African, 16% Amerindian, and 79% European ancestry. Mean (±SD) RGA for African ancestry for self-reported black patients was 0.65 ± 0.19 (range 0.04–0.86); while mean (±SD) RGA for European ancestry for self-reported white patients was 0.77 ± 0.12 (range 0.12–0.88). (Table 4 & Figs. 2A and 2B online) RGA was compared to age, BMI, performance status, stage and grade. However, after adjustment for race, no differences across these groups were statistically significant. (Table 5)
Table 4.
Racial genetic admixture by self-reported race.
| Admixture | Self-reported race | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Black | White | |||||||||||
| n | Mean | Median | SD | Min | Max | N | Mean | Median | SD | Min | Max | |
| African | 70 | 0.65 | 0.71 | 0.19 | 0.04 | 0.86 | 79 | 0.06 | 0.04 | 0.11 | 0.00 | 0.74 |
| American Indian | 70 | 0.15 | 0.14 | 0.06 | 0.04 | 0.50 | 79 | 0.17 | 0.16 | 0.04 | 0.07 | 0.28 |
| European | 70 | 0.20 | 0.15 | 0.17 | 0.05 | 0.83 | 79 | 0.77 | 0.79 | 0.12 | 0.12 | 0.88 |
Table 5.
Racial genetic admixture by prognostic factors.
| n | African admixture | American Indian admixture | European admixture | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | SD | Mean | Median | SD | Mean | Median | SD | ||
| Age (y) | ||||||||||
| <50 | 19 | 0.33 | 0.13 | 0.33 | 0.17 | 0.14 | 0.10 | 0.50 | 0.60 | 0.31 |
| 50 < 60 | 50 | 0.36 | 0.09 | 0.34 | 0.15 | 0.15 | 0.03 | 0.49 | 0.73 | 0.34 |
| 60 < 70 | 44 | 0.34 | 0.10 | 0.34 | 0.16 | 0.16 | 0.04 | 0.50 | 0.73 | 0.33 |
| ≥ 70 | 35 | 0.31 | 0.08 | 0.33 | 0.16 | 0.15 | 0.05 | 0.52 | 0.73 | 0.31 |
| BMI (kg/m2) | ||||||||||
| 18.5–24.9 | 15 | 0.24 | 0.07 | 0.33 | 0.15 | 0.16 | 0.04 | 0.60 | 0.77 | 0.31 |
| 25.0–29.9 | 33 | 0.24 | 0.07 | 0.30 | 0.17 | 0.16 | 0.04 | 0.59 | 0.75 | 0.29 |
| 30.0–34.9 | 43 | 0.30 | 0.10 | 0.32 | 0.17 | 0.16 | 0.07 | 0.53 | 0.73 | 0.31 |
| 35.0–39.9 | 21 | 0.40 | 0.49 | 0.35 | 0.15 | 0.14 | 0.04 | 0.46 | 0.36 | 0.34 |
| ≥40.0 | 36 | 0.49 | 0.67 | 0.33 | 0.14 | 0.14 | 0.04 | 0.37 | 0.20 | 0.32 |
| Performance status | ||||||||||
| 0 | 117 | 0.31 | 0.08 | 0.33 | 0.16 | 0.15 | 0.05 | 0.53 | 0.73 | 0.32 |
| 1 | 29 | 0.44 | 0.64 | 0.34 | 0.16 | 0.15 | 0.05 | 0.40 | 0.25 | 0.32 |
| 2 | 2 | 0.80 | 0.80 | 0.09 | 0.12 | 0.12 | 0.06 | 0.09 | 0.09 | 0.03 |
| Stage | ||||||||||
| 1 | 33 | 0.34 | 0.09 | 0.35 | 0.15 | 0.14 | 0.04 | 0.51 | 0.73 | 0.33 |
| 2 | 38 | 0.45 | 0.64 | 0.34 | 0.15 | 0.15 | 0.04 | 0.41 | 0.22 | 0.33 |
| 3 | 43 | 0.37 | 0.45 | 0.34 | 0.15 | 0.15 | 0.03 | 0.47 | 0.37 | 0.33 |
| 4 | 34 | 0.18 | 0.08 | 0.26 | 0.18 | 0.17 | 0.08 | 0.64 | 0.75 | 0.25 |
| Grade | ||||||||||
| 1 | 35 | 0.32 | 0.06 | 0.34 | 0.16 | 0.15 | 0.07 | 0.52 | 0.74 | 0.33 |
| 2 | 42 | 0.37 | 0.12 | 0.34 | 0.16 | 0.16 | 0.04 | 0.48 | 0.64 | 0.33 |
| 3 | 71 | 0.33 | 0.09 | 0.33 | 0.16 | 0.15 | 0.04 | 0.51 | 0.72 | 0.32 |
Note: after adjusting for race, no differences in admixture across any groups were statistically significant (p > 0.05).
Analysis of PFS by RGA revealed that African ancestry (after stratification by self-reported race and stage) had nonsignificantly worse PFS with HR of 1.11 (95% CI 0.90–1.37) for each 0.10 increase in African admixture. European ancestry was nonsignificantly protective with HR of 0.86 (95% CI 0.70–1.07) for each 0.10 increase in European admixture. Analyses stratified by stage were similar to those stratified by stage and race, as were models adjusted for BMI. (Table 6).
Table 6.
Analyses of progression-free survival by racial genetic admixture.
| Admixturea | HR (95% CI) for 0.10 increase in admixture score | |||
|---|---|---|---|---|
| Stratified by stage | Stratified by stage and adjusted for BMI | Stratified by stage and race | Stratified by stage and race and adjusted for BMI | |
| African | 1.07 (0.98, 1.16) | 1.08 (0.98, 1.18) | 1.11 (0.90, 1.37) | 1.12 (0.91, 1.37) |
| American Indian | 1.08 (0.66, 1.78) | 1.08 (0.65, 1.79) | 1.15 (0.69, 1.92) | 1.13 (0.68, 1.89) |
| European | 0.93 (0.85, 1.02) | 0.92 (0.84, 1.01) | 0.86 (0.69, 1.07) | 0.86 (0.70, 1.07) |
Separate models were run for each admixture score.
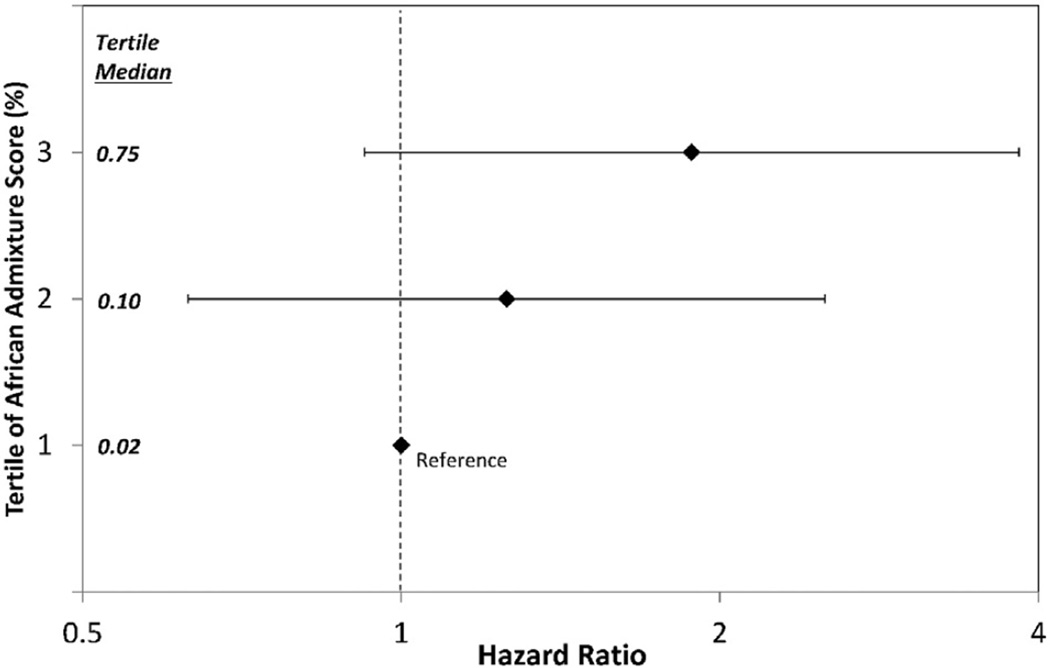
The trend towards increased hazard across the full range of African RGA is shown by analyses using tertiles of African RGA. (Table 7 & Fig. 3) After stratification by stage and race, relative to the lowest tertile (with African RGA of 0%–5%), the HRs (95% CIs) for progression or death were 1.38 (0.64, 2.97) and 2.27 (0.74, 6.95) for patients with African RGA of 6%–66% and 67%–86%, respectively (overall p-value: 0.344). Five-year PFS (95% CIs) were 68% (53%, 80%), 65% (49%, 76%), and 58% (40%, 72%) in tertiles 1, 2, and 3, respectively. Analyses stratified by stage were similar to those stratified by stage and race, as were models adjusted for BMI.
Table 7.
Analyses of progression free survival by tertile of African genetic admixture.
| African admixture (%) |
n (Blacks, Whites) by self report |
HR (95% CI) | |||
|---|---|---|---|---|---|
| Stratified by stage | Stratified by stage and adjusted for BMI |
Stratified by stage and race |
Stratified by stage and race and adjusted for BMI |
||
| 0–5 (reference) | 50 (1, 49) | 1.00 | 1.00 | 1.00 | 1.00 |
| 6–66 | 49 (21, 28) | 1.24 (0.62, 2.48) | 1.26 (0.63, 2.51) | 1.38 (0.64, 2.97) | 1.37 (0.64, 2.94) |
| 67–86 | 50 (48, 2) | 1.81 (0.90, 3.63) | 1.88 (0.92, 3.84) | 2.27 (0.74, 6.95) | 2.23 (0.72, 6.84) |
Note: HRs (95% CI) for BMI (kg/m2) are 0.993 (0.963, 1.023) and 0.994 (0.962, 1.028) for models stratified by stage and stratified by stage and age, respectively.
Fig. 3.
Hazard ratios for progression free survival by the Tertile of African Racial Genetic Admixture Score.
3.3. Analysis of individual SNPs
No individual SNPs were significantly associated with PFS after adjustment for RGA and after Bonferroni correction for multiple testing (all adjusted p-values ≥ 0.702).
4. Discussion
In a retrospective review of advanced/recurrent endometrial cancer, black women had worse survival than white women (median 10.6 vs. 12.2 months, respectively; p < 0.001). This disparity remained when controlling for other factors such as performance status, disease stage, tumor histology, tumor grade and treatment rendered [3]. Our findings in the entire cohort (n = 3045) was similar with 5-year PFS of 83% for white patients compared to 74% for black patients (p < 0.001). One potential weakness of evaluating the entire cohort was that these patients were not enrolled onto therapeutic clinical trials, and differences in treatment could have contributed to this disparity.
Aspects traditionally known to differ between black and white patients have been considered as possible causes of racial/ethnic disparities. Biological factors, such as stage, grade and high-risk histology have been suggested to be the underlying etiology [4,16–19].
Environmental factors such as obesity, body composition, diet, and physical activity have long been considered to be major risk factors for the development of EC and responsible for health disparity [16–18]. Likewise, social factors such as lower socioeconomic status (SES), lack of health insurance, and limited access to medical care has been associated with lower likelihood of surgery and thus responsible for differences in health outcomes for black patients [20–24].
The greatest limitation in studying racial/ethnic disparities is the reliance on the use of race as a categorical self-reported variable. Although there has been controversy regarding the meaning of “race” in the biomedical arena, researchers agree that the true understanding of the etiology of such disparities relies on the decomposition of race as a category into those genetic aspects underlying its classification. Specifically, genetic similarity cannot be inferred simply based on racial categories or phenotypical categories (black or white).
In our analysis, we confirmed this variation with a wide range of racial genetic admixture seen amongst self-reported races. Self-reporting black patients had a mean (±SD) African RGA of 0.65 (±0.19) ranging from0.04–0.86. Similarly, self-reported white patients had mean European RGA of 0.77 (±0.12) ranging from 0.12–0.88. Considering the wide range of RGA in this cohort adds credence to the importance of decomposition of race from categorical variables (black, white) into continuous variables of genotyping racial admixture ancestry. Importantly, our mean for each self-reported race falls within published literature ranges. For example, the mean African admixture in our cohort of 65% was within the range seen in published literature of 42–82%. Likewise, the mean European admixture in white patients of 77% was also within the published reports range of 54–96% [7,8,24–29].
Thus, it is evident that in order to understand and reduce health disparities in EC, there is a need to further investigate the interactions of genetic factors believed to influence racial/ethnic differences. Racial/ethnic differences in complex traits exist even after adjusting for non-biological factors; thereby, supporting the concept that genetic differences and their interaction with environmental factors may underlie population differences in disease risk [29]. Therefore, the goal of this study was to declassify racial/ethnic categorization by investigating the effects of racial genetic admixture on EC outcomes.
In this investigation, we applied a model where racial categorization is decomposed in genetic factors. We observed a trend of worsening PFS across increasing tertiles of African RGA. After stratifying by stage and race, HRs (95% CIs) for PFS for the top two tertiles still described a trend (6%–66% and 67%–86% versus 0%–5%) were 1.38 (0.64, 2.97) and 2.27 (0.74, 6.95), respectively. Sample size and power limitations could have impacted our ability to detect statistically significant results, however this trend did support our hypothesis that genetic definition of race could provide insight to cancer related endpoints.
It is noteworthy that the SNPs evaluated in this manuscript have only been validated to estimate genetic ancestry and are not meant to be indicative of causation of endometrial cancer outcomes. However, it is feasible that these SNPs could highlight specific chromosomal regions of interest where further evaluation could detect causative genetic alterations. Although a much more expansive approach would be needed to perform the required “fine-mapping” of these areas of interest, it could represent a plausible methodology for future directions.
While the association of RGA with PFS was not statistically significant in this small pilot study, HRs of the magnitude seen within could mean real, substantial effects due to the genetics of race. Further examination of these results in more robust cohorts could help elucidate these issues further. Nonetheless, the data within is thought provoking on how we define race/ethnicity in cancer disparities. Considering that this publication examines genetic calculation of race to cancer outcomes, this research is an important first step that coincides with the NCI Strategic Plan for Leading the Nation: Objective #8 to Overcome Cancer Health Disparities. Considering the correlative significance of the genetic calculation of race, further study in this arena is vital in unraveling the etiology of racial disparity in endometrial cancer, improving patient outcomes and hopefully eliminating this racial disparity in the future.
Supplementary Material
HIGHLIGHTS.
Racial genetic admixture offers a different methodology of evaluating racial disparities.
Racial genetic admixture demonstrated a trend with survival in endometrial cancer patients.
Patients with increased African ancestry trended towards worse survival.
Acknowledgments
This study was supported by National Cancer Institute grants to the Gynecologic Oncology Group (GOG) Administrative Office (CA 27469), the GOG Statistical Office (CA 37517), and the NRG Oncology/Gynecologic Oncology Group Grant (1U10 CA180822). The following Gynecologic Oncology Group member institutions participated in the primary treatment studies: Roswell Park Cancer Institute, University of Alabama at Birmingham, Duke University Medical Center, Abington Memorial Hospital, Walter Reed Army Medical Center, Wayne State University, University of Minnesota Medical School, Northwestern Memorial Hospital, University of Mississippi Medical Center, Colorado Gynecologic Oncology Group P.C., University of California at Los Angeles, Fred Hutchinson Cancer Research Center, University of Pennsylvania Cancer Center, Milton S. Hershey Medical Center, University of Cincinnati, University of North Carolina School of Medicine, University of Iowa Hospitals and Clinics, University of Texas Southwestern Medical Center at Dallas, Indiana University School of Medicine, Wake Forest University School of Medicine, University of California Medical Center at Irvine, Rush-Presbyterian-St. Luke's Medical Center, Magee Women's Hospital, University of New Mexico, The Cleveland Clinic Foundation, State University of New York at Stony Brook, Washington University School of Medicine, Memorial Sloan-Kettering Cancer Center, Cooper Hospital/University Medical Center, Columbus Cancer Council, University of Massachusetts Medical School, Fox Chase Cancer Center, Women's Cancer Center, University of Oklahoma, University of Virginia Health Sciences Center, University of Chicago, Mayo Clinic, Case Western Reserve University, Tampa Bay Cancer Consortium, Yale University, University of Wisconsin Hospital, Women and Infants Hospital, The Hospital of Central Connecticut, GYN Oncology of West Michigan, PLLC, Aurora Women's Pavilion of West Allis Memorial Hospital, University of California, San Francisco-Mt. Zion and Community Clinical Oncology Program.
Dr. Paul Goodfellow has a grant from the NCI — NCI P50CA134254.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2015.11.018.
Conflicts of interest
All other co-authors have no conflicts of interest to declare.
Contributor Information
Rodney P. Rocconi, Email: rocconi@health.southalabama.edu.
Heather A. Lankes, Email: hlankes@gogstats.org.
William E. Brady, Email: bbrady@gogstats.org.
Paul J. Goodfellow, Email: paul.goodfellow@osumc.edu.
Nilsa C. Ramirez, Email: nilsa.ramirez@nationwidechildrens.org.
Ronald D. Alvarez, Email: rdalvarez@aol.com.
William Creasman, Email: creasman@musc.edu.
José R. Fernández, Email: jose@uab.edu.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. CA Cancer J. Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Brinton LA, Felix AS, McMeekin DS, Creasman WT, Sherman ME, Mutch D, et al. Etiologic heterogeneity in endometrial cancer: evidence from a Gynecologic Oncology Group trial. Gynecol. Oncol. 2013;129:277–284. doi: 10.1016/j.ygyno.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maxwell GL, Tian C, Risinger J, Brown CL, Rose GS, Thigpen JT, et al. Racial disparity in survival among patients with advanced/recurrent endometrial adenocaqrcinoma: a Gynecologic Oncology Group study. Cancer. 2006;107:2197–2205. doi: 10.1002/cncr.22232. [DOI] [PubMed] [Google Scholar]
- 4.Hill HA, Eley JW, Harlan LC, Greenberg RS, Barrett RJ, II, Chen VW. Racial differences in endometrial cancer survival: the black/white cancer survival study. Obstet Gynecologie. 1996;88:919–926. doi: 10.1016/s0029-7844(96)00341-9. [DOI] [PubMed] [Google Scholar]
- 5.Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Hum. Genet. 2004;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez JR, Allison DB. Understanding racial differences in obesity and metabolic syndrome traits. Nutr. Rev. 2003;61:316–319. doi: 10.1301/nr.2003.sept.316-319. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, et al. Association of African genetic admixture with resting metabolic rate and obesity among women. Obes. Res. 2003;11:904–911. doi: 10.1038/oby.2003.124. [DOI] [PubMed] [Google Scholar]
- 8.Gower BA, Fernandez JR, Beasley TM, Shriver MD, Goran MI. Using genetic admixture to explain racial differences in insulin-related phenotypes. Diabetes. 2003;52:1047–1051. doi: 10.2337/diabetes.52.4.1047. [DOI] [PubMed] [Google Scholar]
- 9.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klimentidis YC, Dulin-Keita A, Casazza K, Willig AL, Allison DB, Fernandez JR. Genetic admixture, social–behavioral factors and body composition are associated with blood pressure differently by racial-ethnic group among children. J. Hum. Hypertens. 2012;26:98–107. doi: 10.1038/jhh.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanis CL, Chakraborty R, Ferrell RE, et al. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am. J. Phys. Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 12.Langholz B, Jiao J. Computational methods for case-cohort studies. Comput. Stat. Data Anal. 2007;51:3737–3748. [Google Scholar]
- 13.Samuelsen SO, Ånestad H, Skrondal A. Stratified case-cohort analysis of general cohort sampling designs. Scand. J. Stat. 2007;34:103–119. [Google Scholar]
- 14.Ohashi J, Tokunaga K. The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J. Hum. Genet. 2001;46:478–482. doi: 10.1007/s100380170048. [DOI] [PubMed] [Google Scholar]
- 15.Zhu XL, Ai ZH, Wang J, Xu YL, Teng YC. Weighted gene co-expression network analysis in identification of endometrial cancer prognosis markers. Asian Pac. J. Cancer Prev. 2012;13:4607–4611. doi: 10.7314/apjcp.2012.13.9.4607. [DOI] [PubMed] [Google Scholar]
- 16.Barrett RJ, II, Harlan LC, Wesley MN, Hill HA, Chen VW, Clayton LA, et al. Endometrial cancer: stage at diagnosis and associated factors in black and white patients. Am. J. Obstet. Gynecol. 1995;173:414–422. doi: 10.1016/0002-9378(95)90261-9. [DOI] [PubMed] [Google Scholar]
- 17.Cronje HS, Fourie S, Doman MJ, Helms JB, Nel JT, Goedhals L. Racial differences in patients with adenocarcinoma of the endometrium. Int. J. Gynaecol. Obstet. 1992;39:213–218. doi: 10.1016/0020-7292(92)90659-7. [DOI] [PubMed] [Google Scholar]
- 18.Bain RP, Greenberg RS, Chung KC. Racial differences in survival of women with endometrial cancer. Am. J. Obstet. Gynecol. 1987;157:914–923. doi: 10.1016/s0002-9378(87)80089-3. [DOI] [PubMed] [Google Scholar]
- 19.Bach PB, Schrag D, Brawley OW, Galaznik A, Yakren S, Begg CB. Survival of blacks and whites after a cancer diagnosis. JAMA. 2002 Apr 24;287(16):2106–2113. doi: 10.1001/jama.287.16.2106. [DOI] [PubMed] [Google Scholar]
- 20.Madison T, Schottenfeld D, Baker V. Cancer of the corpus uteri in white and black women in Michigan, 1985–1994: an analysis of trends in incidence and mortality and their relation to histologic subtype and stage. Cancer. 1998;83:1546–1554. doi: 10.1002/(sici)1097-0142(19981015)83:8<1546::aid-cncr9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 21.Madison T, Schottenfeld D, James SA, Schwartz AG, Gruber SB. Endometrial cancer: socioeconomic status and racial/ethnic differences in stage at diagnosis, treatment, and survival. Am. J. Public Health. 2004;94:2104–2111. doi: 10.2105/ajph.94.12.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coates RJ, Click LA, Harlan LC, Robboy S, Barrett RJ, 2nd, Eley JW, et al. Differences between black and white patients with cancer of the uterine corpus in interval from symptom recognition to initial medical consultation (United States) Cancer Causes Control. 1996;7:328–336. doi: 10.1007/BF00052938. [DOI] [PubMed] [Google Scholar]
- 23.Statistical Abstract of the United States. Washington, DCS: US Bureau of the Census; 1997. [Google Scholar]
- 24.Wright JD, Fiorelli J, Schiff PB, Burke WM, Kansler AL, Cohen CJ, Herzog TJ. Racial disparities for uterine corpus tumors: changes in clinical characteristics and treatment over time. Cancer. 2009 Mar 15;115(6):1276–1285. doi: 10.1002/cncr.24160. [DOI] [PubMed] [Google Scholar]
- 25.Casazza K, Thomas O, Dulin-Keita A, Fernandez JR. Adiposity and genetic admixture, but not race/ethnicity, influence bone mineral content in peripubertal children. J. Bone Miner. Metab. 2010;28:424–432. doi: 10.1007/s00774-009-0143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardel M, Higgins PB, Willig AL, Keita AD, Casazza K, Gower BA, et al. African genetic admixture is associated with body composition and fat distribution in a cross-sectional study of children. Int. J. Obes. 2011;35:60–65. doi: 10.1038/ijo.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casazza K, Willig AL, Gower BA, Nagy TR, Hunter GR, Wallace S, et al. The role of European genetic admixture in the etiology of the insulin resistance syndrome in children: are the effects mediated by fat accumulation? J. Pediatr. 2010;157:50–56. doi: 10.1016/j.jpeds.2010.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardel M, Willig AL, Dulin-Keita A, Casazza K, Beasley TM, Fernandez JR. Parental feeding practices and socioeconomic status are associated with child adiposity in a multi-ethnic sample of children. Appetite. 2012;58:347–353. doi: 10.1016/j.appet.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-comment2007. (comment 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.